1. Introduction
Up to now, two of the most-studied fundamental properties of biomacromolecules such as DNA, proteins, and polysaccharides have been the hydrodynamic and conformational properties [
1,
2]. For these purposes, the intrinsic viscosities and the radii of gyration have been the most often determined [
1,
3,
4]. In the field of biotechnology, changes in the hydrodynamic properties of these kinds of biomolecules are useful in the development and study of targeting pharmaceutical molecules [
5,
6]. Moreover, small fragments of DNA, oligonucleotides, or short biomolecules are important in nanotechnology studies, due to their contribution to several biological processes such as DNA packing around histones, compaction, gene transcription, gene delivery via small volume carriers for gene therapy, among others [
7,
8,
9,
10].
Recently, Pan et al. [
11] reported that zero shear rate viscosity of semi-dilute unentangled DNA solutions have a power law dependence on the scaled concentration
C/C* (where
C is the polymer mass concentration and
C* is the overlap concentration), with an effective exponent depending on the solvent quality parameter
z. In their work, they determined the θ
-temperature of dilute DNA solutions in the presence of salt excess, but they never considered the stiffness of the DNA molecule. Recently, a detailed study of the rheological behavior of high molecular weight DNA (calf-thymus) solutions was reported, as well as evidence of the two critical concentrations of the system—i.e., the overlap and the entanglement concentrations (
C* and
C**, respectively) [
1]. Furthermore, a generalized master curve was proposed from the variation of the specific viscosity at zero shear rate (η
sp,0) as a function of the overlap parameter (
C[η]) in a range of
CDNA[η] values up to 40.
In this paper, a novel representation of the η
sp,0 vs.
C[η] relationship based on the expression previously reported by Kwei et al. [
12] is proposed, taking into account the rheological behavior of the semi-dilute regime with entanglements—i.e., for concentrations over
C** and
CDNA[η] values up to 160. Additionally, chain characteristics of very short molecules (only some tens of base pairs), which are of great importance to molecular or cell biology, [
13,
14,
15] are studied. The η
sp,0 vs.
C[η] relationship was then found to be applicable to DNA chains with a molecular weight lower than the one corresponding to the persistence length of 50 nm, where DNA molecules behave as semi-flexible rods.
2. Materials and Methods
2.1. Materials and Solutions Preparation
Low molecular weight (LMW) DNA from salmon sperm, purchased as lyophilized powder, was purified and then turned into its sodium salt form. The purification process is described in the following section. Anhydrous NaCl was used to prepare a solvent solution at a concentration of 0.1 M. A series of low molecular weight DNA solutions was prepared in the concentration range from 0.5 to 1600 mg/mL. High molecular weight (HMW,
Mw = 6,559,500 g/mol [
1]) DNA samples from calf thymus DNA were used to prepare solutions in a concentration range from 0.01 to 40 mg/mL. A buffer solution prepared with Tris-HCl (100 mM) and EDTA (10 mM) was used to maintain a pH of 7.3. These HMW DNA solutions were prepared with a solvent consisting of a 9:1 ratio of HPLC water and the Tris–HCl/EDTA buffer (TE buffer). All reagents were provided by Sigma-Aldrich Company (Toluca, México). All solutions were prepared with HPLC-grade water. To prevent water evaporation, the vials were closed and sealed with Parafilm
® (Bemis NA, Neenah, WI, USA). All solutions were stored at a temperature of 4 °C in order to prevent DNA degradation and were left for a period of at least one week for stabilization and homogenization.
2.2. Purification of Low Molecular DNA from Salmon Sperm
Low molecular weight lyophilized DNA powder from salmon sperm was firstly dissolved in water, adding a stoichiometric amount of NaOH 1.0 N. A precipitation with ethanol (60%
v/
v) was then performed in the presence of salt excess (NaCl 1.0 M) to exchange the possible divalent counterions and to recover the sodium salt of DNA, which must be water soluble [
16]. Finally, DNA was recovered by solvent exchange with ethanol–water mixtures up to 100% in ethanol, and drying at 40 °C.
2.3. Capillary Measurements
Viscosity measurements of low molecular weight DNA solutions with concentrations between 0.5 and 100 mg/mL were carried out with a capillary viscometer Micro-Ubbelohde (SCHOTT Instruments GmbH, Mainz, Germany) connected with a semi-automatic chronometer ViscoClock (SCHOTT Instruments GmbH, Mainz, Germany) at a temperature of 20 °C. The selected capillary (No. 501 01) has a diameter of 0.53 ± 0.01 mm and a constant K equal to 0.005.
2.4. Rheological Measurements
The rheological behavior of low and high molecular weight DNA solutions was studied through flow measurements by using a DHR-3 rheometer from the TA Instruments Company (New Castle, DE, USA). Three different geometries were used, depending on DNA molecular weight and on the solution concentration: (1) a steel cone with a 60 mm diameter and an angle of 2° was used for low and high molecular weight DNA solutions with concentrations in the dilute regime and in the semi-dilute regime without entanglements (CDNA < C**); (2) a steel cone with a 40 mm diameter and an angle of 2° was used for low molecular weight DNA solutions with concentrations higher than 1000 mg/mL and for high molecular weight DNA solutions with concentrations between 2 mg/mL and 10 mg/mL; and (3) a rough steel cone and plate for high molecular weight DNA solutions with concentrations higher than 10 mg/mL, the cone having a 35 mm diameter and an angle of 2°. Simple shear steady state measurements were performed in a shear rate range from 1 × 10−3 to 1000 s−1, using five points per decade. Each sweep was performed at a temperature of 20 ± 0.1 °C, controlled by a Peltier plate.
3. Results and Discussion
3.1. Low Molecular Weight DNA Sample Characterization
After purification, the commercial low molecular weight DNA from salmon sperm was characterized through UV-Vis and
1H NMR measurements to confirm its structure [
17,
18]. When dissolved in water, the structure of this purified LMW DNA sample corresponds to that of an oligonucleotide in a semi-denatured conformation. The absorbance ratio A
260/A
280 was measured on a purified sample dissolved in NaCl 0.1 M, from which was possible to evaluate its purity (i.e., 1.70 ± 0.01), in good agreement with the literature [
1,
19].
Then, since the DNA molecule bears one formal negative charge per nucleotide, its conformation is certainly sensitive to changes in the ionic strength, in the number of base pairs, and in the molecular weight [
20,
21,
22]. In this manner, the information about intrinsic viscosity is necessary to determine and to understand the hydrodynamic properties of the DNA molecule under specific conditions. Therefore, viscosity capillary measurements were performed in order to determine the intrinsic viscosity of the low molecular weight DNA sample in NaCl 0.1 M at 20 °C. Reduced viscosities were calculated according to Equation (1), and were plotted as a function of DNA concentration (
CDNA) following the Huggins relation [
1,
23].
where
C is the polymer concentration (g/mL), η
red is the reduced viscosity, η
sp is the specific viscosity (corresponding to (η − η
s)/η
s, where η
s is the solvent viscosity), [η] is the intrinsic viscosity (mL/g) and k’ is the Huggins constant.
The intrinsic viscosity, (8.49 mL/g) was obtained from extrapolation to zero concentration. It is worth mentioning that low values of intrinsic viscosities between 7.5 mL/g and 160 mL/g were previously reported for oligonucleotides and oligosaccharides [
2,
24,
25]. A recently established Mark-Houwink relation (Equation (2) for the low molecular weight DNA range—between 10 and 3000 bp (base pair)—was used to estimate the viscometric-average molecular weight of the sample [
2].
Thus, the calculated viscometric-average molecular weight for this sample is equal to 15,000 g/mol, equivalent to around 23 bp (estimated by taking the average weight of a DNA nucleotide in salt solution as 325 g/mol).
Finally, the overlap concentration (C*) was estimated through the deviation from linear behavior in the dilute regime (i.e., 125 mg/mL), which is also in good agreement with the value obtained from the relation to C*~[η]−1.
3.2. Rheological Behavior of Low Molecular Weight DNA
It is well known that long DNA chains like T2, T4, T5, and T7 viruses [
26,
27], and even fragments of whole chains viruses [
28], present a viscoelastic behavior; however, as the chains get smaller, the critical value for the shear rate—characterizing the appearance of non-Newtonian behavior—increases. This transition from Newtonian to non-Newtonian behavior also depends on the polymer concentration. In this work, the viscosity of low molecular weight DNA solutions at low shear rates was studied through flow measurements as a function of DNA concentration. The behavior of the solution remains Newtonian in all the domains of the selected shear rates—i.e., up to 100 s
−1. Then, the variation of the specific viscosity as a function of polymer concentration was analyzed in terms of the overlap parameter (
CDNA[η]) by means of the generalized master curve, initially proposed for hyaluronans with various molecular weights. The importance of this approach is recognized since the intrinsic viscosity values ([η]) also contain information regarding the stiffness of the polymer [
29,
30]. All the data obtained for the specific viscosity determined by capillary and rheological measurements for low molecular weight DNA samples and for the specific viscosity determined in the Newtonian plateau for high molecular DNA samples are plotted together and collapse in the same curve (
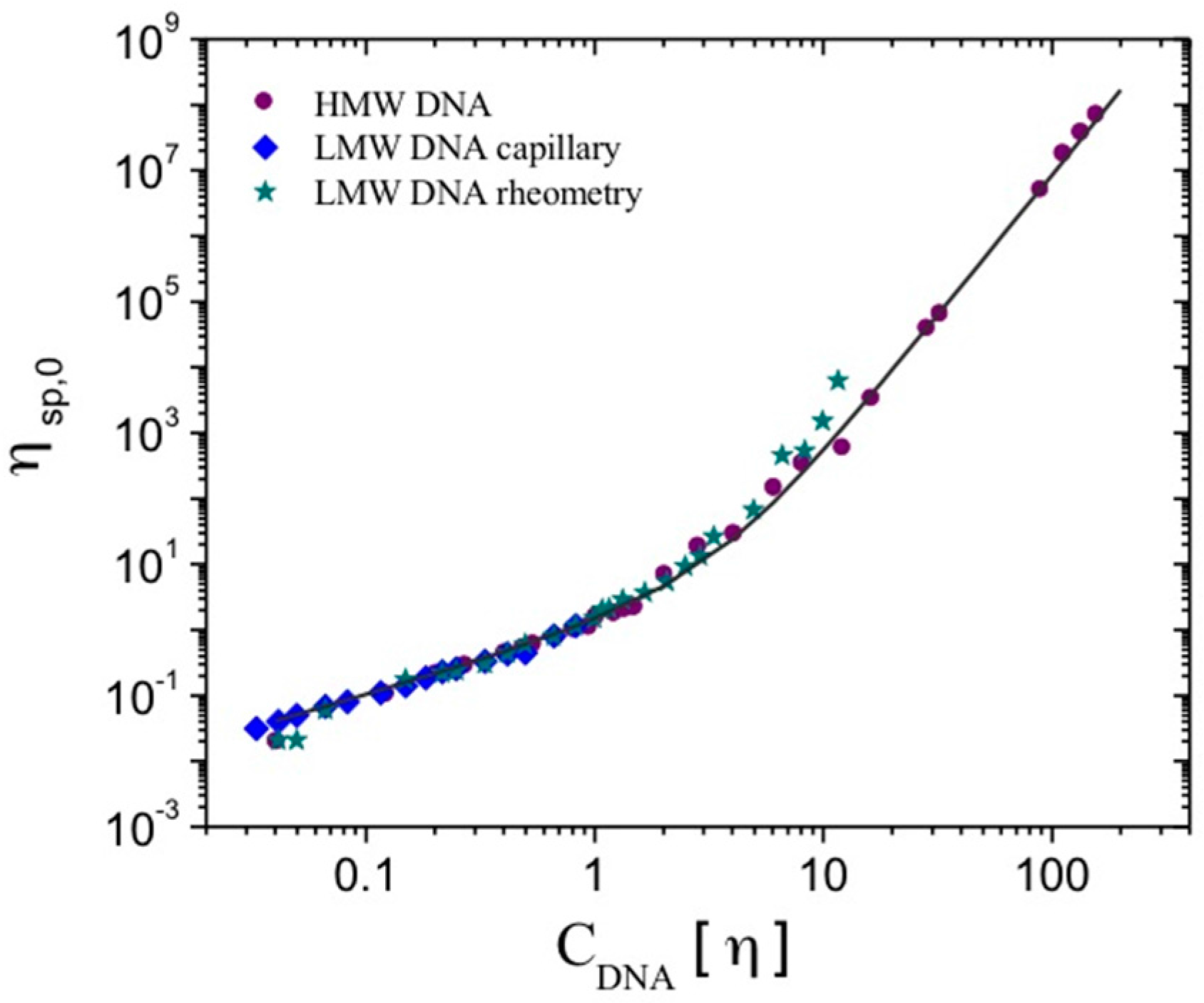
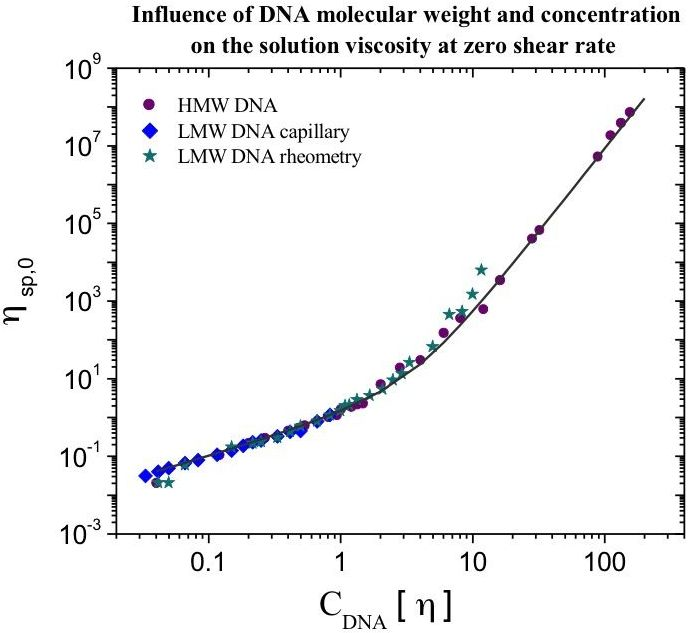
Figure 1).
It should be pointed out that knowing the intrinsic viscosity of the polymer, it becomes possible to calculate the specific viscosity at zero shear rate (ηsp,0) for a given polymer concentration and to estimate the critical concentrations of the system—i.e., the overlap concentration (C*~[η]−1) and the entanglement concentration (C**, corresponding to C[η]~10).
Equation (3) presents a novel representation of the relationship between the specific viscosity and the overlap parameter, which is based on the expression previously proposed by Kwei et al. for hyaluronan samples [
12].
where
k1 represents the Huggins constant, which for several water soluble polymers is equal to 0.4,
k2 = (
k1)
2/2! = 0.08 and
k3 = 0.0213.
In this equation, the fourth term represents the behavior of the semi-dilute entangled regime, with a final slope equal to 4.3 at high polymer concentrations (around
C[η] > 10), instead of the value proposed by Kwei et al. [
12] with
k3 = (
k1)
2/3!. This slope value is in good agreement with the power law established for the regime over the entanglement concentration
C**, for which η/η
Rouse varies as (
C/
C**)
3.4 and
η varies as (
C/
Ce)
4.42 [
31,
32]. Furthermore, the reported behavior of the experimental values for the specific viscosities of hyaluronan and xanthan samples at high polymer concentrations in 0.1 N NaCl followed the slopes of 4.18 and 4.24, respectively [
16,
33].
3.3. Rheological Behavior of High Molecular Weight DNA in the High Concentration Domain
In order to prevent wall-slip phenomena while performing steady state flow measurements of high molecular weight DNA in the concentration domain larger than 10 mg/mL [
34], a cone–plate geometry with roughened base plate and cone was selected. The roughening of the geometry surfaces has been satisfactorily applied by several authors and with different materials, like sandblasting or sandpaper [
35,
36,
37].
At DNA concentrations higher than 10 mg/mL, there is a large non-Newtonian domain, implying the use of a model to describe the rheological curve. Then, the viscosity dependence on shear rate for several DNA solutions with concentrations between 15 and 40 mg/mL was analyzed with the Cross model, given by Equation (4) [
38] (the goodness of the fits having an average
R2 of 0.98 ± 0.01).
where
is the shear rate, η
0 is the zero shear-rate viscosity, η
∞ is the viscosity at infinite shear-rate,
m is a dimensionless parameter related to the degree of shear thinning, and
K has the dimensions of time.
At larger polymer concentrations, the specific viscosity from the zero shear-rate viscosity determined through the Cross model follows the previously-presented master curve up to
CDNA[η] = 160 (
Figure 1). The slope value for specific viscosity as a function of
CDNA[η] for these highly concentrated calf-thymus DNA solutions corresponds to that of the semi-dilute regime with entanglements, and remains 4.3.
4. Conclusions
A commercial low molecular weight DNA sample from salmon sperm was purified, characterized, and then studied in solution in a wide concentration range between 0.5 and 1600 mg/mL. The intrinsic viscosity was found to be equal to 8.49 mL/g, which is in the range of the values of oligonucleotides and oligosaccharides. The viscometric-average molecular weight value for this sample corresponds to 15,000 g/mol. The evidence of the overlap concentration was detected around the concentration of 125 mg/mL. The behavior of these low molecular weight DNA solutions remains Newtonian in all domains of the selected shear rates (i.e., up to 100 s−1).
All of the data for the specific viscosity at zero shear rate obtained for this LMW DNA were plotted together as a function of the overlap parameter, and collapse on the curve obtained for the HMW DNA. The proposed relationship between ηsp,o and CDNA[η] is then valid for DNA chains with molecular weights lower than 1 × 105 g/mol, where chains are in an almost rod-like state, and for DNA chains having higher molecular weights, where molecules behave as worm-like chains.
For high molecular weight DNA solutions from calf-thymus with concentrations up to CDNA[η] = 160, the zero shear-rate viscosity determined through the Cross model follows the improved master curve. Up to C[η] = 160, the slope value remains at 4.3, with a behavior corresponding to that of the semi-dilute regime with entanglements.
Acknowledgments
Lourdes Mónica Bravo-Anaya acknowledges financial support granted by Gabriel Bravo-Galván and Martha Anaya. We acknowledge Hélène Galliard for her valuable help with conducting the experiments.
Author Contributions
Marguerite Rinaudo suggested the work; Lourdes Mónica Bravo-Anaya, Marguerite Rinaudo, Félix Armando Soltero Martínez and Frédéric Pignon conceived and designed the experiments; Lourdes Mónica Bravo-Anaya performed the experiments; Marguerite Rinaudo, Lourdes Mónica Bravo-Anaya and Frédéric Pignon analyzed the data; Félix Armando Soltero Martínez and Frédéric Pignon contributed with reagents; Lourdes Mónica Bravo-Anaya and Marguerite Rinaudo wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| DNA | Deoxyribonucleic Acid |
| LMW | Low Molecular Weight |
| HMW | High Molecular Weight |
| TE | TrisHCl-EDTA |
| bp | Base pair |
References
- Bravo-Anaya, L.M.; Rinaudo, M.; Soltero-Martínez, F.A. Conformation and rheological properties of calf-thymus DNA in solution. Polymers 2016, 8, 1–19. [Google Scholar] [CrossRef]
- Tsortos, A.; Papadakis, G.; Gizeli, E. The intrinsic viscosity of linear DNA. Biopolymers 2011, 95, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Garcia de la Torre, J.; Bloomfield, V.A. Hydrodynamic properties of complex, rigid, biological macromolecules: Theory and applications. Q. Rev. Biophys. 1981, 14, 81–139. [Google Scholar] [CrossRef] [PubMed]
- Berry, G.C. The hydrodynamic and conformational properties of denatured proteins in dilute solutions. Protein Sci. 2010, 19, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, G.; Tsortos, A.; Gizeli, E. Acoustic detection of DNA conformation in genetic assays combined with PCR. Biosens. Bioelectron. 2009, 25, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Arpicco, S.; Milla, P.; Stella, B.; Dosio, F. Hyaluronic acid conjugates as vectors for the active targeting of drugs, genes and nanocomposites in cancer treatment. Molecules 2014, 19, 3193–3230. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.R.; Dohrman, A.F.; Ellison, A.R.; Goncz, K.K.; Gruenert, D.C. Intracellular compartmentalization of DNA fragments in cultured airway epithelial cells mediated by cationic lipids. Pharm. Res. 1999, 16, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Saltzman, W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000, 18, 33–37. [Google Scholar] [PubMed]
- Li, S.D.; Huang, L. Gene therapy progress and prospects: Non-viral gene therapy by systemic delivery. Gene Ther. 2006, 13, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Manjila, S.B.; Baby, J.N.; Bijin, E.N.; Constantine, I.; Pramod, K.; Valsalakumari, J. Novel gene delivery systems. Int. J. Pharm. Investig. 2013, 3, 1–7. [Google Scholar] [PubMed]
- Pan, S.; Nguyen, D.A.; Sridhar, T.; Sunthar, P.; Prakash, J.R. Universal solvent quality crossover of the zero shear rate viscosity of semidilute DNA solutions. J. Rheol. 2014, 58, 339–368. [Google Scholar] [CrossRef]
- Kwei, T.K.; Nakazawa, M.; Matsuoka, S.; Cowman, M.K.; Okamoto, Y. Concentration dependence of solution viscosities of rigid rod polymers. Macromolecules 2000, 33, 235–236. [Google Scholar] [CrossRef]
- Peters, J.P.; Maher, L.J. DNA curvature and flexibility in vitro and in vivo. Q. Rev. Biophys. 2010, 43, 23–63. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.M. Molecular and cellular barriers limiting the effectiveness of antisense oligonucleotides. Biophys. J. 2005, 89, 2286–2295. [Google Scholar] [CrossRef] [PubMed]
- Berriaud, N.; Milas, M.; Rinaudo, M. Characterization and properties of hyaluronic acid (hyaluronan). In Polysaccharides, Structural Diversity and Functional Versatility; Dumitriu, S., Ed.; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Basu, S. Ultraviolet absorption studies on DNA. Biopolymers 1967, 5, 876–878. [Google Scholar] [CrossRef] [PubMed]
- Early, T.A.; Kearns, D.R. 1H nuclear magnetic resonance investigation of flexibility in DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 4165–4169. [Google Scholar] [CrossRef] [PubMed]
- Wilfinger, W.W.; Mackey, K.; Chomczynski, P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques 1997, 22, 478–481. [Google Scholar]
- Evdokimov, Y.M.; Platanov, A.L.; Tikhonenko, A.S.; Varshavsky, Y.M. A compact form of double-stranded DNA in solution. Febs. Lett. 1972, 23, 180–184. [Google Scholar] [CrossRef]
- Frisman, E.V.; Slonitsky, S.V.; Veselkov, A.N. Influence of solvent structure on the conformation of the native DNA molecule. Int. J. Quantum Chem. 1979, 16, 847–855. [Google Scholar] [CrossRef]
- Frank-Kamenetskii, M.D.; Lazurkin, Y.S. Conformational changes in DNA molecules. Annu. Rev. Biophys. Bioeng. 1974, 3, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Pamies, R.; Cifre, G.H.J.; Martínez, C.L.M.; García de la Torre, J. Determination of intrinsic viscosities of macromolecules and nanoparticles. Comparison of single-point and dilution procedures. J. Colloid Polym. Sci. 2008, 286, 1223–1231. [Google Scholar] [CrossRef]
- Heyraud, A.; Rinaudo, M. Gel permeation chromatography of glucose oligomers on polyacrylamide gels, thermodynamic and steric partition mechanisms. J. Chromatogr. 1978, 166, 149–158. [Google Scholar] [CrossRef]
- Mensink, M.A.; Frijlink, H.W.; van der Voort Maarschalk, K.; Hinrichs, W.L.J. Inulin, a flexible oligosaccharide I: Review of its physicochemical characteristics. Carbohydr. Polym. 2015, 130, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Crothers, D.M.; Zimm, B.H. Viscosity and sedimentation of the DNA from bacteriophages T2 and T7 and the relation to molecular weight. J. Mol. Biol. 1965, 12, 525–536. [Google Scholar] [CrossRef]
- Reeg, C.F.; Harrington, R.E. Teady-state opticohydrodynamic properties of DNA: Molecular weight dependence and the internal viscosity problem. Biopolymers 1982, 21, 1315–1332. [Google Scholar] [CrossRef] [PubMed]
- Schumaker, V.N.; Bennett, C. A study of the viscosity of T4r+ DNA using a rotating cylinder viscometer. J. Mol. Biol. 1962, 5, 384–397. [Google Scholar] [CrossRef]
- Rinaudo, M. Polyelectrolyte properties of a plant and animal polysaccharide. Struct. Chem. 2009, 20, 277–289. [Google Scholar] [CrossRef]
- Odijk, T. On the ionic-strength dependence of the intrinsic viscosity of DNA. Biopolymers 1979, 18, 3111–3113. [Google Scholar] [CrossRef] [PubMed]
- Raspaud, E.; Lairez, D.; Adam, M. On the number of blobs per entanglement in semidilute and good solvent solution: Melt influence. Macromolecules 1995, 28, 927–933. [Google Scholar] [CrossRef]
- Musti, R.; Sikorav, J.-L.; Lairez, D.; Jannink, G.; Adam, M. Viscoelastic properties of entangled DNA solutions. C. R. Acad. Sci. 1995, 320, 599–605. [Google Scholar]
- Milas, M.; Rinaudo, M.; Tinland, B. The viscosity dependence on concentration, molecular weight and shear rate of xanthan solutions. Polym. Bull. 1985, 14, 157–164. [Google Scholar] [CrossRef]
- Boukany, P.E.; Hu, Y.T.; Wang, S.-Q. Observations of wall slip and shear banding in an entangled DNA solution. Macromolecules 2008, 41, 2644–2650. [Google Scholar] [CrossRef]
- Nechyporchuk, O.; Belgacem, M.N.; Pignon, F. Rheological properties of micro-/nanofibrillated cellulose suspensions: Wall-slip and shear banding phenomena. Carbohydr. Polym. 2014, 112, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Buscall, R.; McGowan, J.I.; Morton-Jones, A.J. The rheology of concentrated dispersions of weakly attracting colloidal particles with and without wall slip. J. Rheol. 1993, 37, 621–641. [Google Scholar] [CrossRef]
- Pignon, F.; Magnin, A.; Piau, J.-M. Thixotropic colloidal suspensions and flow curves with minimum: Identification of flow regimes and rheometric consequences. J. Rheol. 1996, 40, 573–587. [Google Scholar] [CrossRef]
- Doi, Y.; Gray, F.M.; Buchholz, F.L.; Graham, A.T.; Striegel, A.; Yau, W.W.; Kirkland, J.J.; Bly, D.D.; Gordon, M.J., Jr.; Archer, R.D. Rheology Principles, Measurements, and Applications; Macosko, C.W., Ed.; Wiley-VCH: Weinheim, Germany, 1994. [Google Scholar]
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).