Haploid and Doubled Haploid Techniques in Perennial Ryegrass (Lolium perenne L.) to Advance Research and Breeding
Abstract
:1. Introduction
2. Haploids and Doubled Haploids: Their Production and Use in Breeding and Research
3. Perennial Ryegrass
3.1. Perennial Ryegrass Breeding
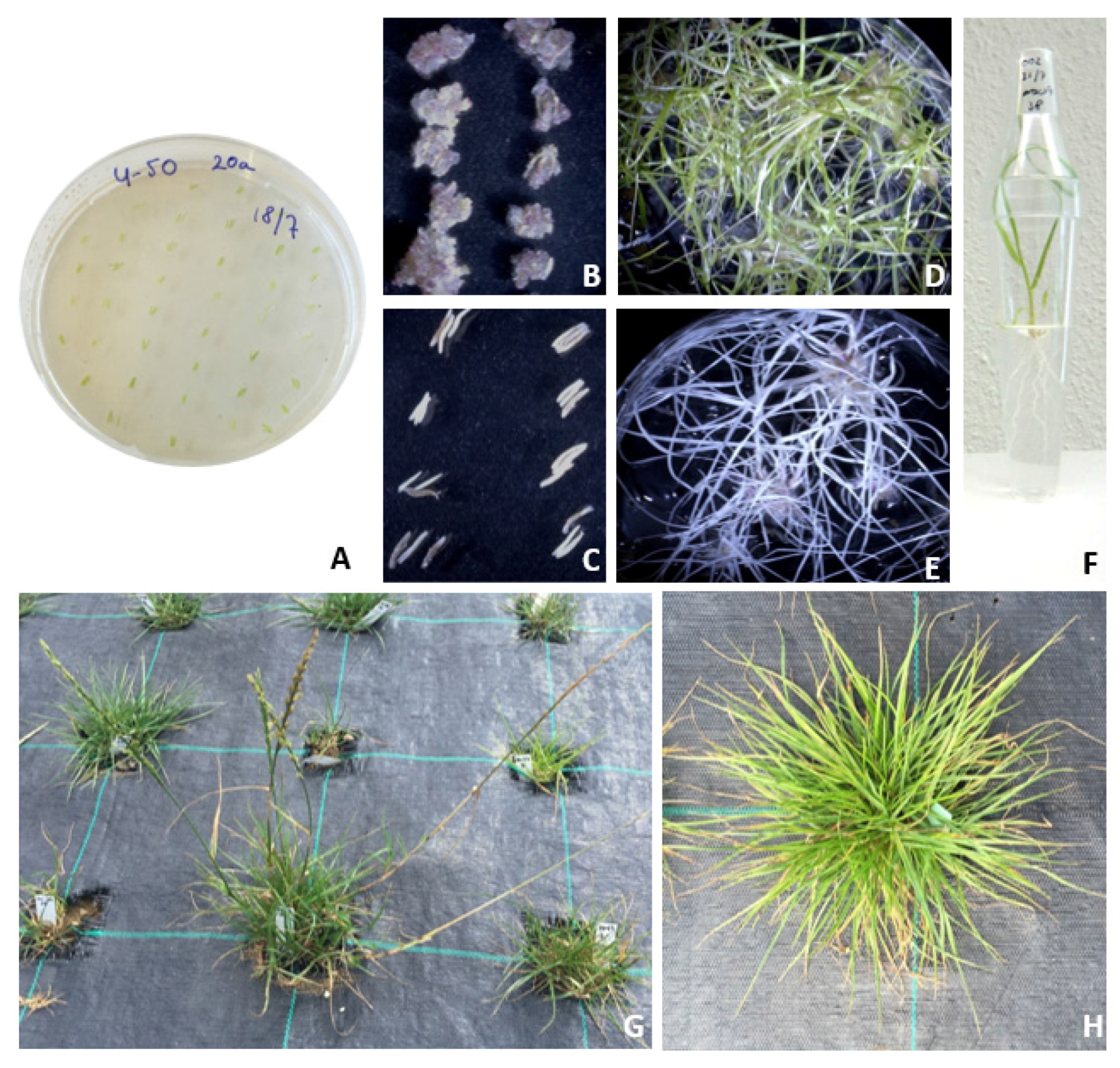
3.2. Doubled Haploids in Perennial Ryegrass
4. Future Applications of Haploid and Doubled Haploid Techniques in Perennial Ryegrass
4.1. Purging Deleterious Alleles
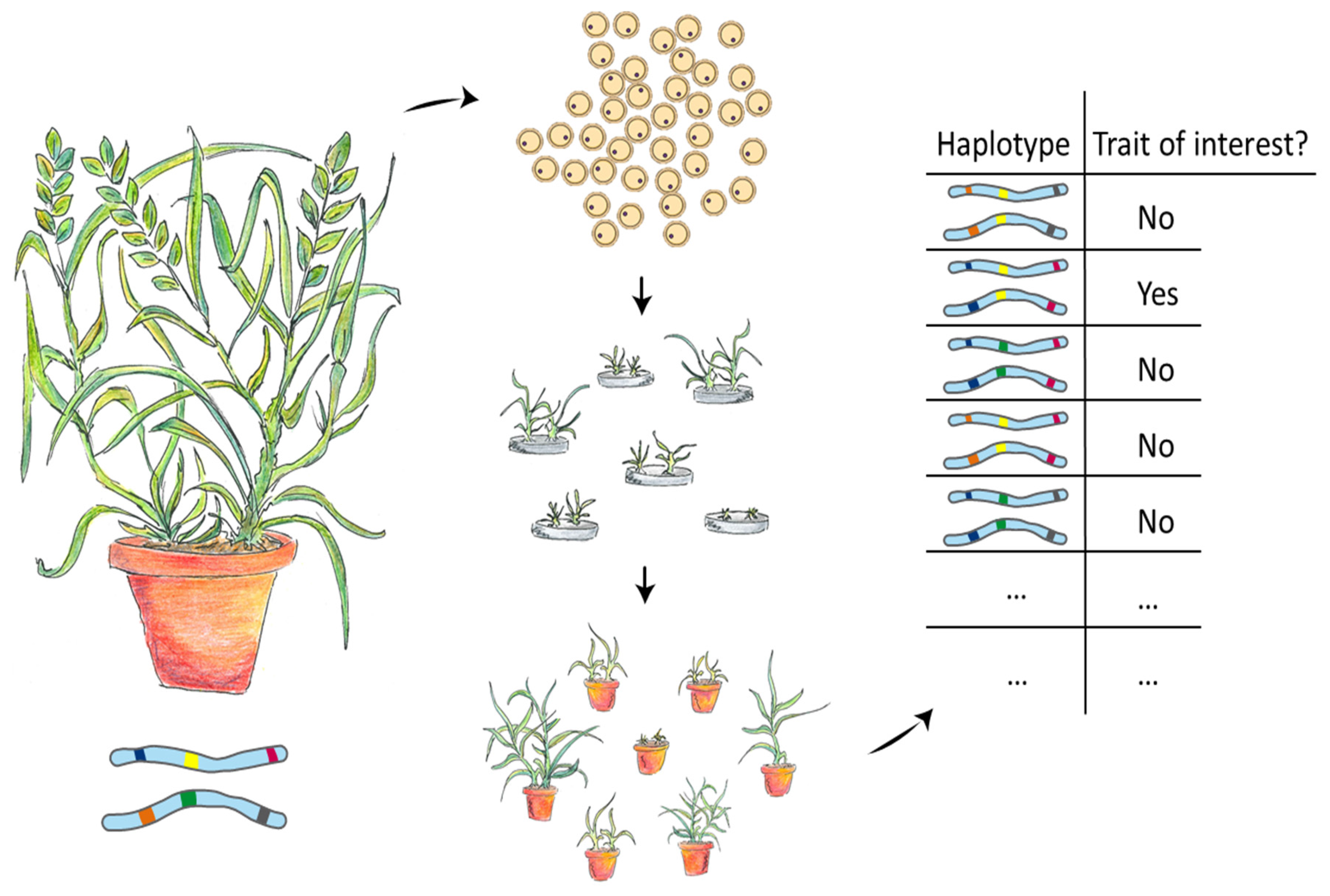
4.2. Doubled Haploids for Genetics and Genomics
4.2.1. Transformation and Mutation
4.2.2. Population Development for Mapping
4.3. Doubled Haploids for Hybrid Breeding
5. Concluding Remarks
Acknowledgments
Conflicts of Interest
Abbreviations
| AC | anther culture |
| DH | doubled haploid |
| CENH3 | centromere-specific histone 3 variant |
| CRISPR | clustered regularly interspaced short palindrome repeats |
| DMSO | dimethyl sulfoxide |
| ELS | embryo-like structures |
| IMC | isolated microspore culture |
| MAS | marker-assisted selection |
| QTL | quantitative trait locus/loci |
| SI | self-incompatible/self-incompatibility |
| TALEN | transcription activator-like effector nuclease |
| TILLING | targeting induced local lesions in genomes |
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.A.; Byerlee, D.; Edmeades, G.O. Crop Yields and Global Food Security: Will Yield Increase Continue to Feed the World? ACIAR Monograph No. 158; Australian Centre for International Agricultural Research: Canberra, Australia, 2014.
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Brummer, C.E.; Barber, W.T.; Collier, S.M.; Cox, T.S.; Johnson, R.; Murray, S.C.; Olsen, R.T.; Pratt, R.C.; Thro, A.M. Plant breeding for harmony between agriculture and the environment. Front. Ecol. Environ. 2011, 9, 561–568. [Google Scholar] [CrossRef]
- Fischer, R.A.; Edmeades, G.O. Breeding and cereal yield progress. Crop Sci. 2010, 50, S85–S98. [Google Scholar] [CrossRef]
- Stamp, P.; Visser, R. The twenty-first century, the century of plant breeding. Euphytica 2012, 186, 585–591. [Google Scholar] [CrossRef]
- Fischer, R.A. Definitions and determination of crop yield, yield gaps, and of rates of change. Field Crops Res. 2015, 182, 9–18. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Dawson, I.K.; Russell, J.; Powell, W.; Steffenson, B.; Thomas, W.T.B.; Waugh, R. Barley: A translational model for adaptation to climate change. New Phytol. 2015, 206, 913–931. [Google Scholar] [CrossRef] [PubMed]
- Germanà, M.A. Gametic embryogenesis and haploid technology as valuable support to plant breeding. Plant Cell Rep. 2011, 30, 839–857. [Google Scholar] [CrossRef] [PubMed]
- Seguí-Simarro, J.M. Androgenesis revisited. Bot. Rev. 2010, 76, 377–404. [Google Scholar] [CrossRef]
- Forster, B.P.; Heberle-Bors, E.; Kasha, K.J.; Touraev, A. The resurgence of haploids in higher plants. Trends Plant Sci. 2007, 12, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M. Haploids in flowering plants: Origins and exploitation. Plant Biotechnol. J. 2010, 8, 377–424. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.L.; Britt, A.B.; Tripathi, L.; Sharma, S.; Upadhyaya, H.D.; Ortiz, R. Haploids: Constraints and opportunities in plant breeding. Biotechnol. Adv. 2015, 33, 812–829. [Google Scholar] [CrossRef] [PubMed]
- Seguí-Simarro, J.M. Editorial: Doubled haploidy in model and recalcitrant species. Front. Plant Sci. 2015, 6, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Geiger, H.H.; Gordillo, G.A. Doubled haploids in hybrid maize breeding. Maydica 2009, 54, 485–499. [Google Scholar]
- Smith, K.F.; Spangenberg, G. Forage breeding for changing environments and production systems: An overview. Crop Pasture Sci. 2014, 65. [Google Scholar] [CrossRef]
- Reheul, D.; De Cauwer, B.; Cougnon, M. The role of forage crops in multifunctional agriculture. In Fodder Crops and Amenity Grasses; Boller, B., Posselt, U., Veronesi, F., Eds.; Springer: New York, NY, USA, 2010; pp. 457–476. [Google Scholar]
- Ferrie, A.M.R.; Caswell, K.L. Isolated microspore culture techniques and recent progress for haploid and doubled haploid plant production. Plant Cell Tissue Organ Cult. 2011, 104, 301–309. [Google Scholar] [CrossRef]
- Seguí-Simarro, J.M.; Nuez, F. How microspores transform into haploid embryos: Changes associated with embryogenesis induction and microspore-derived embryogenesis. Physiol. Plant. 2008, 134, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shariatpanahi, M.E.; Bal, U.; Heberle-Bors, E.; Touraev, A. Stresses applied for the re-programming of plant microspores towards in vitro embryogenesis. Physiol. Plant. 2006, 127, 519–534. [Google Scholar] [CrossRef]
- Islam, S.M.S.; Tuteja, N. Enhancement of androgenesis by abiotic stress and other pretreatments in major crop species. Plant Sci. 2012, 182, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Clarke, H.J.; Small, I.; Siddique, K.H.M. Albinism in plants: A major bottleneck in wide hybridization, androgenesis and doubled haploid culture. CRC. Crit. Rev. Plant Sci. 2009, 28, 393–409. [Google Scholar] [CrossRef]
- Maluszynski, M.; Kasha, K.J.; Forster, B.P.; Szarejko, I. Doubled Haploid Production in Crop Plants: A manual; Maluszynski, M., Kasha, K.J., Forster, B.P., Szarejko, I., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003. [Google Scholar]
- Touraev, A.; Forster, B.P.; Mohan Jain, S. Advances in Haploid Production in Higher Plants; Touraev, A., Forster, B.P., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar]
- Germanà, M.A.; Lambardi, M. In Vitro Embryogenesis in Higher Plants, 1st ed.; Germanà, M.A., Lambardi, M., Eds.; Humana Press: New York, NY, USA, 2016. [Google Scholar]
- Niu, Z.; Jiang, A.; Abu Hammad, W.; Oladzadabbasabadi, A.; Xu, S.S.; Mergoum, M.; Elias, E.M. Review of doubled haploid production in durum and common wheat through wheat × maize hybridization. Plant Breed. 2014, 133, 313–320. [Google Scholar] [CrossRef]
- Chen, J.-F.; Cui, L.; Malik, A.A.; Mbira, K.G. In vitro haploid and dihaploid production via unfertilized ovule culture. Plant Cell Tissue Organ Cult. 2010, 104, 311–319. [Google Scholar] [CrossRef]
- Pink, D.; Bailey, L.; McClement, S.; Hand, P.; Mathas, E.; Buchanan-Wollaston, V.; Astley, D.; King, G.; Teakle, G. Double haploids, markers and QTL analysis in vegetable brassicas. Euphytica 2008, 164, 509–514. [Google Scholar] [CrossRef]
- Tuvesson, S.; Dayteg, C.; Hagberg, P.; Manninen, O.; Tanhuanpää, P.; Tenhola-Roininen, T.; Kiviharju, E.; Weyen, J.; Förster, J.; Schondelmaier, J.; et al. Molecular markers and doubled haploids in European plant breeding programmes. Euphytica 2006, 158, 305–312. [Google Scholar] [CrossRef]
- Devaux, P.; Kasha, K.J. Overview of barley doubled haploid production. In Advances in Haploid Production in Higher Plants; Touraev, A., Forster, B.P., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 47–64. [Google Scholar]
- Mishra, R.; Jwala, G.; Ao, N.R. In Vitro androgenesis in rice: Advantages, constraints and future prospects. Rice Sci. 2016, 23, 57–68. [Google Scholar] [CrossRef]
- Ferrie, A.M.R.; Möllers, C. Haploids and doubled haploids in Brassica spp. for genetic and genomic research. Plant Cell Tissue Organ Cult. 2010, 104, 375–386. [Google Scholar] [CrossRef]
- Lippman, Z.B.; Zamir, D. Heterosis: Revisiting the magic. Trends Genet. 2006, 23, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Yao, H.; Chudalayandi, S.; Vaiman, D.; Veitia, R.A. Heterosis. Plant Cell 2010, 22, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Ferrie, A.M.R. Doubled haploidy as a tool in ornamental breeding. Acta Hortic. 2012, 953, 167–171. [Google Scholar] [CrossRef]
- Obsa, B.T.; Eglinton, J.; Coventry, S.; March, T.; Langridge, P.; Fleury, D. Genetic analysis of developmental and adaptive traits in three doubled haploid populations of barley (Hordeum vulgare L.). Theor. Appl. Genet. 2016, 129, 1139–1151. [Google Scholar] [CrossRef] [PubMed]
- Brew-Appiah, R.A.T.; Ankrah, N.; Liu, W.; Konzak, C.F.; Von Wettstein, D.; Rustgi, S. Generation of doubled haploid transgenic wheat lines by microspore transformation. PLoS ONE 2013, 8, e80155. [Google Scholar] [CrossRef] [PubMed]
- Kumlehn, J.; Serazetdinova, L.; Hensel, G.; Becker, D.; Loerz, H. Genetic transformation of barley (Hordeum vulgare L.) via infection of androgenetic pollen cultures with Agrobacterium tumefaciens. Plant Biotechnol. J. 2006, 4, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Kapusi, E.; Hensel, G.; Coronado, M.J.; Broeders, S.; Marthe, C.; Otto, I.; Kumlehn, J. The elimination of a selectable marker gene in the doubled haploid progeny of co-transformed barley plants. Plant Mol. Biol. 2013, 81, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liu, Z. A new method for generation and screening of Chinese cabbage mutants using isolated microspore culturing and EMS mutagenesis. Euphytica 2016, 207, 23–33. [Google Scholar] [CrossRef]
- Shen, Y.; Pan, G.; Lübberstedt, T. Haploid strategies for functional validation of plant genes. Trends Biotechnol. 2015, 33, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Ferrie, A.M.R.; Caswell, K.L. Applications of doubled haploidy for improving industrial oilseeds. In Industrial Oil Crops; McKeon, T.A., Hayes, D.G., Hildebrand, D.F., Weselake, R.J., Eds.; Elsevier Inc.: London, UK, 2016; pp. 359–378. [Google Scholar]
- Soriano, M.; Li, H.; Boutilier, K. Microspore embryogenesis: Establishment of embryo identity and pattern in culture. Plant Reprod. 2013, 26, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Daghma, D.E.S.; Hensel, G.; Rutten, T.; Melzer, M.; Kumlehn, J. Cellular dynamics during early barley pollen embryogenesis revealed by time-lapse imaging. Front. Plant Sci. 2014, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Seifert, F.; Bössow, S.; Kumlehn, J.; Gnad, H.; Scholten, S. Analysis of wheat microspore embryogenesis induction by transcriptome and small RNA sequencing using the highly responsive cultivar “Svilena”. BMC Plant Biol. 2016, 16, 97. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.F.; Simpson, R.J.; Culvenor, R.A.; Humphreys, M.O.; Prud’Homme, M.P.; Oram, R.N. The effects of ploidy and a phenotype conferring a high water-soluble carbohydrate concentration on carbohydrate accumulation, nutritive value and morphology of perennial ryegrass (Lolium perenne L.). J. Agric. Sci. 2001, 136, 65–74. [Google Scholar] [CrossRef]
- Smith, K.F.; McFarlane, N.M.; Croft, V.M.; Trigg, P.J.; Kearney, G.A. The effects of ploidy and seed mass on the emergence and early vigour of perennial ryegrass (Lolium perenne L.) cultivars. Aust. J. Exp. Agric. 2003, 43, 481–486. [Google Scholar] [CrossRef]
- Nair, R. Developing tetraploid perennial ryegrass (Lolium perenne L.) populations. N. Z. J. Agric. Res. 2004, 47, 45–49. [Google Scholar] [CrossRef]
- Wilkins, P.W. Breeding perennial ryegrass for agriculture. Euphytica 1991, 52, 201–214. [Google Scholar] [CrossRef]
- Sampoux, J.P.; Baudouin, P.; Bayle, B.; Béguier, V.; Bourdon, P.; Chosson, J.F.; Deneufbourg, F.; Galbrun, C.; Ghesquière, M.; Noël, D.; et al. Breeding perennial grasses for forage usage: An experimental assessment of trait changes in diploid perennial ryegrass (Lolium perenne L.) cultivars released in the last four decades. Field Crops Res. 2011, 123, 117–129. [Google Scholar] [CrossRef]
- Humphreys, M.O. Genetic improvement of forage crops—Past, present and future. J. Agric. Sci. 2005, 143, 441–448. [Google Scholar] [CrossRef]
- Cornish, M.A.; Hayward, M.D.; Lawrence, M.J. Self-incompatibility in ryegrass. I. Genetic control in diploid Lolium perenne L. Heredity 1979, 43, 95–106. [Google Scholar] [CrossRef]
- Blackmore, T.; Thorogood, D.; Skøt, L.; Mcmahon, R.; Powell, W.; Hegarty, M. Germplasm dynamics: The role of ecotypic diversity in shaping the patterns of genetic variation in Lolium perenne. Sci. Rep. 2016, 6, 22603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Pembleton, L.W.; Baillie, R.C.; Drayton, M.C.; Hand, M.L.; Bain, M.; Sawbridge, T.I.; Spangenberg, G.C.; Forster, J.W.; Cogan, N.O.I. Development and implementation of a multiplexed single nucleotide polymorphism genotyping tool for differentiation of ryegrass species and cultivars. Mol. Breed. 2014, 33, 435–451. [Google Scholar] [CrossRef]
- Casler, M.D.; Brummer, E.C. Theoretical expected genetic gains for among-and-within-family selection methods in perennial forage crops. Crop Sci. 2008, 48, 890–902. [Google Scholar] [CrossRef]
- Stanis, V.A.; Butenko, R.G. Developing viable haploid plants in anther culture of ryegrass. Dokl. Biol. Sci. 1984, 275, 249–251. [Google Scholar]
- Boppenmeier, J.; Zuchner, S.; Foroughi-Wehr, B. Haploid production from barley yellow dwarf virus resistant clones of Lolium. Plant Breed. 1989, 103, 216–220. [Google Scholar] [CrossRef]
- Olesen, A.; Andersen, S.B.; Due, I.K. Anther culture response in perennial ryegrass (Lolium perenne L.). Plant Breed. 1988, 101, 60–65. [Google Scholar] [CrossRef]
- Opsahl-Ferstad, H.-G.; Bjornstad, A.; Rognli, O.A. Influence of medium and cold pretreatment on androgenetic response in Lolium perenne L. Plant Cell Rep. 1994, 13, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Bante, I.; Sonke, T.; Tandler, R.F.; van den Bruel, A.M.R.; Meyer, E.M. Anther culture of Lolium perenne and Lolium multiflorum. In The Impact of Biotechnology in Agriculture; Sangwan, R., Sangwan-Norreel, B.S., Eds.; Springer: Dordrecht, The Netherlands, 1990; pp. 105–127. [Google Scholar]
- Creemers-Molenaar, J.; Beerepoot, L.J. In Vitro Culture and Micropropagation of Ryegrass (Lolium spp.). In High-Tech and Micropropagation III; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1992; Volume 19, pp. 549–575. [Google Scholar]
- Madsen, S.; Olesen, A.; Dennis, B.; Andersen, S.B. Inheritance of anther-culture response in perennial ryegrass (Lolium perenne L.). Plant Breed. 1995, 114, 165–168. [Google Scholar] [CrossRef]
- Hussain, S.W.; Richardson, K.; Faville, M.; Woodfield, D. Production of haploids and double haploids in annual (Lolium multiflorum) and perennial (L. perenne) ryegrass. In Proceedings of the 13th Australasian Plant Breeding Conference, Christchurch, New Zealand, 18–21 April 2006; Mercer, C.F., Ed.; pp. 531–536.
- Andersen, S.B.; Madsen, S.; Roulund, N.; Halberg, N.; Olesen, A. Haploidy in ryegrass. In In Vritro Haploid Production in Higher Plants; Jain, S.M., Sopory, S.K., Veilleux, R.E., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1997; Volume 4, pp. 133–147. [Google Scholar]
- Halberg, N.; Olesen, A.; Tuvesson, I.K.D.; Andersen, S.B. Genotypes of perennial ryegrass (Lolium perenne L.) with high anther-culture response through hybridization. Plant Breed. 1990, 105, 89–94. [Google Scholar] [CrossRef]
- Opsahl-Ferstad, H.G.; Bjørnstad, N.; Rognli, O.A. Genetic control of androgenetic response in Lolium perenne L. Theor. Appl. Genet. 1994, 89, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Ge, Y.; Mian, R.; Baker, J. Development of highly tissue culture responsive lines of Lolium temulentum by anther culture. Plant Sci. 2005, 168, 203–211. [Google Scholar] [CrossRef]
- Chen, X.-W.; Cistué, L.; Muñoz-Amatriaín, M.; Sanz, M.; Romagosa, I.; Castillo, A.-M.; Vallés, M.-P. Genetic markers for doubled haploid response in barley. Euphytica 2006, 158, 287–294. [Google Scholar] [CrossRef]
- Agache, S.; Bachelier, B.; De Buyser, J.; Henry, Y.; Snape, J. Genetic analysis of anther culture response in wheat using aneuploid, chromosome substitution and translocation lines. Theor. Appl. Genet. 1989, 77, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Torp, A.M.; Andersen, S.B. Albinism in microspore culture. In Advances in Haploid Production in Higher Plants; Touraev, A., Forster, B., Jain, S.M., Eds.; Springer: New York, NY, USA, 2009; pp. 155–160. [Google Scholar]
- Bolibok, H.; Rakoczy-Trojanowska, M. Genetic mapping of QTLs for tissue-culture response in plants. Euphytica 2006, 149, 73–83. [Google Scholar] [CrossRef]
- Nielsen, N.H.; Andersen, S.U.; Stougaard, J.; Jensen, A.; Backes, G.; Jahoor, A. Chromosomal regions associated with the in vitro culture response of wheat (Triticum aestivum L.) microspores. Plant Breed. 2015, 134, 255–263. [Google Scholar] [CrossRef]
- Krzewska, M.; Czyczyło-Mysza, I.; Dubas, E.; Gołebiowska-Pikania, G.; Golemiec, E.; Stojałowski, S.; Chrupek, M.; Zur, I. Quantitative trait loci associated with androgenic responsiveness in triticale (×Triticosecale Wittm.) anther culture. Plant Cell Rep. 2012, 31, 2099–2108. [Google Scholar] [CrossRef] [PubMed]
- Begheyn, R.F.; Vangsgaard, K.; Roulund, N. Efficient doubled haploid production in perennial ryegrass (Lolium perenne L.). In Breeding in a World of Scarcity, Proceedings of the 2015 Meeting of the Section “Forage Crops and Amenity Grasses” of Eucarpia, Ghent, Belgium, 14 June 2016; Springer: Basel, Switzerland; pp. 151–155.
- Melchinger, A.E.; Molenaar, W.S.; Mirdita, V.; Schipprack, W. Colchicine alternatives for chromosome doubling in maize haploids for doubled-haploid production. Crop Sci. 2016, 56, 559–569. [Google Scholar] [CrossRef]
- Opsahl-Ferstad, H.G. Androgenetic Response in Grasses. I. Anther Culture in Perennial Ryegrass (Lolium perenne L.). II. Molecular Studies of Embryogenesis in Barley (Hordeum vulgare L.). Ph.D. Thesis, Agricultural University of Norway, Ås, Norway, 1993. [Google Scholar]
- Madsen, S.; Olesen, A.; Andersen, S.B. Self-fertile doubled haploid plants of perennial ryegrass (Lolium perenne L.). Plant Breed. 1993, 110, 323–327. [Google Scholar] [CrossRef]
- Manzanares, C.; Yates, S.; Ruckle, M.; Nay, M.; Studer, B. TILLING in forage grasses for gene discovery and breeding improvement. New Biotechnol. 2016, 33, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Conaghan, P.; Casler, M.D. A theoretical and practical analysis of the optimum breeding system for perennial ryegrass. Irish J. Agric. Sci. 2011, 50, 47–63. [Google Scholar]
- Wang, Z.Y.; Brummer, E.C. Is genetic engineering ever going to take off in forage, turf and bioenergy crop breeding? Ann. Bot. 2012, 110, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Arias Aguirre, A.; Studer, B.; Frei, U.; Lübberstedt, T. Prospects for hybrid breeding in bioenergy grasses. BioEnergy Res. 2011, 5, 10–19. [Google Scholar] [CrossRef]
- Langridge, P.; Fleury, D. Making the most of “omics” for crop breeding. Trends Biotechnol. 2011, 29, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.K.; Eudes, F. Dimethyl tyrosine conjugated peptide prevents oxidative damage and death of triticale and wheat microspores. Plant Cell Tissue Organ Cult. 2015, 122, 227–237. [Google Scholar] [CrossRef]
- Sriskandarajah, S.; Sameri, M.; Lerceteau-Köhler, E.; Westerbergh, A. Increased recovery of green doubled haploid plants from barley anther culture. Crop Sci. 2015, 55, 2806–2812. [Google Scholar] [CrossRef]
- Echavarri, B.; Cistue, L. Enhancement in androgenesis efficiency in barley (Hordeum vulgare L.) and bread wheat (Triticum aestivum L.) by the addition of dimethyl sulfoxide to the mannitol pretreatment medium. Plant Cell Tissue Organ Cult. 2016, 125, 11–22. [Google Scholar] [CrossRef]
- Devaux, P.; Zivy, M. Protein markers for anther culturability in barley. Theor. Appl. Genet. 1994, 88, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Luo, J.; Chen, W.; Hao, M.; Liu, B.; Yan, Z.; Zhang, B.; Zhang, H.; Zheng, Y.; et al. Synthesizing double haploid hexaploid wheat populations based on a spontaneous alloploidization process. J. Genet. Genom. 2011, 38, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Amatriaín, M.; Castillo, A.M.; Chen, X.W.; Cistué, L.; Vallés, M.P. Identification and validation of QTLs for green plant percentage in barley (Hordeum vulgare L.) anther culture. Mol. Breed. 2008, 22, 119–129. [Google Scholar] [CrossRef]
- Bélanger, S.; Esteves, P.; Clermont, I.; Jean, M.; Belzile, F. Genotyping-by-sequencing on pooled samples and its use in measuring segregation bias during the course of androgenesis in barley. Plant Genome 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Hayward, M.D.; Olesen, A.; Due, I.K.; Jenkins, R.; Morris, P. Segregation of isozyme marker loci amongst androgenetic plants of Lolium perenne L. Plant Breed. 1990, 104, 68–71. [Google Scholar] [CrossRef]
- Kindiger, B. Generation of paternal dihaploids in tall fescue. Grassl. Sci. 2016, 62, 1–5. [Google Scholar] [CrossRef]
- Kindiger, B. Sampling the genetic diversity of tall fescue utilizing gamete selection. In Genetic Diversity in Plants; Caliskan, M., Ed.; InTech: Rijeka, Croatia, 2012; pp. 271–284. [Google Scholar]
- Ravi, M.; Chan, S.W. Centromere-mediated generation of haploid plants. In Plant Centromere Biology; Jiang, J., Birchler, J.A., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 169–181. [Google Scholar]
- Kelliher, T.; Starr, D.; Wang, W.; Mccuiston, J.; Zhong, H.; Nuccio, M.L.; Martin, B. Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize. Front. Plant Sci. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Ashtiyani, R.; Ishii, T.; Niessen, M.; Stein, N.; Heckmann, S.; Gurushidze, M.; Banaei-Moghaddam, A.M.; Fuchs, J.; Schubert, V.; Koch, K.; et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. USA 2015, 112, 11211–11216. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D.; Willis, J.H. The genetics of inbreeding depression. Nat. Rev. Genet. 2009, 10, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Wilde, K.; Burger, H.; Prigge, V.; Presterl, T.; Schmidt, W.; Ouzunova, M.; Geiger, H.H. Testcross performance of doubled-haploid lines developed from European flint maize landraces. Plant Breed. 2010, 129, 181–185. [Google Scholar] [CrossRef]
- Strigens, A.; Schipprack, W.; Reif, J.C.; Melchinger, A.E. Unlocking the genetic diversity of maize landraces with doubled haploids opens new avenues for breeding. PLoS ONE 2013, 8, e57234. [Google Scholar] [CrossRef] [PubMed]
- Jacquard, C.; Nolin, F.; Hécart, C.; Grauda, D.; Rashal, I.; Dhondt-Cordelier, S.; Sangwan, R.S.; Devaux, P.; Mazeyrat-Gourbeyre, F.; Clément, C. Microspore embryogenesis and programmed cell death in barley: Effects of copper on albinism in recalcitrant cultivars. Plant Cell Rep. 2009, 28, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.T.; Tuvesson, I.K.; Andersen, S.B. Nuclear genes affecting percentage of green plants in barley (Hordeum vulgare L.) anther culture. Theor. Appl. Genet. 1991, 82, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Gurushidze, M.; Hensel, G.; Hiekel, S.; Schedel, S.; Valkov, V.; Kumlehn, J. True-breeding targeted gene knock-out in barley using designer TALE-nuclease in haploid cells. PLoS ONE 2014, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Giri, C.C.; Praveena, M. In vitro regeneration, somatic hybridization and genetic transformation studies: An appraisal on biotechnological interventions in grasses. Plant Cell Tissue Organ Cult. 2014, 120, 843–860. [Google Scholar] [CrossRef]
- Eudes, F.; Shim, Y.S.; Jiang, F. Engineering the haploid genome of microspores. Biocatal. Agric. Biotechnol. 2014, 3, 20–23. [Google Scholar] [CrossRef]
- Chugh, A.; Amundsen, E.; Eudes, F. Translocation of cell-penetrating peptides and delivery of their cargoes in triticale microspores. Plant Cell Rep. 2009, 28, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Bilichak, A.; Luu, J.; Eudes, F. Intracellular delivery of fluorescent protein into viable wheat microspores using cationic peptides. Front. Plant Sci. 2015, 6, 666. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Jain, M. The CRISPR-Cas system for plant genome editing: Advances and opportunities. J. Exp. Bot. 2015, 66, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Maluszynski, M.; Ahloowalia, B.S.; Sigurbjörnsson, B. Application of in vivo and in vitro mutation techniques for crop improvement. Euphytica 1995, 85, 303–315. [Google Scholar] [CrossRef]
- Szarejko, I.; Forster, B.P. Doubled haploidy and induced mutation. Euphytica 2007, 158, 359–370. [Google Scholar] [CrossRef]
- Kurowska, M.; Labocha-Pawłowska, A.; Gnizda, D.; Maluszynski, M.; Szarejko, I. Molecular analysis of point mutations in a barley genome exposed to MNU and Gamma rays. Mutat. Res. 2012, 738–739, 52–70. [Google Scholar] [CrossRef] [PubMed]
- Lübberstedt, T.; Frei, U.K. Application of doubled haploids for target gene fixation in backcross programmes of maize. Plant Breed. 2012, 131, 449–452. [Google Scholar] [CrossRef]
- Delourme, R.; Falentin, C.; Fomeju, B.F.; Boillot, M.; Lassalle, G.; André, I.; Duarte, J.; Gauthier, V.; Lucante, N.; Marty, A.; et al. High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genom. 2013, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Cabral, A.L.; Jordan, M.C.; McCartney, C.A.; You, F.M.; Humphreys, D.G.; MacLachlan, R.; Pozniak, C.J. Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2014, 14, 340. [Google Scholar] [CrossRef] [PubMed]
- Sannemann, W.; Huang, B.E.; Mathew, B.; Léon, J. Multi-parent advanced generation inter-cross in barley: High-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 2015, 35, 86. [Google Scholar] [CrossRef]
- Bert, P.F.; Charmet, G.; Sourdille, P.; Hayward, M.D.; Balfourier, F. A high-density molecular map for ryegrass (Lolium perenne) using AFLP markers. Theor. Appl. Genet. 1999, 99, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.S.; Mahoney, N.L.; Hayward, M.D.; Armstead, I.P.; Jones, J.G.; Humphreys, M.O.; King, I.P.; Kishida, T.; Yamada, T.; Balfourier, F.; et al. An enhanced molecular marker based genetic map of perennial ryegrass (Lolium perenne) reveals comparative relationships with other Poaceae genomes. Genome 2002, 45, 282–295. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wang, D.; Guo, Z.; Beavis, W.D. Family-based association mapping in crop species. Theor. Appl. Genet. 2013, 126, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- King, J.; Armstead, I.P.; Donnison, I.S.; Harper, J.A.; Roberts, L.A.; Thomas, H.; Ougham, H.; Thomas, A.; Huang, L.; King, I.P. Introgression mapping in the grasses. Chromosom. Res. 2007, 15, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, M.W.; Gasior, D.; Lesniewska-Bocianowska, A.; Zwierzykowski, Z.; Rapacz, M. Androgenesis as a means of dissecting complex genetic and physiological controls: Selecting useful gene combinations for breeding freezing tolerant grasses. Euphytica 2007, 158, 337–345. [Google Scholar] [CrossRef]
- Petersen, G.; Johansen, B.; Seberg, O. PCR and sequencing from a single pollen grain. Plant Mol. Biol. 1996, 31, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Dreissig, S.; Fuchs, J.; Cápal, P.; Kettles, N.; Byrne, E.; Houben, A. Measuring meiotic crossovers via multi-locus genotyping of single pollen grains in barley. PLoS ONE 2015, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Pan, Y.B.; Chen, R.K. High-throughput procedure for single pollen grain collection and polymerase chain reaction in plants. J. Integr. Plant Biol. 2008, 50, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.A. Tansley review—A unifying theory for general multigenic heterosis: Energy efficiency, protein metabolism, and implications for molecular breeding. New Phytol. 2011, 189, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Hallauer, A.R.; Carena, M.J.; Filho, J.B.M. Inbreeding. In Quantitative Genetics in Maize Breeding; Hallauer, A.R., Carena, M.J., Filho, J.B.M., Eds.; Springer: New York, NY, USA, 2010; pp. 425–475. [Google Scholar]
- Geiger, H.H.; Miedaner, T. Rye breeding. In Cereals; Carena, M.J., Ed.; Springer: New York, NY, USA, 2009; pp. 157–181. [Google Scholar]
- Brummer, E.C. Capturing heterosis in forage crop cultivar development. Crop Sci. 1999, 39, 943–954. [Google Scholar] [CrossRef]
- Riddle, N.C.; Birchler, J.A. Comparative analysis of inbred and hybrid maize at the diploid and tetraploid levels. Theor. Appl. Genet. 2008, 116, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Kopecky, D.; Lukaszewski, A.J.; Gibeault, V. Reduction of ploidy level by androgenesis in intergenic Lolium—Festuca hybrids for turf grass breeding. Crop Sci. 2005, 45, 274–281. [Google Scholar]
- Yang, B.; Thorogood, D.; Armstead, I.P.; Franklin, F.C.H.; Barth, S. Identification of genes expressed during the self-incompatibility response in perennial ryegrass (Lolium perenne L.). Plant Mol. Biol. 2009, 70, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Klaas, M.; Yang, B.; Bosch, M.; Thorogood, D.; Manzanares, C.; Armstead, I.P.; Franklin, F.C.H.; Barth, S. Progress towards elucidating the mechanisms of self-incompatibility in the grasses: Further insights from studies in Lolium. Ann. Bot. 2011, 108, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Manzanares, C.; Barth, S.; Thorogood, D.; Byrne, S.L.; Yates, S.; Czaban, A.; Asp, T.; Yang, B.; Studer, B. A gene encoding a DUF247 domain protein cosegregates with the S self-incompatibility locus in perennial ryegrass. Mol. Biol. Evol. 2016, 33, 870–884. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Studer, B.; Møller, I.M.; Asp, T. Genetics and biology of cytoplasmic male sterility and its applications in forage and turf grass breeding. Plant Breed. 2014, 133, 299–312. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Shinozuka, H.; Wang, J.; Spangenberg, G.C.; Forster, J.W.; Cogan, N.O.I. Design of an F1 hybrid breeding strategy for ryegrasses based on selection of self-incompatibility locus-specific alleles. Front. Plant Sci. 2015, 6, 764. [Google Scholar] [CrossRef] [PubMed]
- Do Canto, J.; Studer, B.; Lubberstedt, T. Overcoming self-incompatibility in grasses: A pathway to hybrid breeding. Theor. Appl. Genet. 2016, 129, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Posselt, U.K. Heterosis in grasses. Czech J. Genet. Plant Breed. 2003, 39, 48–53. [Google Scholar]
| Repeated Self-Fertilization | In Vitro Doubled Haploid Induction | In Vivo Doubled Haploid Induction | |
|---|---|---|---|
| Method available | yes | yes 1 | no |
| Genotype specificity | low | high | unknown |
| Efficiency | low | high 2 | unknown |
| Required skill | low | moderate | low 3 |
| Space required | high | low | high |
| Lab requirements | none | high | low |
| Generations required | 5–6 4 | 1 | 1 |
| Diploid regenerants | 100% | 50%–80% | unknown 5 |
| Obstacles | self-incompatibility inbreeding depression | albinism inbreeding depression | inbreeding depression |
| Side effects of procedure 6 | allows selection every generation | gametoclonal variation somatoclonal variation ploidy level variation segregation distortion | part of inducer genome could integrate |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Begheyn, R.F.; Lübberstedt, T.; Studer, B. Haploid and Doubled Haploid Techniques in Perennial Ryegrass (Lolium perenne L.) to Advance Research and Breeding. Agronomy 2016, 6, 60. https://doi.org/10.3390/agronomy6040060
Begheyn RF, Lübberstedt T, Studer B. Haploid and Doubled Haploid Techniques in Perennial Ryegrass (Lolium perenne L.) to Advance Research and Breeding. Agronomy. 2016; 6(4):60. https://doi.org/10.3390/agronomy6040060
Chicago/Turabian StyleBegheyn, Rachel F., Thomas Lübberstedt, and Bruno Studer. 2016. "Haploid and Doubled Haploid Techniques in Perennial Ryegrass (Lolium perenne L.) to Advance Research and Breeding" Agronomy 6, no. 4: 60. https://doi.org/10.3390/agronomy6040060






