Translocation Renal Cell Carcinoma: An Update on Clinicopathological and Molecular Features
Abstract
:1. Introduction
2. New Category of MiT Family tRCC
3. Clinical Characteristics of MiT Family tRCC
4. Pathological Characteristics of MiT Family tRCC
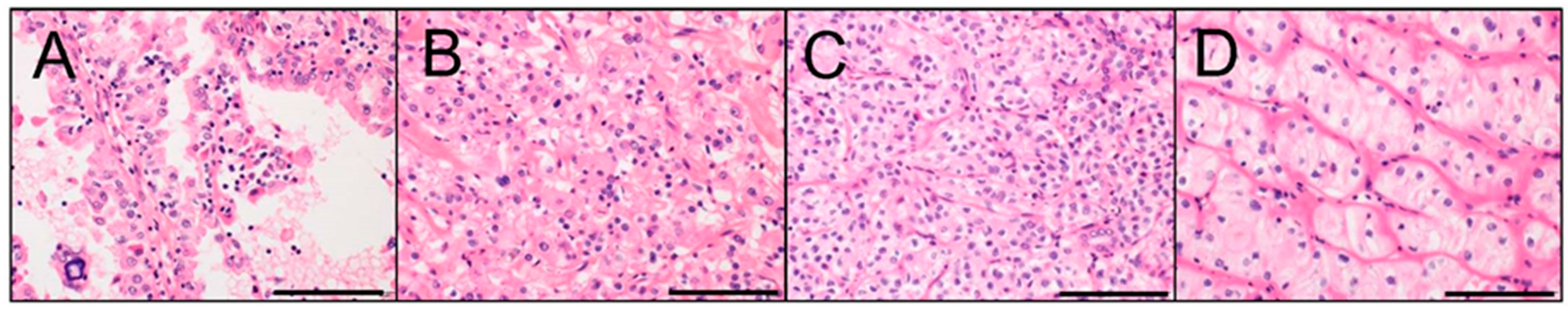
4.1. Xp11 tRCC
4.2. t(6;11) RCC
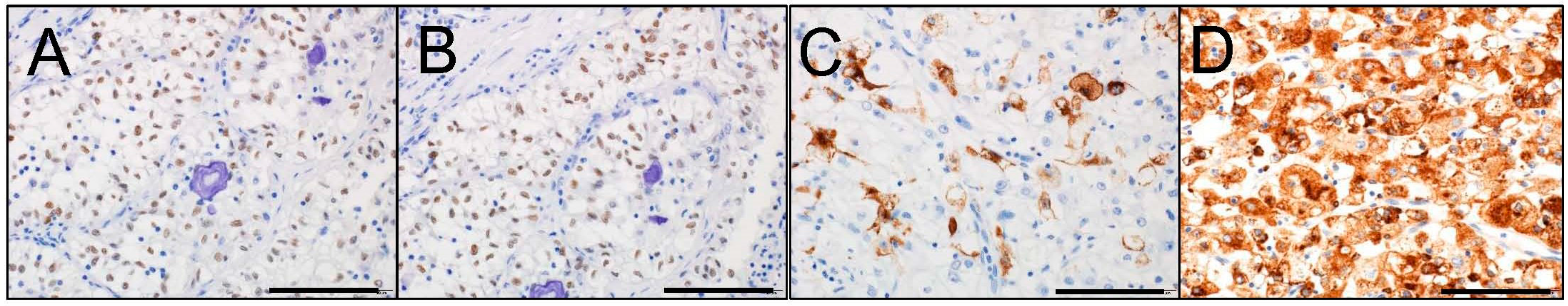
5. Immunohistochemical Characteristics of MiT Family tRCC
6. How to Diagnose MiT Family tRCC
6.1. Immunohistochemistry
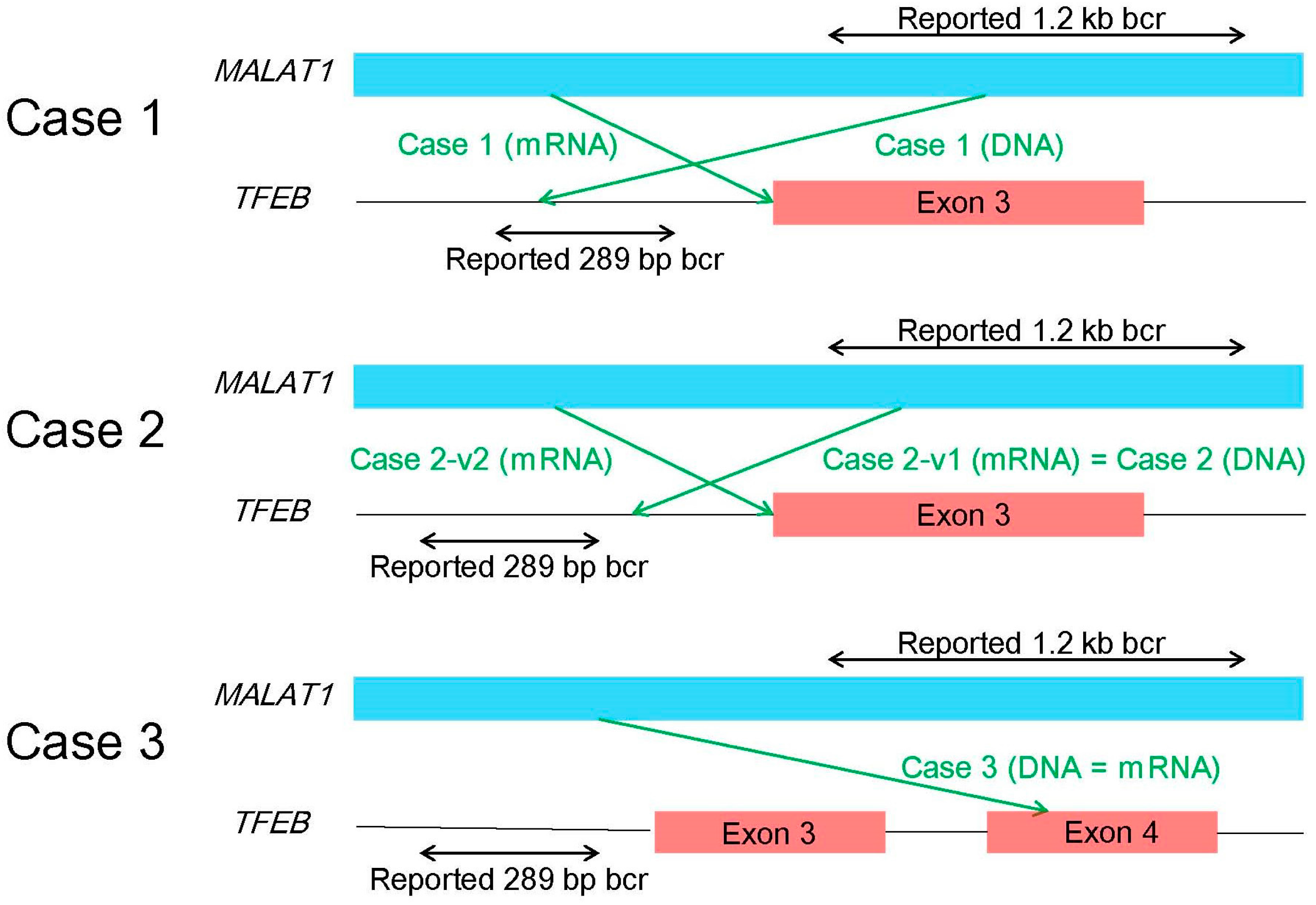
6.2. Break-Apart FISH
6.3. RT-PCR, 5′-RACE, and Karyotyping
7. RCC with Chromosome 6p Amplification
8. ALK-rearranged RCC
9. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| 5′-RACE | 5′-rapid amplification of cDNA ends |
| bHLH | basic helix-loop-helix |
| FFPE | formalin-fixed paraffin-embedded |
| FISH | fluorescence in situ hybridization |
| lncRNA | long non-coding RNA |
| PEComa | perivascular epithelioid cell tumor |
| RCC | renal cell carcinoma |
| RT-PCR | reverse transcriptase-polymerase chain reaction |
| TCGA | The Cancer Genome Atlas |
| tRCC | translocation renal cell carcinoma |
References
- Moch, H.; Humphrey, P.A.; Ulbright, T.M.; Reuter, V.E. WHO Classification of Tumours of the Urinary System and Male Genital Organs, 4th ed.; IARC Press: Lyon, France, 2016. [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar]
- Linehan, W.M.; Spellman, P.T.; Ricketts, C.J.; Creighton, C.J.; Fei, S.S.; Davis, C.; Wheeler, D.A.; Murray, B.A.; Schmidt, L.; Vocke, C.D.; et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016, 374, 135–145. [Google Scholar] [PubMed]
- Davis, C.F.; Ricketts, C.J.; Wang, M.; Yang, L.; Cherniack, A.D.; Shen, H.; Buhay, C.; Kang, H.; Kim, S.C.; Fahey, C.C.; et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014, 26, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Stawiski, E.W.; Pavia-Jimenez, A.; Modrusan, Z.; Kapur, P.; Jaiswal, B.S.; Zhang, N.; Toffessi-Tcheuyap, V.; Nguyen, T.T.; Pahuja, K.B.; et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat. Genet. 2015, 47, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Hakimi, A.A.; Reznik, E.; Lee, C.H.; Creighton, C.J.; Brannon, A.R.; Luna, A.; Aksoy, B.A.; Liu, E.M.; Shen, R.; Lee, W.; et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell 2016, 29, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, M.; Ichikawa, H.; Arai, E.; Chiku, S.; Sakamoto, H.; Fujimoto, H.; Hiramoto, M.; Nammo, T.; Yasuda, K.; Yoshida, T.; et al. Comprehensive exploration of novel chimeric transcripts in clear cell renal cell carcinomas using whole transcriptome analysis. Genes Chromosomes Cancer 2014, 53, 1018–1032. [Google Scholar] [CrossRef] [PubMed]
- Malouf, G.G.; Zhang, J.; Yuan, Y.; Comperat, E.; Roupret, M.; Cussenot, O.; Chen, Y.; Thompson, E.J.; Tannir, N.M.; Weinstein, J.N.; et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Mol. Oncol. 2015, 9, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Rathmell, K.W.; Chen, F.; Creighton, C.J. Genomics of chromophobe renal cell carcinoma: Implications from a rare tumor for pan-cancer studies. Oncoscience 2015, 2, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Christinat, Y.; Krek, W. Integrated genomic analysis identifies subclasses and prognosis signatures of kidney cancer. Oncotarget 2015, 6, 10521–10531. [Google Scholar] [CrossRef] [PubMed]
- Gowrishankar, B.; Przybycin, C.G.; Ma, C.; Nandula, S.V.; Rini, B.; Campbell, S.; Klein, E.; Chaganti, R.S.; Magi-Galluzzi, C.; Houldsworth, J. A genomic algorithm for the molecular classification of common renal cortical neoplasms: Development and validation. J. Urol. 2015, 193, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Eckel-Passow, J.E.; Igel, D.A.; Serie, D.J.; Joseph, R.W.; Ho, T.H.; Cheville, J.C.; Parker, A.S. Assessing the clinical use of clear cell renal cell carcinoma molecular subtypes identified by RNA expression analysis. Urol. Oncol. 2015, 33, 68.e17–68.e23. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.B.; Xu, J.; Skanderup, A.J.; Dong, Y.; Brannon, A.R.; Wang, L.; Won, H.H.; Wang, P.I.; Nanjangud, G.J.; Jungbluth, A.A.; et al. Molecular analysis of aggressive renal cell carcinoma with unclassified histology reveals distinct subsets. Nat. Commun. 2016, 7, 13131. [Google Scholar] [CrossRef] [PubMed]
- De Velasco, G.; Culhane, A.C.; Fay, A.P.; Hakimi, A.A.; Voss, M.H.; Tannir, N.M.; Tamboli, P.; Appleman, L.J.; Bellmunt, J.; Kimryn Rathmell, W.; et al. Molecular subtypes improve prognostic value of international metastatic renal cell carcinoma database consortium prognostic model. Oncologist 2017, 22, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Seles, M.; Hutterer, G.C.; Kiesslich, T.; Pummer, K.; Berindan-Neagoe, I.; Perakis, S.; Schwarzenbacher, D.; Stotz, M.; Gerger, A.; Pichler, M. Current insights into long non-coding RNAs in renal cell carcinoma. Int. J. Mol. Sci. 2016, 17, 573. [Google Scholar] [CrossRef] [PubMed]
- Kedzierska, H.; Poplawski, P.; Hoser, G.; Rybicka, B.; Rodzik, K.; Sokol, E.; Boguslawska, J.; Tanski, Z.; Fogtman, A.; Koblowska, M.; et al. Decreased expression of SRSF2 splicing factor inhibits apoptotic pathways in renal cancer. Int. J. Mol. Sci. 2016, 17, 1598. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Komiyama, T.; Inomoto, C.; Kamiguchi, H.; Kajiwara, H.; Kobayashi, H.; Nakamura, N.; Terachi, T. Mutations in the mitochondrial ND1 gene are associated with postoperative prognosis of localized renal cell carcinoma. Int. J. Mol. Sci. 2016, 17, 2049. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Krause, M.; Samoylenko, A.; Vainio, S. Wnt signaling in renal cell carcinoma. Cancers 2016, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Miyata, Y.; Asai, A.; Mitsunari, K.; Matsuo, T.; Ohba, K.; Mochizuki, Y.; Sakai, H. Met in urological cancers. Cancers 2014, 6, 2387–2403. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Aulmann, S.; Karanjawala, Z.; Fraser, R.B.; Ladanyi, M.; Rodriguez, M.M. Melanotic Xp11 translocation renal cancers: A distinctive neoplasm with overlapping features of PEComa, carcinoma, and melanoma. Am. J. Surg. Pathol. 2009, 33, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Aulmann, S.; Illei, P.B.; Netto, G.J.; Ro, J.; Cho, H.Y.; Dogan, S.; Ladanyi, M.; Martignoni, G.; Goldblum, J.R.; et al. A distinctive subset of PEComas harbors TFE3 gene fusions. Am. J. Surg. Pathol. 2010, 34, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Antonescu, C.R.; Illei, P.B.; Lui, M.Y.; Timmons, C.F.; Newbury, R.; Reuter, V.E.; Garvin, A.J.; Perez-Atayde, A.R.; Fletcher, J.A.; et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am. J. Pathol. 2001, 159, 179–192. [Google Scholar] [CrossRef]
- Argani, P.; Antonescu, C.R.; Couturier, J.; Fournet, J.C.; Sciot, R.; Debiec-Rychter, M.; Hutchinson, B.; Reuter, V.E.; Boccon-Gibod, L.; Timmons, C.; et al. PRCC-TFE3 renal carcinomas: Morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21). Am. J. Surg. Pathol. 2002, 26, 1553–1566. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Hawkins, A.; Griffin, C.A.; Goldstein, J.D.; Haas, M.; Beckwith, J.B.; Mankinen, C.B.; Perlman, E.J. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am. J. Pathol. 2001, 158, 2089–2096. [Google Scholar] [CrossRef]
- Malouf, G.G.; Su, X.; Yao, H.; Gao, J.; Xiong, L.; He, Q.; Comperat, E.; Couturier, J.; Molinie, V.; Escudier, B.; et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin. Cancer Res. 2014, 20, 4129–4140. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Reuter, V.E.; Zhang, L.; Sung, Y.S.; Ning, Y.; Epstein, J.I.; Netto, G.J.; Antonescu, C.R. TFEB-amplified renal cell carcinomas: An aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am. J. Surg. Pathol. 2016, 40, 1484–1495. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.R.; Grignon, D.J.; Cheng, L.; Favazza, L.; Gondim, D.D.; Carskadon, S.; Gupta, N.S.; Chitale, D.A.; Kalyana-Sundaram, S.; Palanisamy, N. Renal cell carcinoma with chromosome 6p amplification including the TFEB gene: A novel mechanism of tumor pathogenesis? Am. J. Surg. Pathol. 2017, 41, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Johnson, S.H.; Vasmatzis, G.; Porath, B.; Rustin, J.G.; Rao, P.; Costello, B.A.; Leibovich, B.C.; Thompson, R.H.; Cheville, J.C.; et al. TFEB-VEGFA (6p21.1) co-amplified renal cell carcinoma: A distinct entity with potential implications for clinical management. Mod. Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kmetec, A.; Jeruc, J. Xp 11.2 translocation renal carcinoma in young adults; recently classified distinct subtype. Radiol. Oncol. 2014, 48, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Sukov, W.R.; Hodge, J.C.; Lohse, C.M.; Leibovich, B.C.; Thompson, R.H.; Pearce, K.E.; Wiktor, A.E.; Cheville, J.C. TFE3 rearrangements in adult renal cell carcinoma: Clinical and pathologic features with outcome in a large series of consecutively treated patients. Am. J. Surg. Pathol. 2012, 36, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Fujiwara, M.; Togashi, Y.; Nomura, K.; Mukai, H.; Fujii, Y.; Yamamoto, S.; Yonese, J.; Fukui, I.; Ishikawa, Y. Diverse fusion patterns and heterogeneous clinicopathologic features of renal cell carcinoma with t(6;11) translocation. Am. J. Surg. Pathol. 2012, 36, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.E.; Illei, P.B.; Allaf, M.; Gonzalez, N.; Morris, K.; Hicks, J.; Demarzo, A.; Reuter, V.E.; Amin, M.B.; Epstein, J.I.; et al. t(6;11) renal cell carcinoma (RCC): Expanded immunohistochemical profile emphasizing novel RCC markers and report of 10 new genetically confirmed cases. Am. J. Surg. Pathol. 2014, 38, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Yonescu, R.; Morsberger, L.; Morris, K.; Netto, G.J.; Smith, N.; Gonzalez, N.; Illei, P.B.; Ladanyi, M.; Griffin, C.A. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am. J. Surg. Pathol. 2012, 36, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.L.; Eble, J.N.; Subhawong, A.P.; Martignoni, G.; Zhong, M.; Ladanyi, M.; Epstein, J.I.; Netto, G.J.; Argani, P. Clinical heterogeneity of Xp11 translocation renal cell carcinoma: Impact of fusion subtype, age, and stage. Mod. Pathol. 2014, 27, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Lae, M.; Ballard, E.T.; Amin, M.; Manivel, C.; Hutchinson, B.; Reuter, V.E.; Ladanyi, M. Translocation carcinomas of the kidney after chemotherapy in childhood. J. Clin. Oncol. 2006, 24, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Hicks, J.; De Marzo, A.M.; Albadine, R.; Illei, P.B.; Ladanyi, M.; Reuter, V.E.; Netto, G.J. Xp11 translocation renal cell carcinoma (RCC): Extended immunohistochemical profile emphasizing novel RCC markers. Am. J. Surg. Pathol. 2010, 34, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.R.; Eble, J.N.; Palanisamy, N. Sclerosing TFEB-rearrangement renal cell carcinoma: A recurring histologic pattern. Hum. Pathol. 2017, 62, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Lal, P.; Hutchinson, B.; Lui, M.Y.; Reuter, V.E.; Ladanyi, M. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: A sensitive and specific immunohistochemical assay. Am. J. Surg. Pathol. 2003, 27, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Rao, Q.; Williamson, S.R.; Zhang, S.; Eble, J.N.; Grignon, D.J.; Wang, M.; Zhou, X.J.; Huang, W.; Tan, P.H.; Maclennan, G.T.; et al. TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: Expanding the morphologic spectrum. Am. J. Surg. Pathol. 2013, 37, 804–815. [Google Scholar] [CrossRef] [PubMed]
- Camparo, P.; Vasiliu, V.; Molinie, V.; Couturier, J.; Dykema, K.J.; Petillo, D.; Furge, K.A.; Comperat, E.M.; Lae, M.; Bouvier, R.; et al. Renal translocation carcinomas: Clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am. J. Surg. Pathol. 2008, 32, 656–670. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, G.; Gobbo, S.; Camparo, P.; Brunelli, M.; Munari, E.; Segala, D.; Pea, M.; Bonetti, F.; Illei, P.B.; Netto, G.J.; et al. Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod. Pathol. 2011, 24, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Zhong, M.; Reuter, V.E.; Fallon, J.T.; Epstein, J.I.; Netto, G.J.; Antonescu, C.R. TFE3-Fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am. J. Surg. Pathol. 2016, 40, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, G.; Pea, M.; Gobbo, S.; Brunelli, M.; Bonetti, F.; Segala, D.; Pan, C.C.; Netto, G.; Doglioni, C.; Hes, O.; et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod. Pathol. 2009, 22, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Macher-Goeppinger, S.; Roth, W.; Wagener, N.; Hohenfellner, M.; Penzel, R.; Haferkamp, A.; Schirmacher, P.; Aulmann, S. Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Mod. Pathol. 2012, 25, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Cajaiba, M.M.; Jennings, L.J.; Rohan, S.M.; Perez-Atayde, A.R.; Marino-Enriquez, A.; Fletcher, J.A.; Geller, J.I.; Leuer, K.M.; Bridge, J.A.; Perlman, E.J. ALK-rearranged renal cell carcinomas in children. Genes Chromosomes Cancer 2016, 55, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Lae, M.; Hutchinson, B.; Reuter, V.E.; Collins, M.H.; Perentesis, J.; Tomaszewski, J.E.; Brooks, J.S.; Acs, G.; Bridge, J.A.; et al. Renal carcinomas with the t(6;11)(p21;q12): Clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am. J. Surg. Pathol. 2005, 29, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Green, W.M.; Yonescu, R.; Morsberger, L.; Morris, K.; Netto, G.J.; Epstein, J.I.; Illei, P.B.; Allaf, M.; Ladanyi, M.; Griffin, C.A.; et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am. J. Surg. Pathol. 2013, 37, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Argani, P.; Zhang, L.; Reuter, V.E.; Tickoo, S.K.; Antonescu, C.R. RBM10-TFE3 renal cell carcinoma: A potential diagnostic pitfall due to cryptic intrachromosomal Xp11.2 inversion resulting in false-negative TFE3 FISH. Am. J. Surg. Pathol. 2017, 41, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.Y.; Wang, Z.; Chen, N.; Gan, H.L.; Teng, X.D.; Shi, S.S.; Wang, X.; Wei, X.; Ye, S.B.; Li, R.; et al. Xp11.2 translocation renal cell carcinoma with NONO-TFE3 gene fusion: Morphology, prognosis, and potential pitfall in detecting TFE3 gene rearrangement. Mod. Pathol. 2017, 30, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Debelenko, L.V.; Raimondi, S.C.; Daw, N.; Shivakumar, B.R.; Huang, D.; Nelson, M.; Bridge, J.A. Renal cell carcinoma with novel VCL-ALK fusion: New representative of ALK-associated tumor spectrum. Mod. Pathol. 2011, 24, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Marino-Enriquez, A.; Ou, W.B.; Weldon, C.B.; Fletcher, J.A.; Perez-Atayde, A.R. ALK rearrangement in sickle cell trait-associated renal medullary carcinoma. Genes Chromosomes Cancer 2011, 50, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.E.; Deyrup, A.T.; Marino-Enriquez, A.; Fletcher, J.A.; Bridge, J.A.; Illei, P.B.; Netto, G.J.; Argani, P. VCL-ALK renal cell carcinoma in children with sickle-cell trait: The eighth sickle-cell nephropathy? Am. J. Surg. Pathol. 2014, 38, 858–863. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, E.; Togashi, Y.; Kuroda, N.; Sakata, S.; Hatano, S.; Asaka, R.; Yuasa, T.; Yonese, J.; Kitagawa, M.; Mano, H.; et al. Identification of anaplastic lymphoma kinase fusions in renal cancer: Large-scale immunohistochemical screening by the intercalated antibody-enhanced polymer method. Cancer 2012, 118, 4427–4436. [Google Scholar] [CrossRef] [PubMed]
- Kusano, H.; Togashi, Y.; Akiba, J.; Moriya, F.; Baba, K.; Matsuzaki, N.; Yuba, Y.; Shiraishi, Y.; Kanamaru, H.; Kuroda, N.; et al. Two cases of renal cell carcinoma harboring a novel STRN-ALK fusion gene. Am. J. Surg. Pathol. 2016, 40, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Sukov, W.R.; Hodge, J.C.; Lohse, C.M.; Akre, M.K.; Leibovich, B.C.; Thompson, R.H.; Cheville, J.C. ALK alterations in adult renal cell carcinoma: Frequency, clinicopathologic features and outcome in a large series of consecutively treated patients. Mod. Pathol. 2012, 25, 1516–1525. [Google Scholar] [CrossRef] [PubMed]
- Butrynski, J.E.; D'Adamo, D.R.; Hornick, J.L.; Dal Cin, P.; Antonescu, C.R.; Jhanwar, S.C.; Ladanyi, M.; Capelletti, M.; Rodig, S.J.; Ramaiya, N.; et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N. Engl. J. Med. 2010, 363, 1727–1733. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.J.; Mok, T.; Kim, D.W.; Wu, Y.L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Camidge, D.R.; Shaw, A.T.; Gadgeel, S.; Ahn, J.S.; Kim, D.W.; Ou, S.I.; Perol, M.; Dziadziuszko, R.; Rosell, R.; et al. Alectinib versus Crizotinib in untreated ALK-positive non-small-cell lung cancer. N. Engl. J. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, R.; Wu, Y.; Song, M.; Li, J.; Yang, Q.; Chen, X.; Bao, J.; Zhao, Q. L1198F mutation resensitizes crizotinib to ALK by altering the conformation of inhibitor and ATP binding sites. Int. J. Mol. Sci. 2017, 18, 482. [Google Scholar] [CrossRef] [PubMed]
- Nenadic, I.; Staber, J.; Dreier, S.; Simons, G.; Schildgen, V.; Brockmann, M.; Schildgen, O. Cost saving opportunities in NSCLC therapy by optimized diagnostics. Cancers 2017, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, S.I. Targeted therapies in non-small cell lung cancer-beyond EGFR and ALK. Cancers 2015, 7, 930–949. [Google Scholar] [CrossRef] [PubMed]
| RCC subtypes | TFE3 | TFEB | Cathepsin K | HMB45 | Melan A | CAIX | CK7 | AMACR |
|---|---|---|---|---|---|---|---|---|
| Xp11 tRCC | + | ‒ | +/‒ | ‒/f+ | f+/‒ | ‒/f+ | ‒ | + |
| t(6;11) RCC | ‒ | + | + | +/‒ | + | ‒/f+ | ‒ | + |
| Clear cell RCC | ‒ | ‒ | ‒ | ‒ | ‒ | + | ‒ | ‒ |
| Papillary RCC | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | + | +/‒ |
| Chromophobe RCC | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | + | ‒ |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inamura, K. Translocation Renal Cell Carcinoma: An Update on Clinicopathological and Molecular Features. Cancers 2017, 9, 111. https://doi.org/10.3390/cancers9090111
Inamura K. Translocation Renal Cell Carcinoma: An Update on Clinicopathological and Molecular Features. Cancers. 2017; 9(9):111. https://doi.org/10.3390/cancers9090111
Chicago/Turabian StyleInamura, Kentaro. 2017. "Translocation Renal Cell Carcinoma: An Update on Clinicopathological and Molecular Features" Cancers 9, no. 9: 111. https://doi.org/10.3390/cancers9090111






