NRT2.4 and NRT2.5 Are Two Half-Size Transporters from the Chlamydomonas NRT2 Family
Abstract
:1. Introduction
2. Results
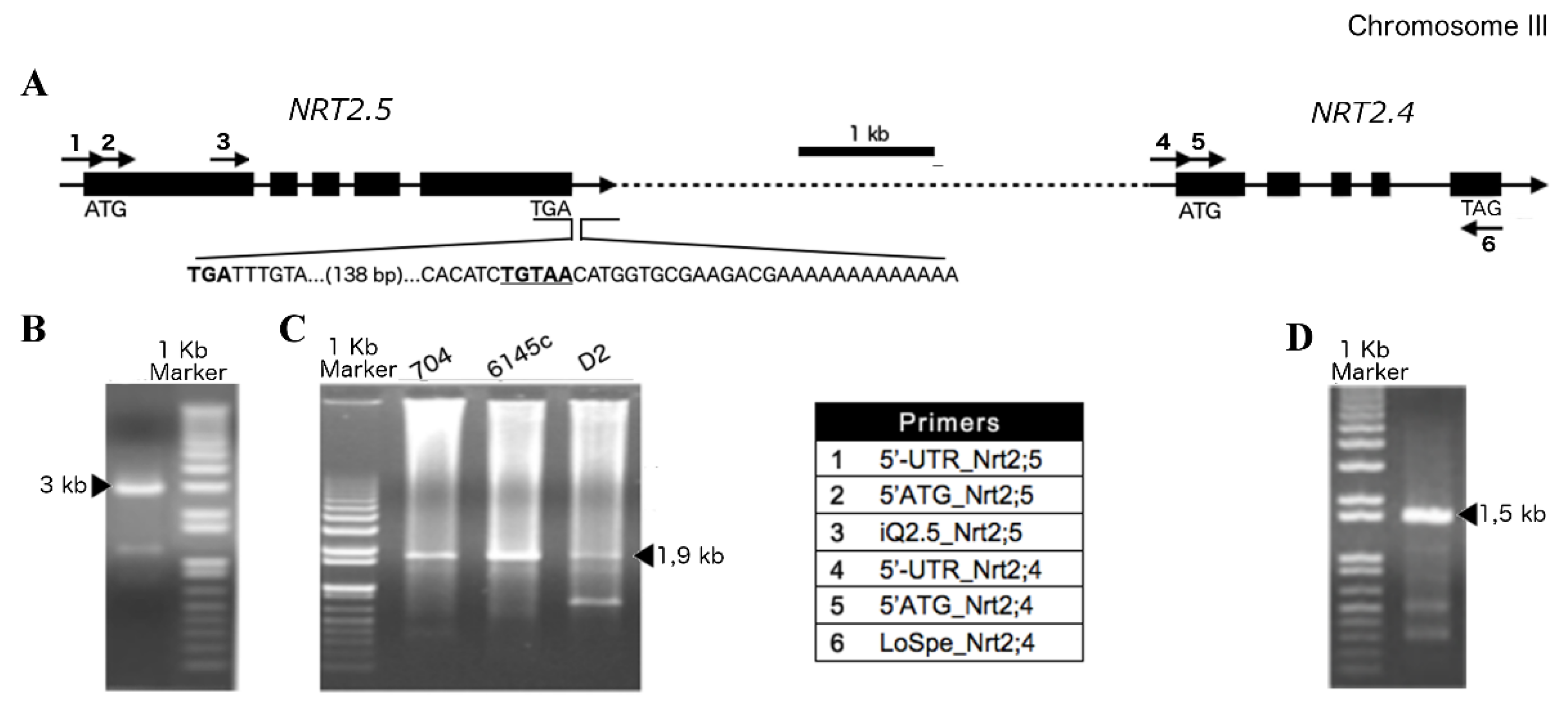
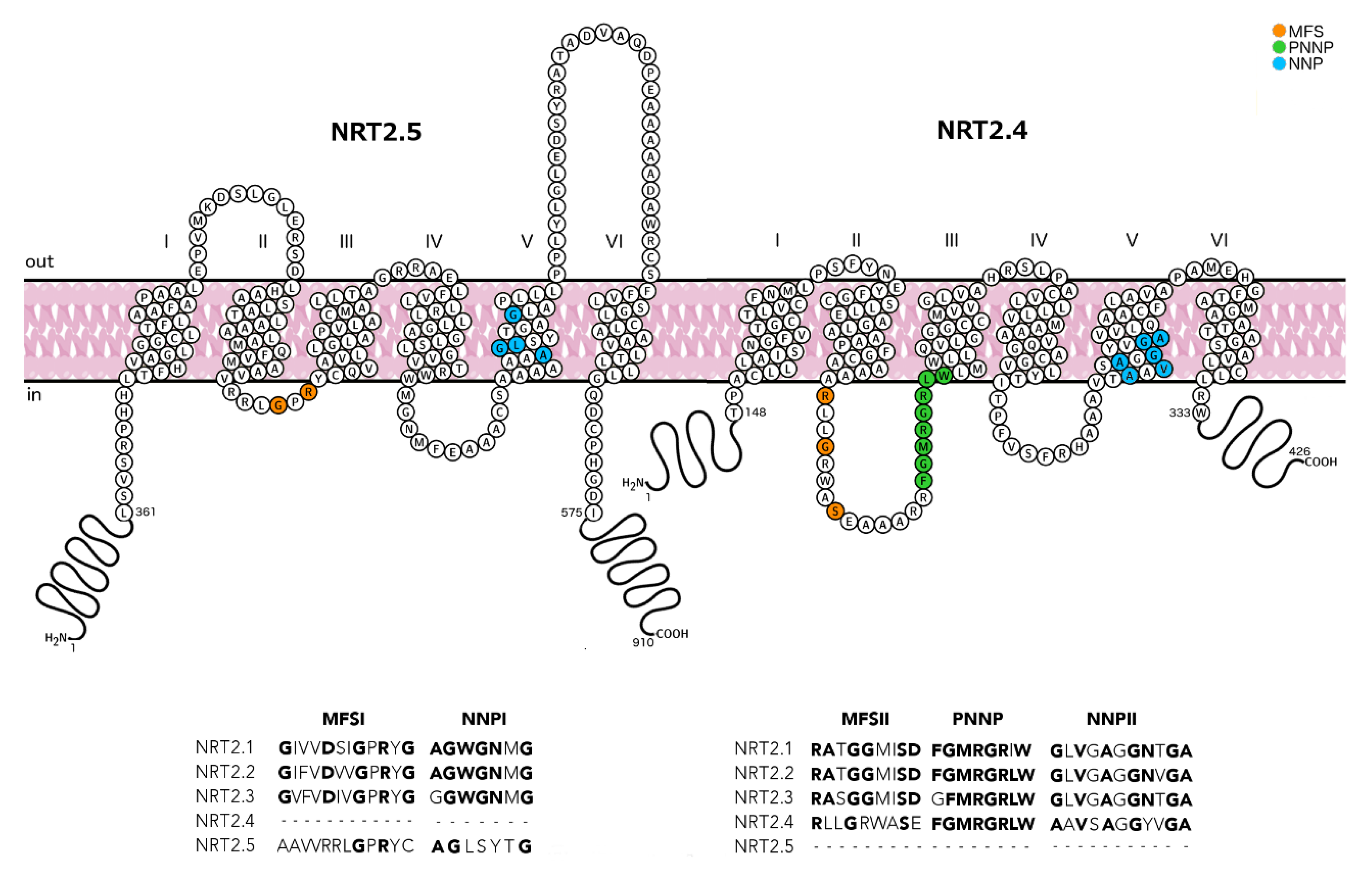
2.1. Half-Size Transporters Encoded by NRT2.4 and NRT2.5
2.2. Expression of NRT2.4 and NRT2.5 is Not Regulated by Nitrogen
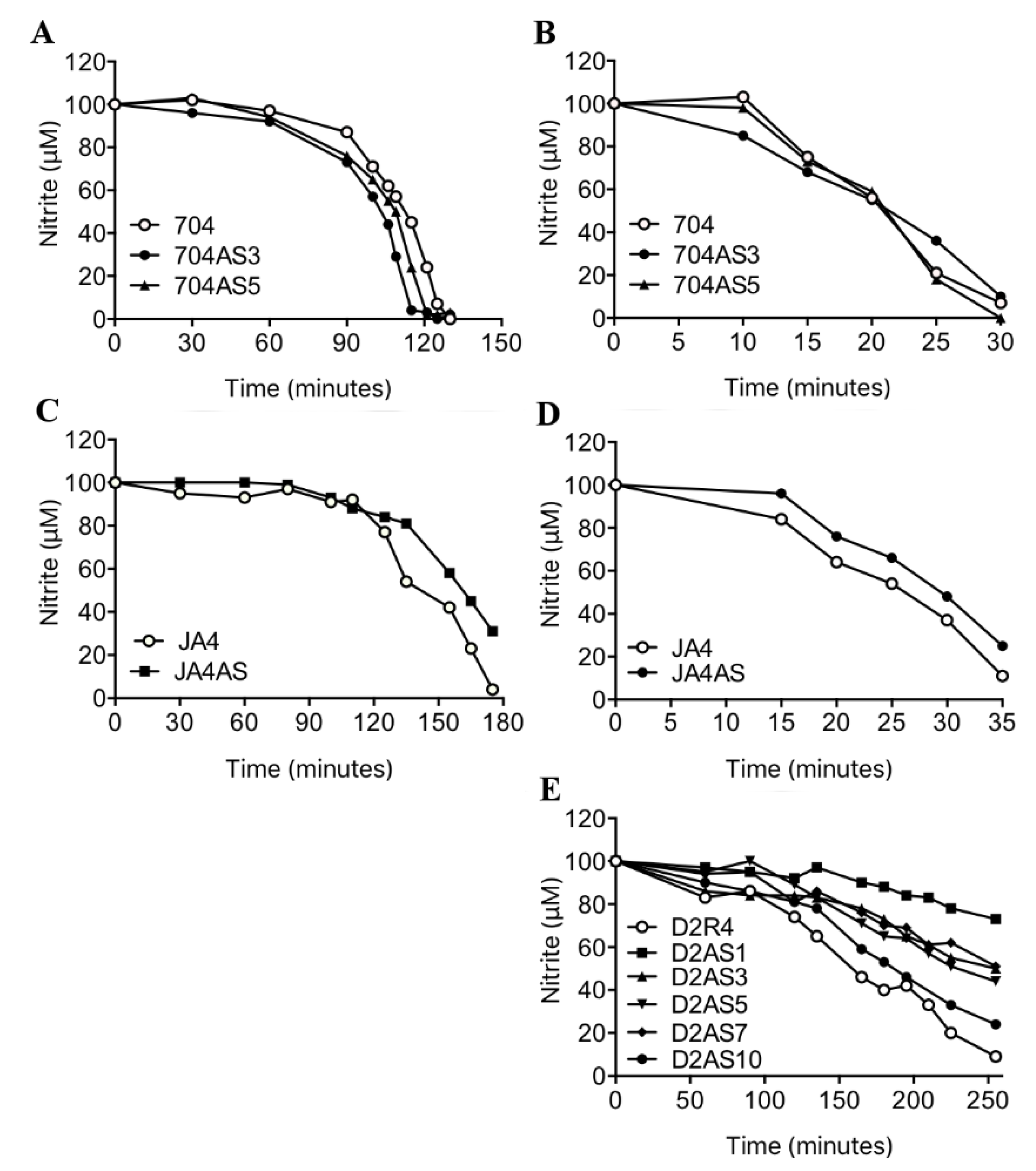
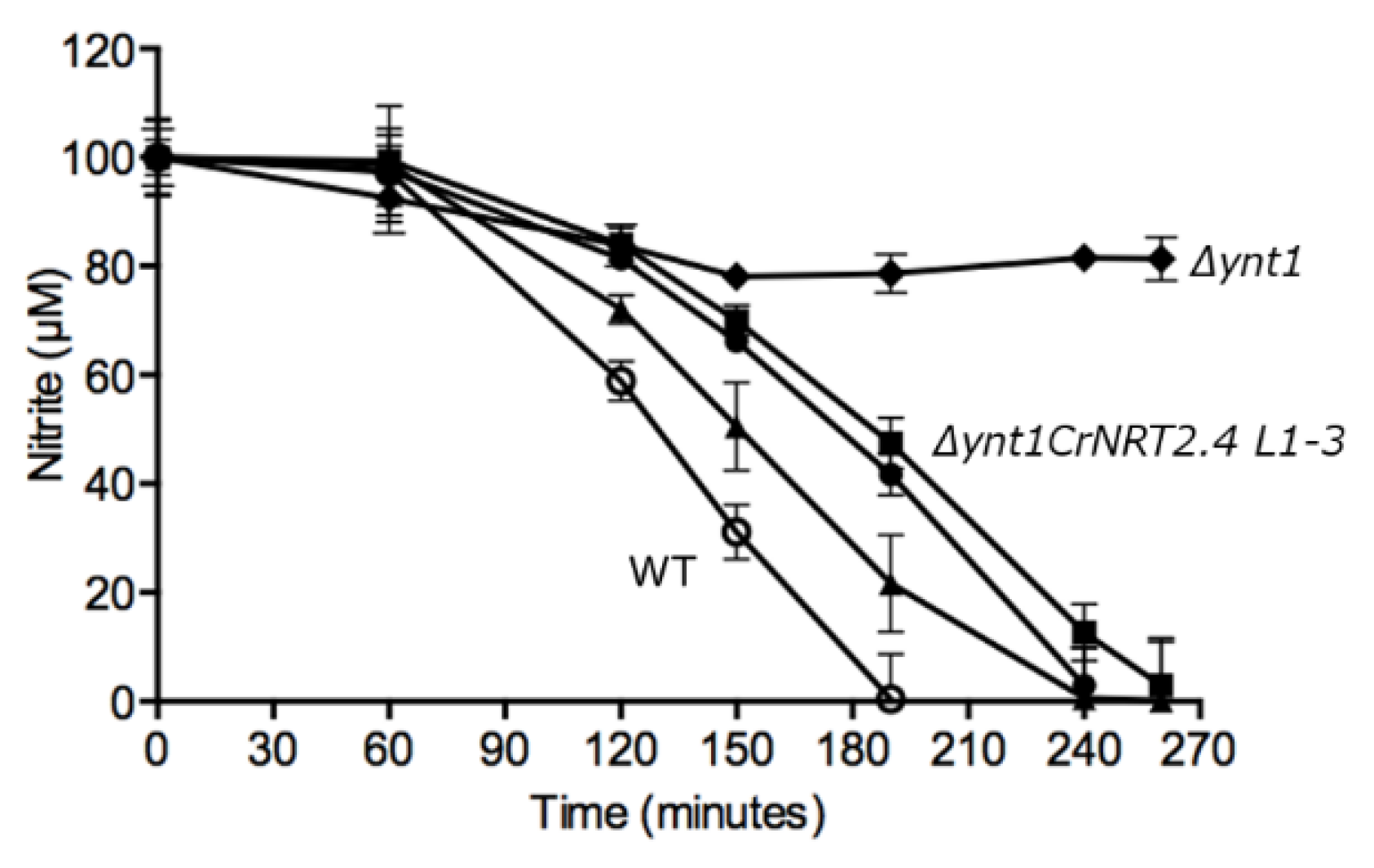
2.3. NRT2.4 is a Nitrite Transporter
3. Discussion
4. Experimental Section
4.1. Strains and Growth Conditions
4.2. Silencing of NRT2.4
4.3. Expression of CrNRT2.4 in Hansenula Polymorpha
4.4. cDNA Synthesis, Isolation of cDNA for NRT2.4 and NRT2.5 and Quantification of Gene Expression
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Bouguyon, E.; Gojon, A.; Nacry, P. Nitrate sensing and signaling in plants. Semin. Cell Dev. Biol. 2012, 23, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Ruffel, S.; Krouk, G.; Coruzzi, G.M. A systems view of responses to nutritional cues in Arabidopsis: Toward a paradigm shift for predictive network modeling. Plant Physiol. 2010, 152, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A.; David, L.C.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.S.; Chaillou, S.; Ferrario-Méry, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Fernández, E.; Galván, A. Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 2007, 58, 2279–2287. [Google Scholar] [CrossRef] [PubMed]
- Fernández, E.; Galván, A. Nitrate assimilation in Chlamydomonas. Eukaryot. Cell 2008, 7, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Luque, E.; Chamizo-Ampudia, A.; Llamas, A.; Galván, A.; Fernández, E. Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 2015, 6, 899. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, Y.; Wang, J.; Cheng, C.; Huang, W.; Lu, P.; Xu, Y.-N.; Wang, P.; Yan, N.; Shi, Y. Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel. Nature 2009, 462, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Tovell, N.; Clegg, S.; Trimmer, M.; Cole, J. A single channel for nitrate uptake, nitrite export and nitrite uptake by Escherichia coli NarU and a role for NirC in nitrite export and uptake. Biochem. J. 2009, 417, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, W.; Siddiqi, Y.; Symington, V.F.; Kinghorn, J.R.; Unkles, S.E.; Glass, A.D.M. Nitrite transport is mediated by the nitrite-specific high-affinity NitA transporter and by nitrate transporters NrtA, NrtB in Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, E.; González-Montelongo, R.; Giraldez, T.; de la Rosa, D.A.; Siverio, J.M. Molecular components of nitrate and nitrite efflux in yeast. Eukaryot. Cell 2014, 13, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Rexach, J.; Fernández, E.; Galván, A. The Chlamydomonas reinhardtii Nar1 gene encodes a chloroplast membrane protein involved in nitrite transport. Plant Cell 2000, 12, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Mariscal, V.; Moulin, P.; Orsel, M.; Miller, A.J.; Fernández, E.; Galván, A. Differential regulation of the Chlamydomonas Nar1 gene family by carbon and nitrogen. Protist 2006, 157, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Spalding, M.H. Acclimation to very low CO2: Contribution of limiting CO2 inducible proteins, LCIB and LCIA, to inorganic carbon uptake in Chlamydomonas reinhardtii. Plant Physiol. 2014, 166, 2040–2050. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Sato, E.; Iguchi, H.; Fukuda, Y.; Fukuzawa, H. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2015, 112, 7315–7320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsel, M.; Krapp, A.; Daniel-Vedele, F. Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 2002, 129, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Vidmar, J.J.; Glass, A.D.M. Regulation of NRT1 and NRT2 Gene Families of Arabidopsis thaliana: Responses to Nitrate Provision. Plant Cell Physiol. 2003, 44, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Unkles, S.E.; Hawker, K.L.; Grieve, C.; Campbell, E.I.; Montague, P.; Kinghorn, J.R. crnA encodes a nitrate transporter in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1991, 88, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Unkles, S.E.; Zhou, D.; Siddiqi, M.Y.; Kinghorn, J.R.; Glass, A.D.M. Apparent genetic redundancy facilitates ecological plasticity for nitrate transport. EMBO J. 2001, 20, 6246–6255. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.; Galván, A.; Fernández, E. Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 1994, 5, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.D.; González, C.; Ávila, J.; Brito, N.; Siverio, J.M. The YNT1 gene encoding the nitrate transporter in the yeast Hansenula polymorpha is clustered with genes YNI1 and YNR1 encoding nitrite reductase and nitrate reductase, and its disruption causes inability to grow in nitrate. Biochem. J. 1997, 321, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.J.; Ubbink-Kok, T.; Molenaar, D.; Konings, W.N.; Driessen, A.J.M. Nark is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol. Microbiol. 1994, 12, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H. Major Facilitator Superfamily. Microbiol. Mol. Biol. 1998, 62, 1–34. [Google Scholar]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H. The Major Facilitator Superfamily (MFS) Revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 2000, 1465, 219–235. [Google Scholar] [CrossRef]
- Clegg, S.; Feng, Y.; Griffiths, L.; Cole, J.A. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: Two nitrate and three nitrite transporters. Mol. Microbiol. 2002, 44, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Fernández, E.; Galván, A.; Miller, A.J. A high affinity nitrate transport system from Chlamydomonas requires two gene products. FEBS Lett. 2000, 466, 225–227. [Google Scholar] [CrossRef]
- Okamoto, M.; Kumar, A.; Li, W.; Wang, Y.; Siddiqi, M.Y.; Crawford, N.M.; Glass, A.D. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006, 140, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Orsel, M.; Chopin, F.; Leleu, O.; Smith, S.J.; Krapp, A.; Daniel-Vedele, F.; Miller, A.J. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein–protein interaction. Plant Physiol. 2006, 142, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Zhou, J.J.; Li, Z.; Miller, A.J. A two-component high-affinity nitrate uptake system in barley. Plant J. 2005, 41, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Kotur, Z.; Mackenzie, N.; Ramesh, S.; Tyerman, S.D.; Kaiser, B.N.; Glass, A.D.M. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 2012, 194, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Yong, Z.; Kotur, Z.; Glass, A.D.M. Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 2010, 63, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Galván, A.; Quesada, A.; Fernández, E. Nitrate and nitrate are transported by different specific transport systems and by a bispecific transporter in Chlamydomonas reinhardtii. J. Biol. Chem. 1996, 271, 2088–2092. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Karabika, E.; Kinghorn, J.R.; Glass, A.D.; Unkles, S.E.; Rouch, D.A. High-affinity nitrate/nitrite transporters NrtA and NrtB of Aspergillus nidulans exhibit high specificity and different inhibitor sensitivity. Microbiology 2015, 161, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Available online: www.tcdb.org (accessed on 25 October 2015).
- Available online: http://bioinformatics.oxfordjournals.org/content/30/6/884.full.pdf+html (accessed on 27 October 2015).
- Rexach, J.; Montero, B.; Fernández, E.; Galván, A. Differential regulation of the high affinity nitrite transport systems III and IV in Chlamydomonas reinhardtii. J. Biol. Chem. 1999, 274, 27801–27806. [Google Scholar] [CrossRef] [PubMed]
- Molnar, A.; Bassett, A.; Thuenemann, E.; Schwach, F.; Karkare, S.; Ossow-ski, S.; Weigel, D.; Baulcombe, D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009, 58, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Machín, F.; Medina, B.; Navarro, F.J.; Pérez, M.D.; Veenhuis, M.; Tejera, P.; Lorenzo, H.; Lancha, A.; Siverio, J.M. The role of Ynt1 in nitrate and nitrite transport in the yeast Hansenula polymorpha. Yeast 2004, 3, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://genome.jgi.doe.gov/Chlre4/Chlre4.home.html. (accessed on 23 October 2015).
- Available online: http://phytozome.jgi.doe.gov/pz/portal.html. (accessed on 27 October 2015).
- Quesada, A.; Gómez, I.; Fernández, E. Clustering of the nitrite reductase gene and a light-regulated gene with nitrate assimilation loci in Chlamydomonas reinhardtii. Planta 1998, 206, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.; Llamas, A.; Schnell, R.A.; Higuera, J.J.; González-Ballester, D.; Lefebvre, P.A.; Fernández, E.; Galván, A. Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell 2007, 19, 3491–3503. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.T.; Guerra, E.; Fernández, E.; Galván, A. Nitrite reductase mutants as an approach to understanding nitrate assimilation in Chlamydomonas reinhardtii. Plant Physiol. 2000, 122, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Schulze, W.; Thaminy, S.; Stagljar, I.; Frommer, W.B.; Ward, J.M. Intra- and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure 2002, 10, 763–772. [Google Scholar] [CrossRef]
- Schmollinger, S.; Mühlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-Sparing Mechanisms in Chlamydomonas Affect the Transcriptome, the Proteome, and Photosynthetic Metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Feng, H.; Tan, Y.; Xu, Y.; Miao, Q.; Xu, G. A putative 6 trans-membrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J. Integr Plant Biol. 2015. Available online: www.wileyonlinelibrary.com/journal/jipb (accessed on 25 September 2015). [Google Scholar] [CrossRef] [PubMed]
- Higuera, J.J.; Fernández, E.; Galván, A. Chlamydomonas NZF1, a tandem-repeated zinc finger factor involved in nitrate signalling by controlling the regulatory gene NIT2. Plant Cell Environ. 2014, 37, 2139–2160. [Google Scholar] [CrossRef] [PubMed]
- Rexach, J.; Llamas, A.; Fernández, E.; Galván, A. The activity of the high-affinity nitrate transport system I (NRT2;1, NAR2) is responsible for the efficient signalling of nitrate assimilation genes in Chlamydomonas reinhardtii. Planta 2002, 215, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Llamas, A.; Igeño, M.I.; Galván, A.; Fernández, E. Nitrate signalling on the nitrate reductase gene promoter depends directly on the activity of the nitrate transport systems in Chlamydomonas. Plant J. 2002, 30, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Harris, E. The Chlamydomonas Sourcebook; Academic Press: New York, NY, USA, 2009. [Google Scholar]
- Available online: http://wmd3.weigelworld.org/cgi- bin/webapp.cgi?page=Home;project=stdwmd (accessed on 25 July 2015).
- Kindle, K.L. High-frecuency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef] [PubMed]
- González-Ballester, D.; de Montaigu, A.; Higuera, J.J.; Galván, A.; Fernández, E. Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol. 2005, 137, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Martín, Y.; Navarro, F.J.; Siverio, J.M. Functional characterization of the Arabidopsis thaliana nitrate transporter CHL1 in the yeast Hansenula polymorpha. Plant Mol. Biol. 2008, 68, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Available online: www.bio-rad.com (accessed on 28 July 2015).
- González-Ballester, D.; Camargo, A.; Fernández, E. Ammonium transporter genes in Chlamydomonas: The nitrate-specific regulatory gene Nit2 is involved in Amt1;1 expression. Plant Mol. Biol. 2004, 56, 863–878. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Higuera, J.J.; Calatrava, V.; González, Z.; Mariscal, V.; Siverio, J.M.; Fernández, E.; Galván, A. NRT2.4 and NRT2.5 Are Two Half-Size Transporters from the Chlamydomonas NRT2 Family. Agronomy 2016, 6, 20. https://doi.org/10.3390/agronomy6010020
Higuera JJ, Calatrava V, González Z, Mariscal V, Siverio JM, Fernández E, Galván A. NRT2.4 and NRT2.5 Are Two Half-Size Transporters from the Chlamydomonas NRT2 Family. Agronomy. 2016; 6(1):20. https://doi.org/10.3390/agronomy6010020
Chicago/Turabian StyleHiguera, Jose Javier, Victoria Calatrava, Zaira González, Vicente Mariscal, Jose Manuel Siverio, Emilio Fernández, and Aurora Galván. 2016. "NRT2.4 and NRT2.5 Are Two Half-Size Transporters from the Chlamydomonas NRT2 Family" Agronomy 6, no. 1: 20. https://doi.org/10.3390/agronomy6010020





