Asymmetric Ketone Reduction by Immobilized Rhodotorula mucilaginosa
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Immobilized Cells
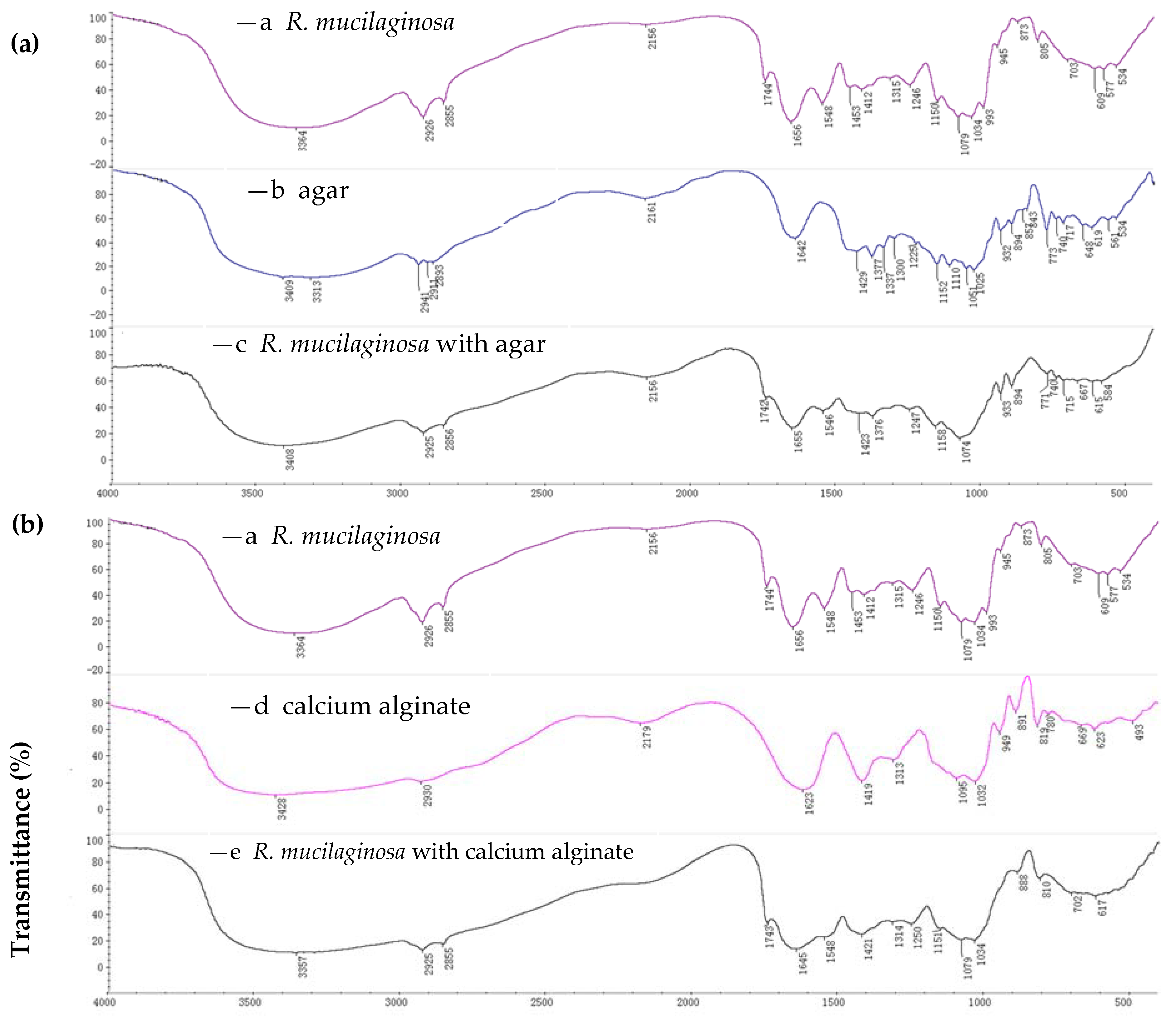
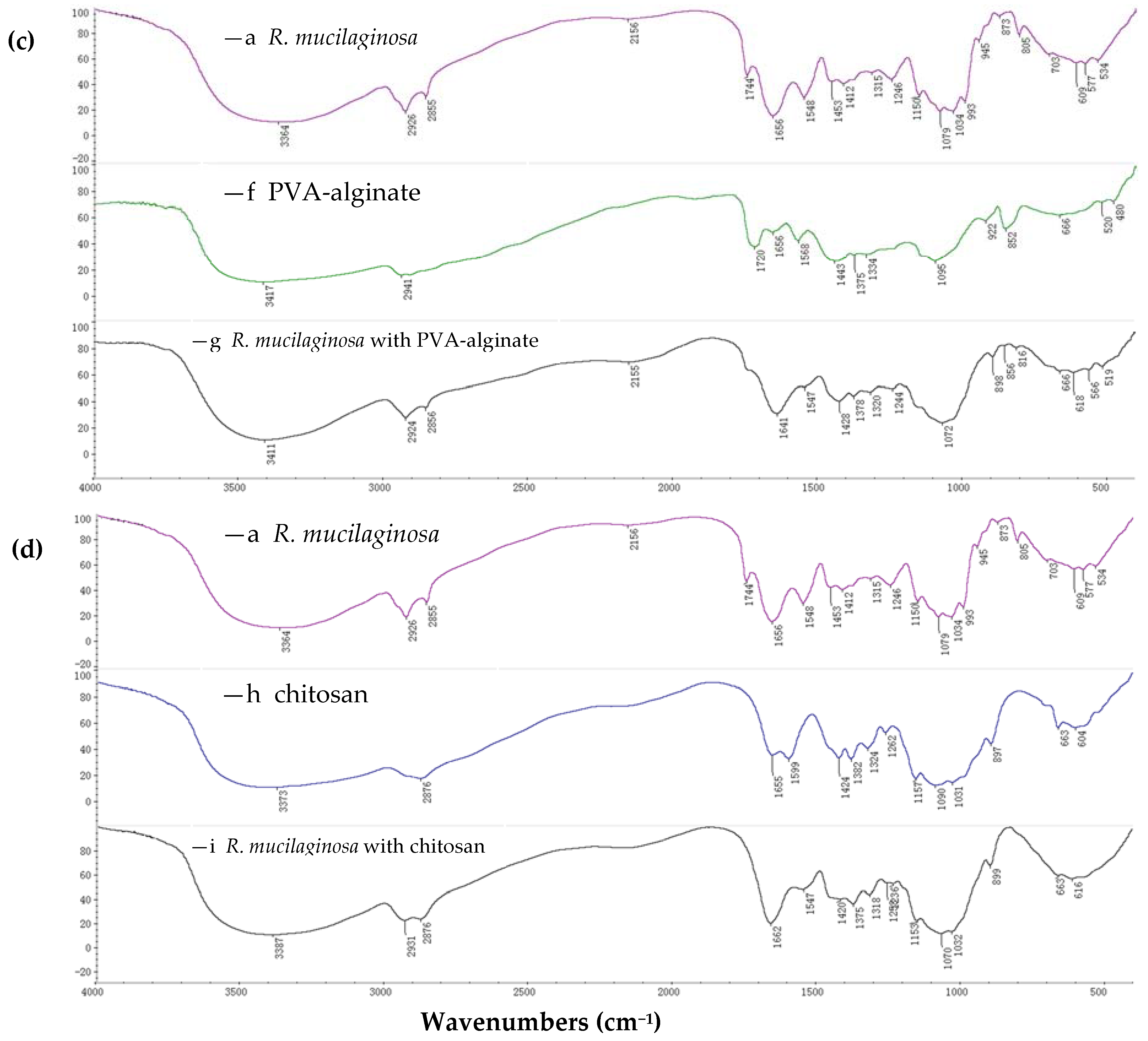
2.1.1. FTIR Spectroscopy
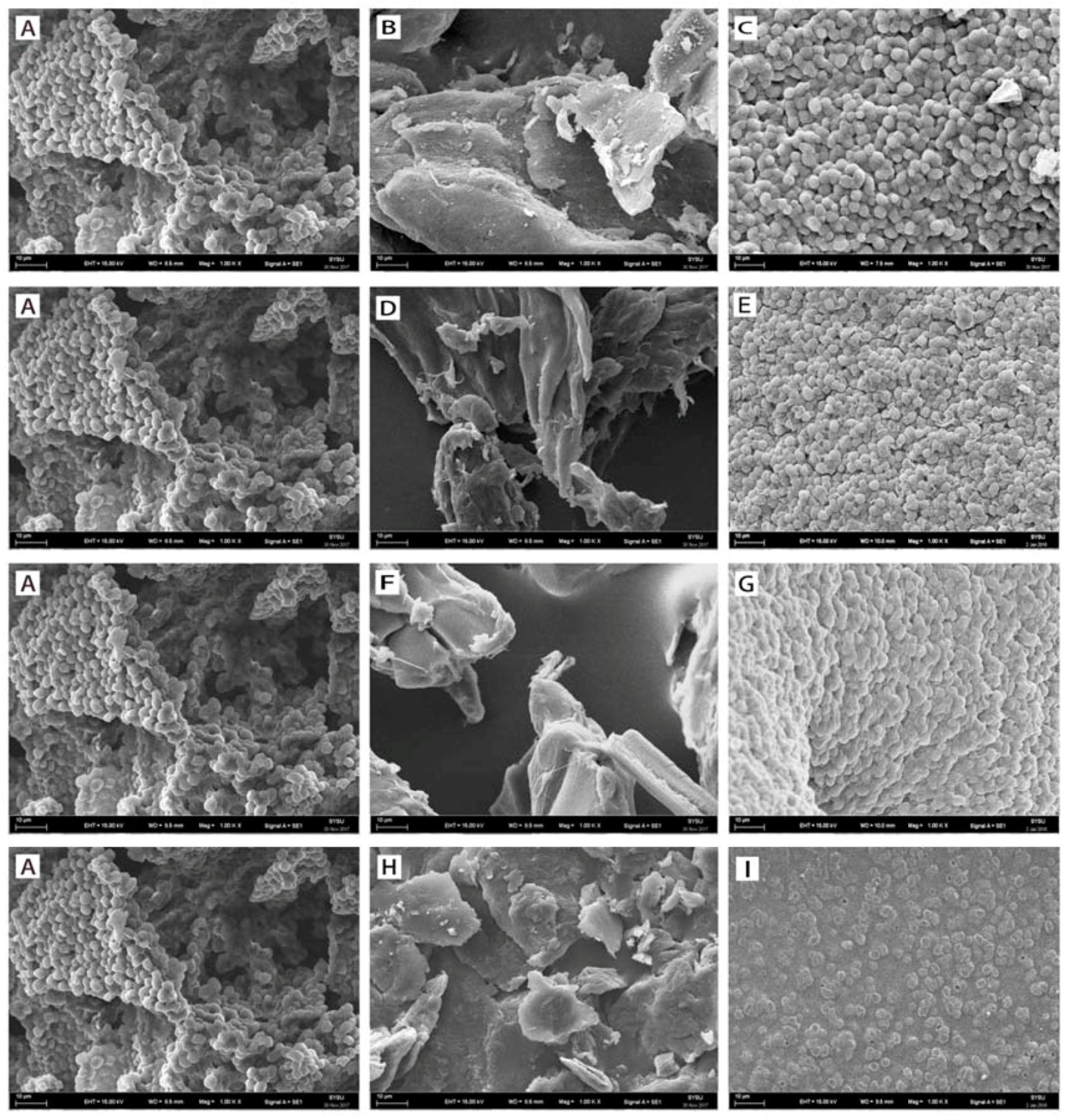
2.1.2. SEM Spectroscopy
2.2. Catalytic Activity of Immobilized Cells for the Reduction of Various Ketones
2.3. Stability of Immobilized Cells on Agar
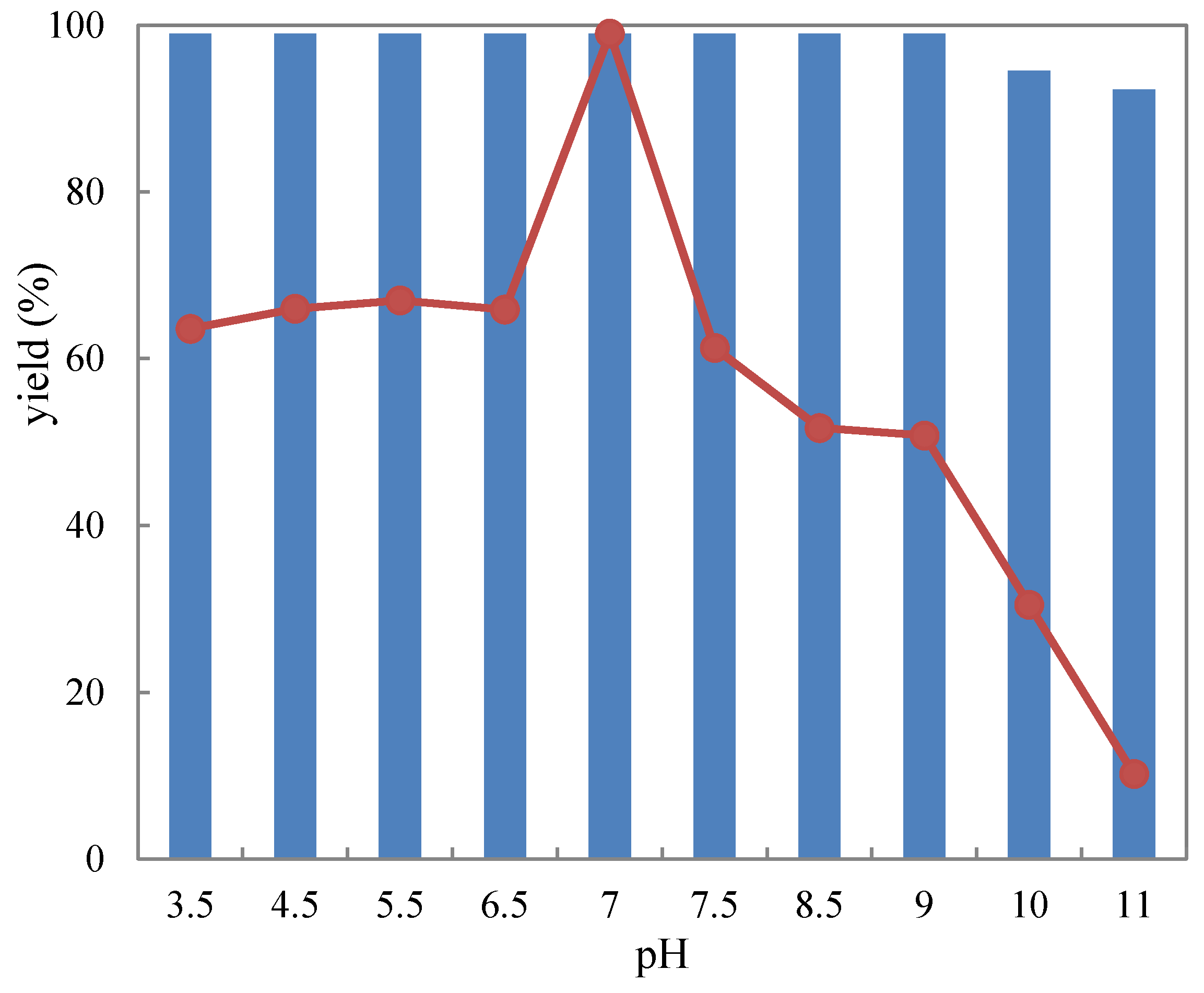
2.3.1. pH Tolerance
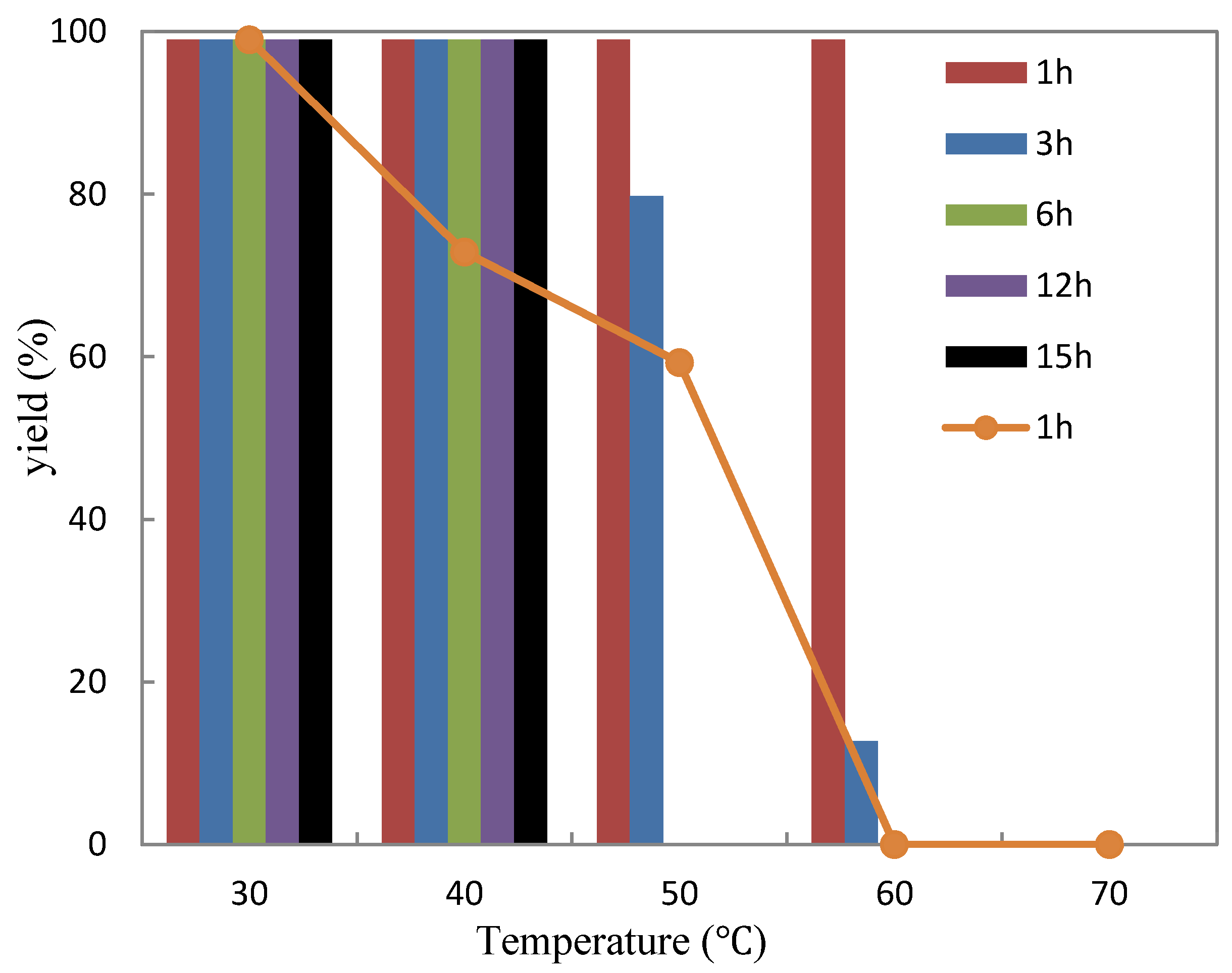
2.3.2. Thermostability
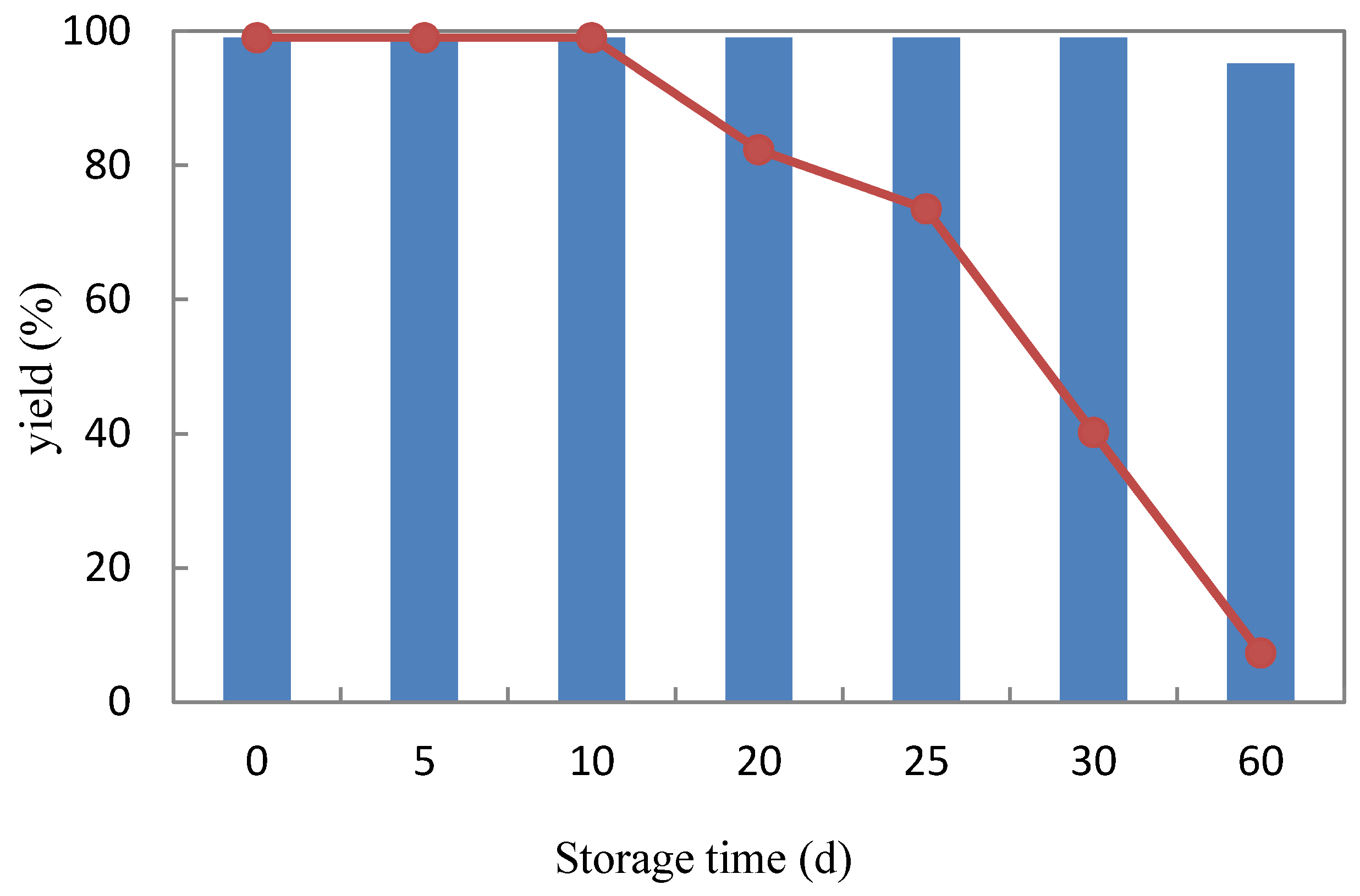
2.3.3. Storage Stability
2.3.4. Recyclability
3. Materials and Methods
3.1. General Methods
3.2. Microorganism and Culture Conditions
3.3. Immobilization of R. mucilaginosa GIM 2.157 by Various Matrices
3.3.1. On Agar
3.3.2. On Calcium Alginate
3.3.3. On PVA-Alginate
3.3.4. On Chitosan
3.4. Scanning Electron Microscopy
3.5. Bioconversion of Ketones with Immobilized Cells
3.6. pH Profile
3.7. Thermostability
3.8. Storage Stability
3.9. Reusability
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kataoka, M.; Kita, K.; Wada, M.; Yasohara, Y.; Hasegawa, J.; Shimizu, S. Novel bioreduction system for the production of chiral alcohols. Appl. Microbiol. Biotechnol. 2003, 62, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Nealon, C.M.; Musa, M.M.; Patel, J.M.; Phillips, R.S. Controlling substrate specificity and stereospecificity of alcohol dehydrogenases. ACS Catal. 2015, 2, 2100–2114. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, Y.; Xiao, R. Redesigning alcohol dehydrogenases/reductases for more efficient biosynthesis of enantiopure isomers. Biotechnol. Adv. 2015, 33, 1671–1684. [Google Scholar] [CrossRef] [PubMed]
- Gyarmati, J.; Hajdu, C.; Dinya, Z.; Micskei, K.; Pályi, G. Asymmetric induction by amino acid ligands in chromium(II)-assisted reduction of ketones. J. Organomet. Chem. 1999, 586, 106–109. [Google Scholar] [CrossRef]
- Micskei, K.; Holczknecht, O.; Hajdu, C.; Patonay, T.; Marchis, V.; Meo, M.; Zucchi, C.; Pályi, G. Asymmetric synthesis of amino acids by Cr(II) complexes of natural amino acids. J. Organomet. Chem. 2003, 682, 143–148. [Google Scholar] [CrossRef]
- Micskei, K.; Patonay, T.; Caglioti, L.; Pályi, G. Amino acid ligand chirality for enantioselective syntheses. Chem. Biodivers. 2010, 7, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Kroutil, W.; Mang, H.; Edegger, K.; Faber, K. Recent advances in the biocatalytic reduction of ketones and oxidation of sec-alcohols. Curr. Opin. Chem. Biol. 2004, 8, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Stefaan, M.A.; De, W.; Theo, S.; Hans, E.S.; Oliver, M. Biocatalytic reductions: From lab curiosity to “first choice”. Acc. Chem. Res. 2007, 40, 1260–1266. [Google Scholar]
- Rocha, L.C.; de Souza, A.L.; Filho, U.P.R.; Filho, S.P.C.; Sette, L.D.; Porto, A.L.M. Immobilization of marine fungi on silica gel, silica xerogel and chitosan for biocatalytic reduction of ketones. J. Mol. Catal. B Enzym. 2012, 84, 160–165. [Google Scholar] [CrossRef]
- Chen, B.-S.; Liu, H.; de Souza, F.Z.R.; Liu, L. Organic solvent-tolerant marine microorganisms as catalysts for kinetic resolution of cyclic β-hydroxy ketones. Mar. Biotechnol. 2017, 19, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; de Souza, F.Z.R.; Liu, L.; Chen, B.-S. The use of marine-derived fungi for preparation of enantiomerically pure alcohols. Appl. Microbiol. Biotechnol. 2018, 102, 1317–1330. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, B.-S.; de Souza, F.Z.R.; Liu, L. A comparative study on asymmetric reduction of ketones using the growing and resting cells of marine-derived fungi. Mar. Drugs 2018, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-H.; Wang, X.-T.; Lou, W.-Y.; Li, Y.; Wu, H.; Zong, M.-H.; Smith, T.J.; Chen, X.-D. Immobilization of Acetobacter sp. CCTCC M209061 for efficient asymmetric reduction of ketones and biocatalyst recycling. Microb. Cell Fact. 2012, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Lusta, K.A.; Chung, I.K.; Sul, I.W.; Park, H.S.; Shin, D.I. Immobilization of fungus Aspergillus sp. by a novel cryogel technique for production of extracellular hydrolytic enzymes. Process Biochem. 2000, 35, 1177–1182. [Google Scholar] [CrossRef]
- Sanchez, S.; Demain, A.L. Enzymes and bioconversions of industrial, pharmaceutical, and biotechnological significance. Org. Process Res. 2011, 15, 224–230. [Google Scholar] [CrossRef]
- Rocha, L.C.; Seleghim, M.H.R.; Comasseto, J.V.; Sette, L.D.; Porto, A.L.M. Stereoselective bioreduction of α-azido ketones by whole cells of marine-derived fungi. Mar. Biotechnol. 2015, 17, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Prabhu, P.; Lee, J.K. Alginate immobilization of recombinant Escherichia coli whole cells harboring L-arabinose isomerase for L-ribulose production. Bioprocess. Biosyst. Eng. 2010, 33, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Huang, K.L.; Jiang, Y.R.; Ding, P. Production of (R)-mandelic acid by immobilized cells of Saccharomyces cerevisiae on chitosan carrier. Process Biochem. 2007, 42, 1465–1469. [Google Scholar] [CrossRef]
- Idris, A.; Suzana, W. Effect of calcium alginate concentration, bead diameter, initial pH and temperature on lactic acid production from pineapple waste using immobilized Lactobacillus delbrueckii. Process Biochem. 2006, 41, 1117–1123. [Google Scholar] [CrossRef]
- Wang, W.; Huang, X.J.; Cao, J.D.; Lan, P.; Wu, W. Immobilization of calcium alginate sulfates on polysulfone ultrafiltration membranes for selective adsorption of low-density lipoprotein. Acta Biomater. 2014, 10, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Lin, H.Y.; Chen, Z.; Megharaj, M.; Naidu, R. Biodegradation of crystal violet using Burkholderia vietnamiensis C09V immobilized on PVA–calcium alginate–kaolin gel beads. Ecotoxiol. Environ. Saf. 2012, 83, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, B. Application of chitin- and chitosan-based materials for enzyme immobilizations: A review. Enzyme Microb. Technol. 2004, 35, 126–139. [Google Scholar] [CrossRef]
- Nunes, M.A.P.; Vila-Real, H.; Fernandes, P.C.B.; Ribeiro, M.H.L. Immobilization of naringinase in PVA–alginate matrix using an innovative technique. Appl. Biochem. Biotechnol. 2010, 160, 2129–2147. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, J.; Kang, X.; Wang, C.; Wang, D.; Liu, J.; Aksay, I.A.; Lin, Y. Glucose biosensor based on immobilization of glucose oxidase in platinum nanoparticles/graphene/chitosan nanocomposite film. Talanta 2009, 80, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.C.; Tseng, R.L.; Juang, R.S. Enhanced abilities of highly swollen chitosan beads for color removal and tyrosinase immobilization. J. Hazard. Mater. 2001, 81, 167–177. [Google Scholar] [CrossRef]
- Kumirska, J.; Czerwicka, M.; Kaczyński, Z.; Bychowska, A.; Brzozowski, K.; Thöming, J.; Stepnowski, P. Application of spectroscopic methods for structural analysis of chitin and chitosan. Mar. Drugs 2010, 8, 1567–1636. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.E.D.; Morris, D.J.; Wills, M. Asymmetric hydrogenation of ketones using Ir(III) complexes of N-alkyl-N′-tosyl-1,2-ethanediamine ligands. Tetrahedron Lett. 2009, 50, 688–692. [Google Scholar] [CrossRef]
- Yearick, K.; Wolf, C. Catalytic enantioselective addition of diethylzinc to trifluoromethyl ketones. Org. Lett. 2008, 10, 4391. [Google Scholar]


| Reduction Products | Biocatalysts | ||||
|---|---|---|---|---|---|
| Free Cells | R. mucilaginosa Cells Immobilized on | ||||
| Agar | Calcium Alginate | PVA-Alginate | Chitosan | ||
| 2a | 99 (99, S) | 99 (99, S) | 99 (99, S) | 99 (99, S) | 0 |
| 2b | 99 (99, S) | 99 (89, S) | 86 (99, S) | 89 (94, S) | 0 |
| 2c | 99 (99, S) | 73 (68, S) | 71 (19, S) | 51 (54, S) | 0 |
| 2d | 99 (99, S) | 99 (99, S) | 89 (99, S) | 97 (99, S) | 0 |
| 2e | 99 (99, S) | 99 (99, S) | 94 (89, S) | 99 (99, S) | 0 |
| 2f | 99 (99, S) | 94 (83, S) | 91 (94, S) | 59 (77, S) | 0 |
| 2g | 99 (99, S) | 99 (91, S) | 94 (97, S) | 75 (66, S) | 0 |
| 2h | 99 (99, S) | 99 (99, S) | 93 (99, S) | 94 (99, S) | 0 |
| 2i | 0 | 0 | 0 | 0 | 0 |
| 2j | 0 | 0 | 0 | 0 | 0 |
| 2k | 99 (99, S) | 99 (89, S) | 99 (85, S) | 77 (84, S) | 88 (91, R) |
| 2l | 99 (99, R) | 99 (73, R) | 99 (82, R) | 23 (21, R) | 72 (84, S) |
| 2m | 0 | 0 | 0 | 0 | 0 |
| 2n | 0 | 0 | 0 | 0 | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Duan, W.-D.; De Souza, F.Z.R.; Liu, L.; Chen, B.-S. Asymmetric Ketone Reduction by Immobilized Rhodotorula mucilaginosa. Catalysts 2018, 8, 165. https://doi.org/10.3390/catal8040165
Liu H, Duan W-D, De Souza FZR, Liu L, Chen B-S. Asymmetric Ketone Reduction by Immobilized Rhodotorula mucilaginosa. Catalysts. 2018; 8(4):165. https://doi.org/10.3390/catal8040165
Chicago/Turabian StyleLiu, Hui, Wen-Di Duan, Fayene Zeferino Ribeiro De Souza, Lan Liu, and Bi-Shuang Chen. 2018. "Asymmetric Ketone Reduction by Immobilized Rhodotorula mucilaginosa" Catalysts 8, no. 4: 165. https://doi.org/10.3390/catal8040165





