Specific Depletion of Leukemic Stem Cells: Can MicroRNAs Make the Difference?
Abstract
:1. Introduction
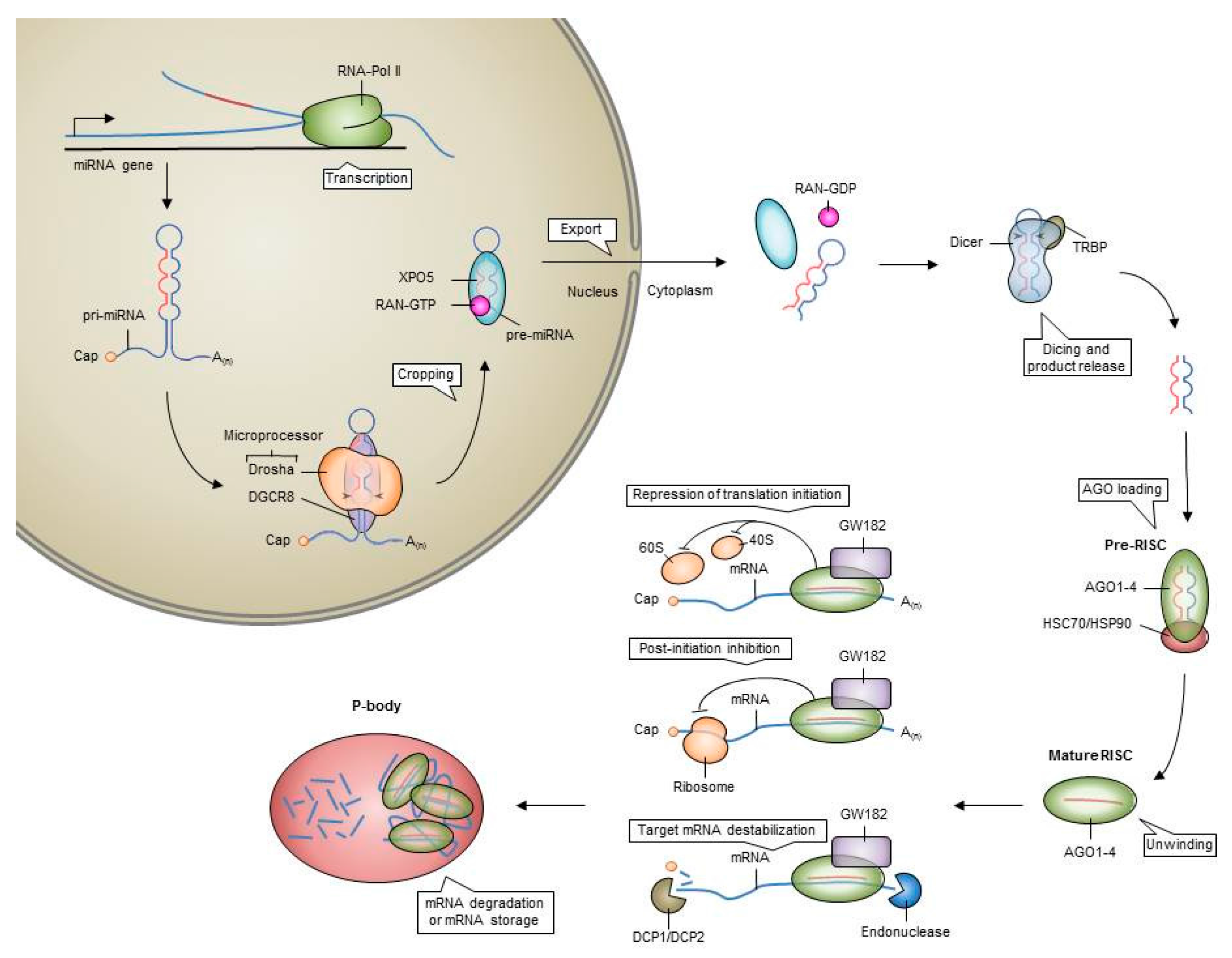
2. MicroRNAs
3. MicroRNAs in Healthy HSCs
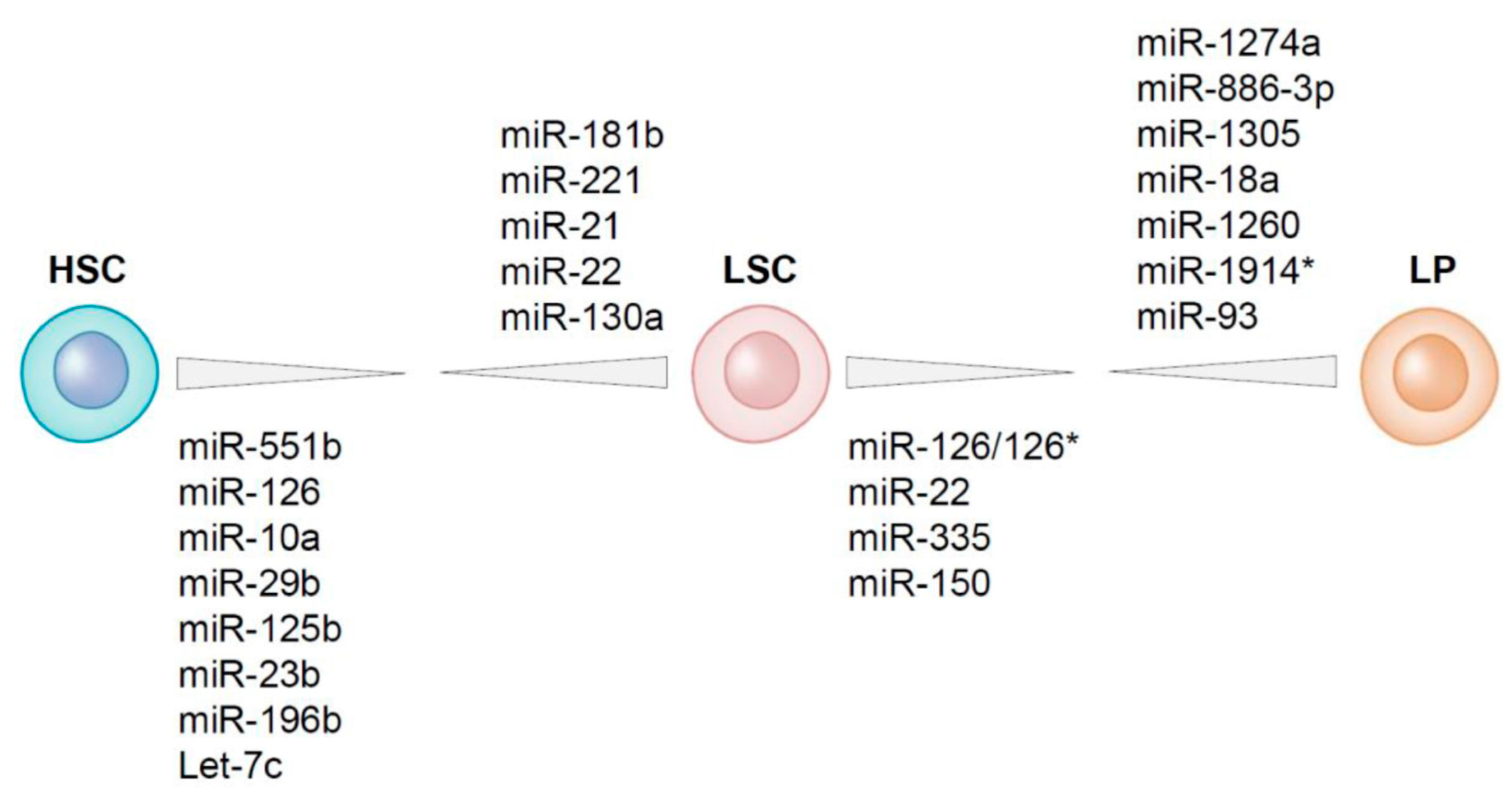
4. Differential Expression of MicroRNAs between LSCs and HSCs Residing within the AML Bone Marrow
5. MicroRNAs Differentially Expressed between LSCs and Leukemic Progenitors
6. Therapeutic Approaches to Specifically Eliminate LSCs: Sensitization to Chemotherapy
7. The Delivery of MiRNA Modulators to AML LSCs within the Leukemic Bone Marrow
8. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Rowe, J.M.; Kim, H.T.; Cassileth, P.A.; Lazarus, H.M.; Litzow, M.R.; Wiernik, P.H.; Tallman, M.S. Adult patients with acute myeloid leukemia who achieve complete remission after 1 or 2 cycles of induction have a similar prognosis: A report on 1980 patients registered to 6 studies conducted by the Eastern Cooperative Oncology Group. Cancer 2010, 116, 5012–5021. [Google Scholar] [CrossRef] [PubMed]
- Van Rhenen, A.; Feller, N.; Kelder, A.; Westra, A.H.; Rombouts, E.; Zweegman, S.; van der Pol, M.A.; Waisfisz, Q.; Ossenkoppele, G.J.; Schuurhuis, G.J. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin. Cancer Res. 2005, 11, 6520–6527. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, F.; Yoshida, S.; Saito, Y.; Hijikata, A.; Kitamura, H.; Tanaka, S.; Nakamura, R.; Tanaka, T.; Tomiyama, H.; Saito, N.; et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 2007, 25, 1315–1321. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Taussig, D.C.; Miraki-Moud, F.; Anjos-Afonso, F.; Pearce, D.J.; Allen, K.; Ridler, C.; Lillington, D.; Oakervee, H.; Cavenagh, J.; Agrawal, S.G.; et al. Anti-CD38 antibody-mediated clearance of human repopulating cells masks the heterogeneity of leukemia-initiating cells. Blood 2008, 112, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Taussig, D.C.; Vargaftig, J.; Miraki-Moud, F.; Griessinger, E.; Sharrock, K.; Luke, T.; Lillington, D.; Oakervee, H.; Cavenagh, J.; Agrawal, S.G.; et al. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(-) fraction. Blood 2010, 115, 1976–1984. [Google Scholar] [CrossRef] [PubMed]
- Farge, T.; Saland, E.; de Toni, F.; Aroua, N.; Hosseini, M.; Perry, R.; Bosc, C.; Sugita, M.; Stuani, L.; Fraisse, M.; et al. Chemotherapy Resistant Human Acute Myeloid Leukemia Cells are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017. [Google Scholar] [CrossRef] [PubMed]
- Pabst, C.; Bergeron, A.; Lavallee, V.-P.; Yeh, J.; Gendron, P.; Norddahl, G.L.; Krosl, J.; Boivin, I.; Deneault, E.; Simard, J.; et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016, 127, 2018–2127. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.-C.; LaMere, M.; Stevens, B.M.; Ashton, J.M.; Myers, J.R.; O’Dwyer, K.M.; Liesveld, J.L.; Mendler, J.H.; Guzman, M.; Morrissette, J.D.; et al. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood 2016, 128, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.W.K.; Mitchell, A.; Kennedy, J.A.; Chen, W.C.; McLeod, J.; Ibrahimova, N.; Arruda, A.; Popescu, A.; Gupta, V.; Schimmer, A.D.; et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016, 540, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Terwijn, M.; Zeijlemaker, W.; Kelder, A.; Rutten, A.P.; Snel, A.N.; Scholten, W.J.; Pabst, T.; Verhoef, G.; Löwenberg, B.; Zweegman, S.; et al. Leukemic stem cell frequency: A strong biomarker for clinical outcome in acute myeloid leukemia. PLoS ONE 2014, 9, e107587. [Google Scholar] [CrossRef] [PubMed]
- Kikushige, Y.; Shima, T.; Takayanagi, S.; Urata, S.; Miyamoto, T.; Iwasaki, H.; Takenaka, K.; Teshima, T.; Tanaka, T.; Inagaki, Y.; et al. TIM-3 Is a Promising Target to Selectively Kill Acute Myeloid Leukemia Stem Cells. Cell Stem Cell 2010, 7, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Van Rhenen, A.; Moshaver, B.; Kelder, A.; Feller, N.; Nieuwint, A.W.M.; Zweegman, S.; Ossenkoppele, G.J.; Schuurhuis, G.J. Aberrant marker expression patterns on the CD34+CD38− stem cell compartment in acute myeloid leukemia allows to distinguish the malignant from the normal stem cell compartment both at diagnosis and in remission. Leukemia 2007, 21, 1700–1707. [Google Scholar] [CrossRef] [PubMed]
- Van Rhenen, A.; van Dongen, G.A.M.S.; Kelder, A.; Rombouts, E.J.; Feller, N.; Moshaver, B.; Stigter-van Walsum, M.; Zweegman, S.; Ossenkoppele, G.J.; Jan Schuurhuis, G. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood 2007, 110, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Schuurhuis, G.J.; Meel, M.H.; Wouters, F.; Min, L.A.; Terwijn, M.; de Jonge, N.A.; Kelder, A.; Snel, A.N.; Zweegman, S.; Ossenkoppele, G.J.; et al. Normal hematopoietic stem cells within the AML bone marrow have a distinct and higher ALDH activity level than co-existing leukemic stem cells. PLoS ONE 2013, 8, e78897. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Plevritis, S.K.; Majeti, R.; Alizadeh, A.A. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA 2010, 304, 2706–2715. [Google Scholar] [CrossRef] [PubMed]
- Gal, H.; Amariglio, N.; Trakhtenbrot, L.; Jacob-Hirsh, J.; Margalit, O.; Avigdor, A.; Nagler, A.; Tavor, S.; Ein-Dor, L.; Lapidot, T.; et al. Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia 2006, 20, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Eppert, K.; Takenaka, K.; Lechman, E.R.; Waldron, L.; Nilsson, B.; van Galen, P.; Metzeler, K.H.; Poeppl, A.; Ling, V.; Beyene, J.; et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011, 17, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Kitamura, H.; Hijikata, A.; Tomizawa-Murasawa, M.; Tanaka, S.; Takagi, S.; Uchida, N.; Suzuki, N.; Sone, A.; Najima, Y.; et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci. Transl. Med. 2010, 2, 17ra9. [Google Scholar] [CrossRef] [PubMed]
- Majeti, R.; Becker, M.W.; Tian, Q.; Lee, T.-L.M.; Yan, X.; Liu, R.; Chiang, J.-H.; Hood, L.; Clarke, M.F.; Weissman, I.L. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 3396–3401. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, H.J.M.; Woolthuis, C.M.; Vos, A.Z.; Mulder, A.; van den Berg, E.; Kluin, P.M.; van der Weide, K.; de Bont, E.S.J.M.; Huls, G.; Vellenga, E.; et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 2011, 25, 1825–1833. [Google Scholar] [CrossRef] [PubMed]
- Huan, J.; Hornick, N.I.; Goloviznina, N.A.; Kamimae- Lanning, A.N.; David, L.L.; Wilmarth, P.A.; Mori, T.; Chevillet, J.R.; Narla, A.; Roberts, C.T.; et al. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia 2015, 29, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Hornick, N.I.; Doron, B.; Abdelhamed, S.; Huan, J.; Harrington, C.A.; Shen, R.; Cambronne, X.A.; Chakkaramakkil Verghese, S.; Kurre, P. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci. Signal 2016, 9, ra88. [Google Scholar] [CrossRef] [PubMed]
- Konopleva, M.Y.; Jordan, C.T. Leukemia stem cells and microenvironment: Biology and therapeutic targeting. J. Clin. Oncol. 2011, 29, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-A.; Shim, J.-S.; Lee, G.-Y.; Yim, H.W.; Kim, T.-M.; Kim, M.; Leem, S.-H.; Lee, J.-W.; Min, C.-K.; Oh, I.-H. Microenvironmental remodeling as a parameter and prognostic factor of heterogeneous leukemogenesis in acute myelogenous leukemia. Cancer Res. 2015, 75, 2222–2231. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C.P.; Tan, R.; Zhou, B.; Yue, S.-B.; Schaffert, S.; Biggs, J.R.; Doyonnas, R.; Lo, M.-C.; Perry, J.M.; Renault, V.M.; et al. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011, 21, 798–810. [Google Scholar] [CrossRef] [PubMed]
- Gangaraju, V.K.; Lin, H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009, 10, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Wojtowicz, E.E.; Lechman, E.R.; Hermans, K.G.; Schoof, E.M.; Wienholds, E.; Isserlin, R.; van Veelen, P.A.; Broekhuis, M.J.C.; Janssen, G.M.C.; Trotman-Grant, A.; et al. Ectopic miR-125a Expression Induces Long-Term Repopulating Stem Cell Capacity in Mouse and Human Hematopoietic Progenitors. Cell Stem Cell 2016, 19, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Lu, J.; Schlanger, R.; Zhang, H.; Wang, J.Y.; Fox, M.C.; Purton, L.E.; Fleming, H.H.; Cobb, B.; Merkenschlager, M.; et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. USA 2010, 107, 14229–14234. [Google Scholar] [CrossRef] [PubMed]
- Felli, N.; Fontana, L.; Pelosi, E.; Botta, R.; Bonci, D.; Facchiano, F.; Liuzzi, F.; Lulli, V.; Morsilli, O.; Santoro, S.; et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. USA 2005, 102, 18081–18086. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Cammarata, G.; Augugliaro, L.; Salemi, D.; Agueli, C.; La Rosa, M.; Dagnino, L.; Civiletto, G.; Messana, F.; Marfia, A.; Bica, M.G.; et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am. J. Hematol. 2010, 85, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lu, J.; Sun, M.; Mi, S.; Zhang, H.; Luo, R.T.; Chen, P.; Wang, Y.; Yan, M.; Qian, Z.; et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc. Natl. Acad. Sci. USA 2008, 105, 15535–15540. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Garofalo, M.; Martelli, M.P.; Briesewitz, R.; Wang, L.; Fernandez-Cymering, C.; Volinia, S.; Liu, C.-G.; Schnittger, S.; Haferlach, T.; et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. USA 2008, 105, 3945–3950. [Google Scholar] [CrossRef] [PubMed]
- De Leeuw, D.C.; van den Ancker, W.; Denkers, F.; de Menezes, R.X.; Westers, T.M.; Ossenkoppele, G.J.; van de Loosdrecht, A.A.; Smit, L. MicroRNA profiling can classify acute leukemias of ambiguous lineage as either acute myeloid leukemia or acute lymphoid leukemia. Clin. Cancer Res. 2013, 19, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Rubnitz, J.E.; Onciu, M.; Pounds, S.; Shurtleff, S.; Cao, X.; Raimondi, S.C.; Behm, F.G.; Campana, D.; Razzouk, B.I.; Ribeiro, R.C.; et al. Acute mixed lineage leukemia in children: The experience of St Jude Children’s Research Hospital. Blood 2009, 113, 5083–5089. [Google Scholar] [CrossRef] [PubMed]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef] [PubMed]
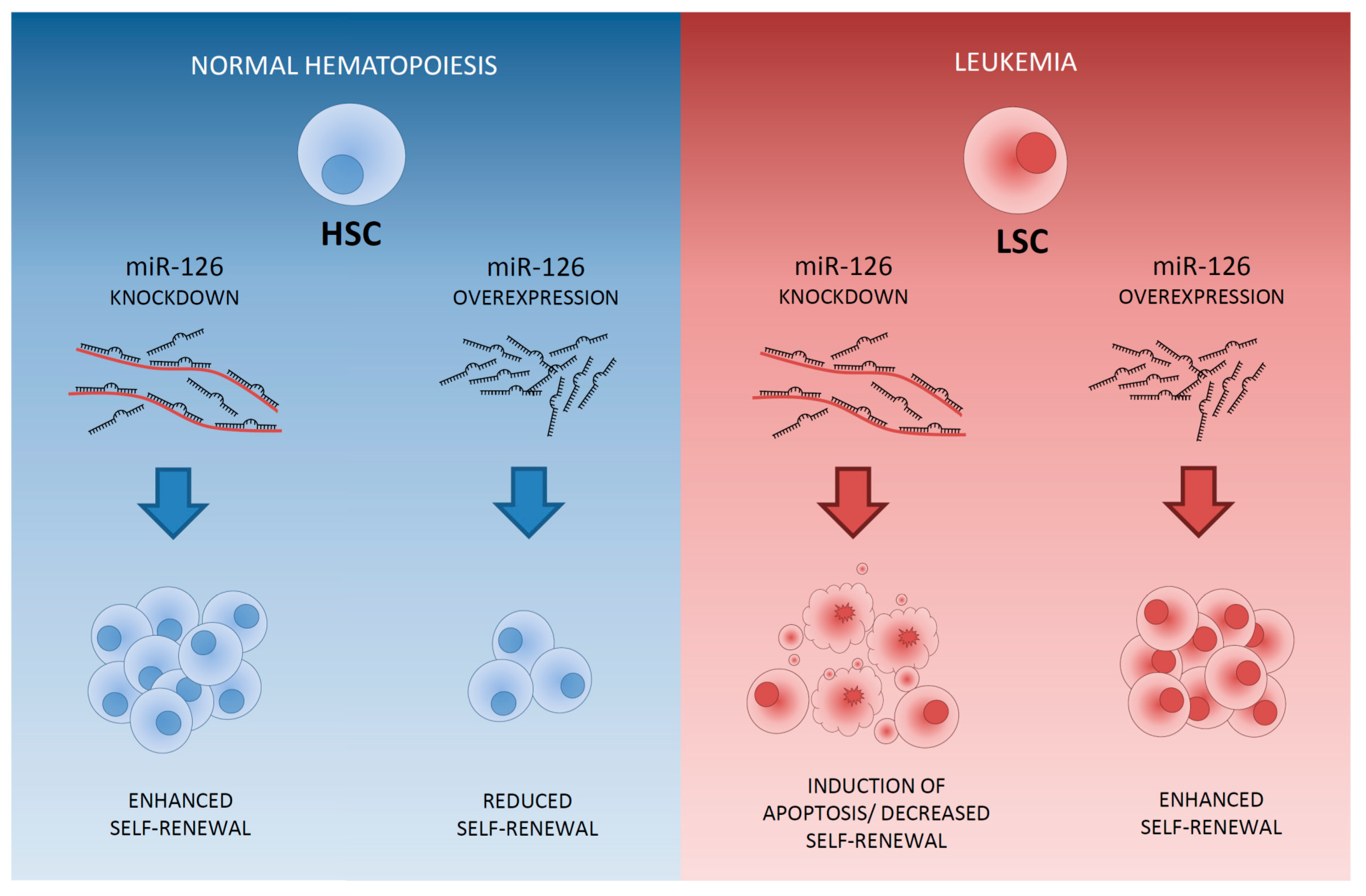
- De Leeuw, D.C.; Denkers, F.; Olthof, M.C.; Rutten, A.P.; Pouwels, W.; Schuurhuis, G.J.; Ossenkoppele, G.J.; Smit, L. Attenuation of microRNA-126 expression that drives CD34+38− stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014, 74, 2094–2105. [Google Scholar] [CrossRef] [PubMed]
- De Leeuw, D.C.; Verhagen, H.J.M.P.; Denkers, F.; Kavelaars, F.G.; Valk, P.J.M.; Schuurhuis, G.J.; Ossenkoppele, G.J.; Smit, L. MicroRNA-551b is highly expressed in hematopoietic stem cells and a biomarker for relapse and poor prognosis in acute myeloid leukemia. Leukemia 2016, 30, 742–746. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Notta, F.; Eppert, K.; Nguyen, L.T.; Ohashi, P.S.; Dick, J.E. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 2010, 11, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Majeti, R.; Park, C.Y.; Weissman, I.L. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 2007, 1, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Notta, F.; Doulatov, S.; Laurenti, E.; Poeppl, A.; Jurisica, I.; Dick, J.E. Isolation of Single Human Hematopoietic Stem Cells Capable of Long-Term Multilineage Engraftment. Science 2011, 333, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Edvardsson, L.; Dykes, J.; Olofsson, T. Isolation and characterization of human myeloid progenitor populations—TpoR as discriminator between common myeloid and megakaryocyte/erythroid progenitors. Exp. Hematol. 2006, 34, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Manz, M.G.; Miyamoto, T.; Akashi, K.; Weissman, I.L. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl. Acad. Sci. USA 2002, 99, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Summers, Y.J.; Heyworth, C.M.; de Wynter, E.A.; Hart, C.A.; Chang, J.; Testa, N.G. AC133+ G0 Cells from Cord Blood Show a High Incidence of Long-Term Culture-Initiating Cells and a Capacity for More Than 100 Million-Fold Amplification of Colony-Forming Cells In Vitro. Stem Cells 2004, 22, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Yasui, K.; Yamashita, N.; Horie, Y.; Yamada, T.; Tani, Y.; Shibata, H.; Nakano, T. In Vitro Proliferation Potential of AC133 Positive Cells in Peripheral Blood. Stem Cells 2000, 18, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Liao, R.; Sun, J.; Zhang, L.; Lou, G.; Chen, M.; Zhou, D.; Chen, Z.; Zhang, S. MicroRNAs play a role in the development of human hematopoietic stem cells. J. Cell Biochem. 2008, 104, 805–817. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Gibson, W.S.J.; Balazs, A.B.; Baltimore, D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA 2010, 107, 14235–14240. [Google Scholar] [CrossRef] [PubMed]
- Georgantas, R.W.; Hildreth, R.; Morisot, S.; Alder, J.; Liu, C.; Heimfeld, S.; Calin, G.A.; Croce, C.M.; Civin, C.I. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA 2007, 104, 2750–2755. [Google Scholar] [CrossRef] [PubMed]
- Merkerova, M.; Vasikova, A.; Belickova, M.; Bruchova, H. MicroRNA expression profiles in umbilical cord blood cell lineages. Stem Cells Dev. 2010, 19, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Wang, E.; Ren, J.; Childs, R.; Shin, J.W.; Khuu, H.; Marincola, F.M.; Stroncek, D.F. Differentiation of two types of mobilized peripheral blood stem cells by microRNA and cDNA expression analysis. J. Transl. Med. 2008, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Bissels, U.; Wild, S.; Tomiuk, S.; Hafner, M.; Scheel, H.; Mihailovic, A.; Choi, Y.-H.; Tuschl, T.; Bosio, A. Combined characterization of microRNA and mRNA profiles delineates early differentiation pathways of CD133+ and CD34+ hematopoietic stem and progenitor cells. Stem Cells 2011, 29, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-C.; Park, C.Y.; Bhagat, G.; Zhang, J.; Wang, Y.; Fan, J.-B.; Liu, M.; Zou, Y.; Weissman, I.L.; Gu, H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J. Exp. Med. 2010, 207, 475–489. [Google Scholar] [CrossRef] [PubMed]
- Ooi, A.G.L.; Sahoo, D.; Adorno, M.; Wang, Y.; Weissman, I.L.; Park, C.Y. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. USA 2010, 107, 21505–21510. [Google Scholar]
- Hu, W.; Dooley, J.; Chung, S.S.; Chandramohan, D.; Cimmino, L.; Mukherjee, S.; Mason, C.E.; de Strooper, B.; Liston, A.; Park, C.Y. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood 2015, 125, 2206–2216. [Google Scholar] [CrossRef] [PubMed]
- Gerrits, A.; Walasek, M.A.; Olthof, S.; Weersing, E.; Ritsema, M.; Zwart, E.; van Os, R.; Bystrykh, L.V.; de Haan, G. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood 2012, 119, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Gentner, B.; Visigalli, I.; Hiramatsu, H.; Lechman, E.; Ungari, S.; Giustacchini, A.; Schira, G.; Amendola, M.; Quattrini, A.; Martino, S.; et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci. Transl. Med. 2010, 2, 58ra84. [Google Scholar] [CrossRef] [PubMed]
- Lechman, E.R.; Gentner, B.; Ng, S.W.K; Schoof, E.M.; van Galen, P.; Kennedy, J.A.; Nucera, S.; Ciceri, F.; Kaufmann, K.B.; Takayama, N.; et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell 2016, 29, 214–228. [Google Scholar] [CrossRef] [PubMed]
- Copley, M.R.; Babovic, S.; Benz, C.; Knapp, D.J.H.F.; Beer, P.A.; Kent, D.G.; Wohrer, S.; Treloar, D.Q.; Day, C.; Rowe, K.; et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 2013, 15, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Huang, X.; Timani, K.A.; Broxmeyer, H.E.; He, J.J. MicroRNA-124 Targets Tip110 Expression and Regulates Hematopoiesis. Stem Cells Dev. 2015, 24, 2009–2017. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, M.; Nguyen, D.; Chen, C.; Shields, L.; Lodish, H.F. MicroRNA-125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica 2012, 97, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Le, M.T.N.; Shyh-Chang, N.; Khaw, S.L.; Chin, L.; Teh, C.; Tay, J.; O’Day, E.; Korzh, V.; Yang, H.; Lal, A.; et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 2011, 7, e1002242. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, M.; Harris, M.H.; Zhou, B.; Lodish, H.F. MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 21558–21563. [Google Scholar] [CrossRef] [PubMed]
- Surdziel, E.; Cabanski, M.; Dallmann, I.; Lyszkiewicz, M.; Krueger, A.; Ganser, A.; Scherr, M.; Eder, M. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood 2011, 117, 4338–4348. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; So, A.Y.-L.; Mehta, A.; Minisandram, A.; Sinha, N.; Jonsson, V.D.; Rao, D.S.; O’Connell, R.M.; Baltimore, D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc. Natl. Acad. Sci. USA 2012, 109, 4233–4238. [Google Scholar] [CrossRef] [PubMed]
- Jinlong, S.; Lin, F.; Yonghui, L.; Li, Y.; Weidong, W. Identification of let-7a-2-3p or/and miR-188-5p as Prognostic Biomarkers in Cytogenetically Normal Acute Myeloid Leukemia. PLoS ONE 2015, 10, e0118099. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-Y.; Zhang, X.-J.; Feng, D.-D.; Zhang, H.; Zeng, C.-W.; Han, B.-W.; Zhou, A.-D.; Qu, L.-H.; Xu, L.; Chen, Y.-Q. miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic malignancies. J. Biol. Chem. 2011, 286, 38253–38263. [Google Scholar] [CrossRef] [PubMed]
- Emmrich, S.; Rasche, M.; Schöning, J.; Reimer, C.; Keihani, S.; Maroz, A.; Xie, Y.; Li, Z.; Schambach, A.; Reinhardt, D.; et al. miR-99a/100~125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFβ and Wnt signaling. Genes Dev. 2014, 28, 858–874. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.-F.; Hu, Y.-L.; Uttarwar, L.; Passegue, E.; Largman, C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol. Cell Biol. 2008, 28, 4609–4619. [Google Scholar] [CrossRef] [PubMed]
- Lechman, E.R.; Gentner, B.; van Galen, P.; Giustacchini, A.; Saini, M.; Boccalatte, F.E.; Hiramatsu, H.; Restuccia, U.; Bachi, A.; Voisin, V.; et al. Attenuation of miR-126 Activity Expands HSC In Vivo without Exhaustion. Cell Stem Cell 2012, 11, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Zhao, J.L.; Sinha, N.; Marinov, G.K.; Mann, M.; Kowalczyk, M.S.; alimidi, R.P.; Du, X.; Erikci, E.; Regev, A.; et al. The MicroRNA-132 and MicroRNA-212 Cluster Regulates Hematopoietic Stem Cell Maintenance and Survival with Age by Buffering FOXO3 Expression. Immunity 2015, 42, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Starczynowski, D.T.; Kuchenbauer, F.; Wegrzyn, J.; Rouhi, A.; Petriv, O.; Hansen, C.L.; Humphries, R.K.; Karsan, A. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp. Hematol. 2011, 39, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Starczynowski, D.T.; Kuchenbauer, F.; Argiropoulos, B.; Sung, S.; Morin, R.; Muranyi, A.; Humphries, R.K.; Karsan, A. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 2010, 16, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.L.; Rao, D.S.; O’Connell, R.M.; Garcia-Flores, Y.; Baltimore, D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. Elife 2013, 2, e00537. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Li, Z.; Chen, P.; He, C.; Cao, D.; Elkahloun, A.; Lu, J.; Pelloso, L.A.; Wunderlich, M.; Huang, H.; et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 3710–3715. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Vecchiarelli-Federico, L.M.; Li, Y.-J.; Egan, S.E.; Spaner, D.; Hough, M.R.; Ben-David, Y. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood 2012, 119, 4486–4498. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M.; Kutok, J.L.; Rajewsky, K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008, 9, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, H.; Chen, P.; He, M.; Li, Y.; Arnovitz, S.; Jiang, X.; He, C.; Hyjek, E.; Zhang, J.; et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012, 3, 688. [Google Scholar] [CrossRef] [PubMed]
- Yekta, S.; Shih, I.-H.; Bartel, D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004, 304, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Rich, A.; Dahl, R. MiR-24 promotes the survival of hematopoietic cells. PLoS ONE 2013, 8, e55406. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Merchan, A.; Cerrato, C.; Luengo, G.; Dominguez, O.; Piris, M.A.; Serrano, M.; Gonzalez, S. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle. 2010, 9, 3277–3285. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.M.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E.; et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Meenhuis, A.; van Veelen, P.A.; de Looper, H.; van Boxtel, N.; van den Berge, I.J.; Sun, S.M.; Taskesen, E.; Stern, P.; de Ru, A.H.; van Adrichem, A.J.; et al. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood 2011, 118, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Neilson, J.R.; Zheng, G.X.Y.; Burge, C.B.; Sharp, P.A. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007, 21, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-J.; Chau, J.; Ebert, P.J.R.; Sylvester, G.; Min, H.; Liu, G.; Braich, R.; Manoharan, M.; Soutschek, J.; Skare, P.; et al. miR-181a Is an Intrinsic Modulator of T Cell Sensitivity and Selection. Cell 2007, 129, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-S.; Gong, J.-N.; Yu, J.; Wang, F.; Zhang, X.-H.; Yin, X.-L.; Tan, Z.-Q.; Luo, Z.-M.; Yang, G.-H.; Shen, C.; et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 2012, 119, 4992–5004. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Boldin, M.P.; Taganov, K.D.; Nicoll, J.; Paquette, R.L.; Baltimore, D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008, 205, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.T.; Häger, M.; Glenthøj, A.; Asmar, F.; Clemmensen, S.N.; Mora-Jensen, H.; Borregaard, N.; Cowland, J.B. miRNA-130a regulates C/EBP-ε expression during granulopoiesis. Blood 2014, 123, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Pelosi, E.; Greco, P.; Racanicchi, S.; Testa, U.; Liuzzi, F.; Croce, C.M.; Brunetti, E.; Grignani, F.; Peschle, C. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 2007, 9, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Zardo, G.; Ciolfi, A.; Vian, L.; Starnes, L.M.; Billi, M.; Racanicchi, S.; Maresca, C.; Fazi, F.; Travaglini, L.; Noguera, N.; et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 2012, 119, 4034–4046. [Google Scholar] [CrossRef] [PubMed]
- Fazi, F.; Rosa, A.; Fatica, A.; Gelmetti, V.; De Marchis, M.L.; Nervi, C.; Bozzoni, I. A Minicircuitry Comprised of MicroRNA-223 and Transcription Factors NFI-A and C/EBPα Regulates Human Granulopoiesis. Cell 2005, 123, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Pulikkan, J.A.; Dengler, V.; Peramangalam, P.S.; Peer Zada, A.A.; Müller-Tidow, C.; Bohlander, S.K.; Tenen, D.G.; Behre, G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood 2010, 115, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Iwama, A.; Satake, M.; Kohu, K. MicroRNA-27 enhances differentiation of myeloblasts into granulocytes by post-transcriptionally downregulating Runx1. Br. J. Haematol. 2009, 145, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Katzerke, C.; Madan, V.; Gerloff, D.; Bräuer-Hartmann, D.; Hartmann, J.-U.; Wurm, A.A.; Müller-Tidow, C.; Schnittger, S.; Tenen, D.G.; Niederwieser, D.; et al. Transcription factor C/EBPα-induced microRNA-30c inactivates Notch1 during granulopoiesis and is downregulated in acute myeloid leukemia. Blood 2013, 122, 2433–2442. [Google Scholar] [CrossRef] [PubMed]
- Pulikkan, J.A.; Peramangalam, P.S.; Dengler, V.; Ho, P.A.; Preudhomme, C.; Meshinchi, S.; Christopeit, M.; Nibourel, O.; Müller-Tidow, C.; Bohlander, S.K.; et al. C/EBPα regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood 2010, 116, 5638–5649. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Ballarino, M.; Sorrentino, A.; Sthandier, O.; De Angelis, F.G.; Marchioni, M.; Masella, B.; Guarini, A.; Fatica, A.; Peschle, C.; et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 19849–19854. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Bulgarelli, J.; Ruberti, S.; Rontauroli, S.; Sacchi, G.; Norfo, R.; Pennucci, V.; Zini, R.; Salati, S.; Prudente, Z.; et al. MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF. Cell Death Differ. 2015, 22, 1906–1921. [Google Scholar] [CrossRef] [PubMed]
- Kamat, V.; Paluru, P.; Myint, M.; French, D.L.; Gadue, P.; Diamond, S.L. MicroRNA screen of human embryonic stem cell differentiation reveals miR-105 as an enhancer of megakaryopoiesis from adult CD34+ cells. Stem Cells 2014, 32, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Hu, C.; Arnovitz, S.; Bugno, J.; Yu, M.; Zuo, Z.; Chen, P.; Huang, H.; Ulrich, B.; Gurbuxani, S.; et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat. Commun. 2016, 7, 11452. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Chen, M.-T.; Zhang, X.-H.; Yin, X.-L.; Ning, H.-M.; Su, R.; Lin, H.-S.; Song, L.; Wang, F.; Ma, Y.-N.; et al. The PU.1-Modulated MicroRNA-22 Is a Regulator of Monocyte/Macrophage Differentiation and Acute Myeloid Leukemia. PLoS Genet. 2016, 12, e1006259. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes-Sternberg, C.; Meerson, A.; Shaked, I.; Soreq, H. MicroRNA modulation of megakaryoblast fate involves cholinergic signaling. Leuk. Res. 2006, 30, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.; Gao, L.; McClellan, S.; Finan, M.A.; Butler, T.W.; Owen, L.B.; Piazza, G.A.; Xi, Y. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2012, 19, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Klusmann, J.-H.; Li, Z.; Böhmer, K.; Maroz, A.; Koch, M.L.; Emmrich, S.; Godinho, F.J.; Orkin, S.H.; Reinhardt, D. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010, 24, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Grabher, C.; Payne, E.M.; Johnston, A.B.; Bolli, N.; Lechman, E.; Dick, J.E.; Kanki, J.P.; Look, A.T. Zebrafish microRNA-126 determines hematopoietic cell fate through c-Myb. Leukemia 2011, 25, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.; Narla, A.; Nonami, A.; Mullally, A.; Dimitrova, N.; Ball, B.; McAuley, J.R.; Poveromo, L.; Kutok, J.L.; Galili, N.; et al. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q- syndrome. Blood 2011, 118, 4666–4673. [Google Scholar] [CrossRef] [PubMed]
- Labbaye, C.; Spinello, I.; Quaranta, M.T.; Pelosi, E.; Pasquini, L.; Petrucci, E.; Biffoni, M.; Nuzzolo, E.R.; Billi, M.; Foà, R.; et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 2008, 10, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Kalota, A.; Jin, S.; Gewirtz, A.M. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood 2009, 113, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Guo, S.; Ebert, B.L.; Zhang, H.; Peng, X.; Bosco, J.; Pretz, J.; Schlanger, R.; Wang, J.Y.; Mak, R.H.; et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev. Cell 2008, 14, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Barroga, C.F.; Pham, H.; Kaushansky, K. Thrombopoietin regulates c-Myb expression by modulating micro RNA 150 expression. Exp. Hematol. 2008, 36, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Romania, P.; Lulli, V.; Pelosi, E.; Biffoni, M.; Peschle, C.; Marziali, G. MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors. Br. J. Haematol. 2008, 143, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bai, H.; Zhang, Z.; Li, W.; Dong, L.; Wei, X.; Ma, Y.; Zhang, J.; Yu, J.; Sun, G.; et al. The up-regulation of miR-199b-5p in erythroid differentiation is associated with GATA-1 and NF-E2. Mol. Cells 2014, 37, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.-Y.; Wang, F.; Yu, J.; Yang, G.-H.; Liu, X.-L.; Zhang, J.-W. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J. Cell. Mol. Med. 2009, 13, 4551–4559. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, D.; Wang, F.; Li, T.; Dong, L.; Liu, H.; Ma, Y.; Jiang, F.; Yin, H.; Yan, W.; et al. A comprehensive analysis of GATA-1-regulated miRNAs reveals miR-23a to be a positive modulator of erythropoiesis. Nucleic Acids Res. 2013, 41, 4129–4143. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, Y.; Guo, L.; Dong, L.; Liu, H.; Yin, H.; Zhang, Z.; Li, Y.; Liu, C.; Ma, Y.; et al. A regulatory circuit comprising GATA1/2 switch and microRNA-27a/24 promotes erythropoiesis. Nucleic Acids Res. 2014, 42, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Tenedini, E.; Roncaglia, E.; Ferrari, F.; Orlandi, C.; Bianchi, E.; Bicciato, S.; Tagliafico, E.; Ferrari, S. Integrated analysis of microRNA and mRNA expression profiles in physiological myelopoiesis: Role of hsa-mir-299-5p in CD34+ progenitor cells commitment. Cell Death Dis. 2010, 1, e28. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.; Gutman, D.; Meire, E.; Cáceres, M.; Rigoutsos, I.; Bentwich, Z.; Lieberman, J. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood 2009, 114, 2181–2192. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yu, J.; Yang, G.-H.; Wang, X.-S.; Zhang, J.-W. Regulation of erythroid differentiation by miR-376a and its targets. Cell Res. 2011, 21, 1196–1209. [Google Scholar] [CrossRef] [PubMed]
- Dore, L.C.; Amigo, J.D.; Dos Santos, C.O.; Zhang, Z.; Gai, X.; Tobias, J.W.; Yu, D.; Klein, A.M.; Dorman, C.; Wu, W.; et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3333–3338. [Google Scholar] [CrossRef] [PubMed]
- Pase, L.; Layton, J.E.; Kloosterman, W.P.; Carradice, D.; Waterhouse, P.M.; Lieschke, G.J. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood 2009, 113, 1794–1804. [Google Scholar] [CrossRef] [PubMed]
- Kamesaki, H.; Michaud, G.Y.; Irving, S.G.; Suwabe, N.; Kamesaki, S.; Okuma, M.; Cossman, J. TPA-induced arrest of erythroid differentiation is coupled with downregulation of GATA-1 and upregulation of GATA-2 in an erythroid cell line SAM-1. Blood 1996, 87, 999–1005. [Google Scholar] [PubMed]
- Lulli, V.; Romania, P.; Morsilli, O.; Cianciulli, P.; Gabbianelli, M.; Testa, U.; Giuliani, A.; Marziali, G. MicroRNA-486-3p regulates γ-globin expression in human erythroid cells by directly modulating BCL11A. PLoS ONE 2013, 8, e60436. [Google Scholar] [CrossRef] [PubMed]
- Zhai, P.-F.; Wang, F.; Su, R.; Lin, H.-S.; Jiang, C.-L.; Yang, G.-H.; Yu, J.; Zhang, J.-W. The regulatory roles of microRNA-146b-5p and its target platelet-derived growth factor receptor α (PDGFRA) in erythropoiesis and megakaryocytopoiesis. J. Biol. Chem. 2014, 289, 22600–22613. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Volinia, S.; Liu, C.-G.; Fernandez-Cymering, C.; Palumbo, T.; Pichiorri, F.; Fabbri, M.; Coombes, K.; Alder, H.; Nakamura, T.; et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008, 111, 3183–3189. [Google Scholar] [CrossRef] [PubMed]
- Feller, N.; van der Velden, V.H.J.; Brooimans, R.A.; Boeckx, N.; Preijers, F.; Kelder, A.; de Greef, I.; Westra, G.; te Marvelde, J.G.; Aerts, P.; et al. Defining consensus leukemia-associated immunophenotypes for detection of minimal residual disease in acute myeloid leukemia in a multicenter setting. Blood Cancer J. 2013, 3, e129. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.T.; Upchurch, D.; Szilvassy, S.J.; Guzman, M.L.; Howard, D.S.; Pettigrew, A.L.; Meyerrose, T.; Rossi, R.; Grimes, B.; Rizzieri, D.A.; et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 2000, 14, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Wouters, R.; Cucchi, D.; Kaspers, G.J.L.; Schuurhuis, G.J.; Cloos, J. Relevance of leukemic stem cells in acute myeloid leukemia: Heterogeneity and influence on disease monitoring, prognosis and treatment design. Expert Rev. Hematol. 2014, 7, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Liu, Y.; Wang, Y.; Zhang, Y.; Luo, Q.; Man, X.; Wei, F.; Yu, X. Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med. Oncol. 2016, 33, 126. [Google Scholar] [CrossRef] [PubMed]
- Chaluvally-Raghavan, P.; Jeong, K.J.; Pradeep, S.; Silva, A.M.; Yu, S.; Liu, W.; Moss, T.; Rodriguez-Aguayo, C.; Zhang, D.; Ram, P.; et al. Direct Upregulation of STAT3 by MicroRNA-551b-3p Deregulates Growth and Metastasis of Ovarian Cancer. Cell Rep. 2016, 15, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wells, A.; Padilla, M.T.; Kato, K.; Kim, K.C.; Lin, Y. A signaling pathway consisting of miR-551b, catalase and MUC1 contributes to acquired apoptosis resistance and chemoresistance. Carcinogenesis 2014, 35, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Heaphy, C.E.A.; Havelange, V.; Fabbri, M.; Volinia, S.; Tsao, T.; Zanesi, N.; Kornblau, S.M.; Marcucci, G.; Calin, G.A.; et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009, 114, 5331–5341. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Schwind, S.; Yu, B.; Santhanam, R.; Wang, H.; Hoellerbauer, P.; Mims, A.; Klisovic, R.; Walker, A.R.; Chan, K.K.; et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: A novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 2013, 19, 2355–2367. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.A.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef] [PubMed]
- Blum, W.; Garzon, R.; Klisovic, R.B.; Schwind, S.; Walker, A.; Geyer, S.; Liu, S.; Havelange, V.; Becker, H.; Schaaf, L.; et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc. Natl. Acad. Sci. USA 2010, 107, 7473–7478. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.R.; Higgins, R.R.; Grutkoski, P.S.; Bousquet, M.; Quelen, C.; Bartholomaus, L.M.; Brousset, P. Myeloid neoplasm with translocation t(2;11)(p21;q23-24), elevated microRNA 125b-1, and JAK2 exon 12 mutation. Br. J. Haematol. 2015, 169, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, M.; Quelen, C.; Rosati, R.; Mansat-De Mas, V.; La Starza, R.; Bastard, C.; Lippert, E.; Talmant, P.; Lafage-Pochitaloff, M.; Leroux, D.; et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 2008, 205, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Radmacher, M.D.; Maharry, K.; Mrózek, K.; Ruppert, A.S.; Paschka, P.; Vukosavljevic, T.; Whitman, S.P.; Baldus, C.D.; Langer, C.; et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008, 358, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Vukosavljevic, T.; Paschka, P.; Whitman, S.P.; Langer, C.; Baldus, C.D.; Liu, C.-G.; et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2008, 26, 5078–5087. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, H.; Li, Y.; Jiang, X.; Chen, P.; Arnovitz, S.; Radmacher, M.D.; Maharry, K.; Elkahloun, A.; Yang, X.; et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood 2012, 119, 2314–2324. [Google Scholar] [CrossRef] [PubMed]
- Isken, F.; Steffen, B.; Merk, S.; Dugas, M.; Markus, B.; Tidow, N.; Zühlsdorf, M.; Illmer, T.; Thiede, C.; Berdel, W.E.; et al. Identification of acute myeloid leukaemia associated microRNA expression patterns. Br. J. Haematol. 2008, 140, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Roscigno, G.; Quintavalle, C.; Donnarumma, E.; Puoti, I.; Diaz-Lagares, A.; Iaboni, M.; Fiore, D.; Russo, V.; Todaro, M.; Romano, G.; et al. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget 2010, 1, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, L.; Ischenko, I.; Bao, Q.; Schwarz, B.; Nieß, H.; Wang, Y.; Renner, A.; Mysliwietz, J.; Jauch, K.-W.; et al. Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target Oncol. 2015, 10, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Riccioni, R.; Lulli, V.; Castelli, G.; Biffoni, M.; Tiberio, R.; Pelosi, E.; Lo-Coco, F.; Testa, U. miR-21 is overexpressed in NPM1-mutant acute myeloid leukemias. Leuk. Res. 2015, 39, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Zhu, X.; Li, Y.; Dong, D.; Yao, J.; Lin, C.; Huang, K.; Hu, H.; Fei, J. miRNA-21 regulates arsenic-induced anti-leukemia activity in myelogenous cell lines. Med. Oncol. 2011, 28, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, X.; Gu, J.; Hu, H.; Dong, D.; Yao, J.; Lin, C.; Fei, J. Anti-miR-21 oligonucleotide enhances chemosensitivity of leukemic HL60 cells to arabinosylcytosine by inducing apoptosis. Hematology 2010, 15, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Xu, R.; Cao, Z.; Wei, D.; Wang, C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011, 585, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, Y.; Gu, J.; Zhu, X.; Dong, D.; Yao, J.; Lin, C.; Fei, J. Antisense oligonucleotide against miR-21 inhibits migration and induces apoptosis in leukemic K562 cells. Leuk. Lymphoma 2010, 51, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Schopman, N.C.T.; Heynen, S.; Haasnoot, J.; Berkhout, B. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 2010, 7, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-J.; Liu, G.-H.; Ye, Y.-W.; Fu, Y.; Zhang, X.-F. The role of microRNA-1274a in the tumorigenesis of gastric cancer: Accelerating cancer cell proliferation and migration via directly targeting FOXO4. Biochem. Biophys. Res. Commun. 2015, 459, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S. A Novel Type of Non-coding RNA, nc886, Implicated in Tumor Sensing and Suppression. Genom. Inform. 2015, 13, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kunkeaw, N.; Jeon, S.H.; Lee, I.; Johnson, B.H.; Kang, G.-Y.; Bang, J.Y.; Park, H.S.; Leelayuwat, C.; Lee, Y.S. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA 2011, 17, 1076–1089. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Lee, K.; Jang, H.-J.; Lee, G.K.; Park, J.-L.; Kim, S.-Y.; Kim, S.-B.; Johnson, B.H.; Zo, J.I.; Lee, J.-S.; et al. Epigenetic silencing of the non-coding RNA nc886 provokes oncogenes during human esophageal tumorigenesis. Oncotarget 2014, 5, 3472–3481. [Google Scholar] [CrossRef] [PubMed]
- Oler, A.J.; Alla, R.K.; Roberts, D.N.; Wong, A.; Hollenhorst, P.C.; Chandler, K.J.; Cassiday, P.A.; Nelson, C.A.; Hagedorn, C.H.; Graves, B.J.; et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010, 17, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Treppendahl, M.B.; Qiu, X.; Søgaard, A.; Yang, X.; Nandrup-Bus, C.; Hother, C.; Andersen, M.K.; Kjeldsen, L.; Möllgaard, L.; Hellström-Lindberg, E.; et al. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood 2012, 119, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Helbo, A.S.; Treppendahl, M.; Aslan, D.; Dimopoulos, K.; Nandrup-Bus, C.; Holm, M.S.; Andersen, M.K.; Liang, G.; Kristensen, L.S.; Grønbæk, K. Hypermethylation of the VTRNA1-3 Promoter is Associated with Poor Outcome in Lower Risk Myelodysplastic Syndrome Patients. Genes 2015, 6, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Song, Y.; Bi, N.; Shen, J.; Liu, W.; Fan, J.; Sun, G.; Tong, T.; He, J.; Shi, Y.; et al. DNA methylation-mediated repression of miR-886–3p predicts poor outcome of human small cell lung cancer. Cancer Res. 2013, 73, 3326–3335. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Collin, J.; Zhu, L.; Montaner, D.; Armstrong, L.; Neganova, I.; Lako, M. A Novel Role for miR-1305 in Regulation of Pluripotency-Differentiation Balance, Cell Cycle, and Apoptosis in Human Pluripotent Stem Cells. Stem Cells 2016, 34, 2306–2317. [Google Scholar] [CrossRef] [PubMed]
- Dorrance, A.M.; Neviani, P.; Ferenchak, G.J.; Huang, X.; Nicolet, D.; Maharry, K.S.; Ozer, H.G.; Hoellarbauer, P.; Khalife, J.; Hill, E.B.; et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia 2015, 29, 2143–2153. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, P.; Su, R.; Li, Y.; Hu, C.; Wang, Y.; Arnovitz, S.; He, M.; Gurbuxani, S.; Zuo, Z.; et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood Am. Soc. Hematol. 2015, 126, 2005–2015. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.H.; Valdez, R.; Theisen, B.K.; Guo, W.; Ferguson, D.O.; Wu, H.; Arnovitz, S.; He, M.; Gurbuxani, S.; Zuo, Z.; et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 2006, 441, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with “antagomirs”. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.C. Inhibition of microRNA with antisense oligonucleotides. Methods 2008, 44, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Vester, B.; Wengel, J. LNA (locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. [Google Scholar] [CrossRef] [PubMed]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U.; et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Wengel, J. LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol. 2003, 21, 74–81. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Gao, X.-M.; Winbanks, C.E.; Boey, E.J.H.; Tham, Y.K.; Kiriazis, H.; Gregorevic, P.; Obad, S.; Kauppinen, S.; Du, X.-J.; et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl. Acad. Sci. USA 2012, 109, 17615–17620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Roccaro, A.M.; Rombaoa, C.; Flores, L.; Obad, S.; Fernandes, S.M.; Sacco, A.; Liu, Y.; Ngo, H.; Quang, P.; et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood 2012, 120, 1678–1686. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Bader, A.G.; Brown, D.; Winkler, M. The promise of microRNA replacement therapy. Cancer Res. 2010, 70, 7027–7030. [Google Scholar] [CrossRef] [PubMed]
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2010, 29, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.-W.; Chang, T.-C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Fayyaz, S.; Shatynska-Mytsyk, I.; Javed, Z.; Jabeen, S.; Yaylim, I.; Javed, Z.; Jabeen, S.; Yaylim, I.; Gasparri, M.L.; et al. Is miR-34a a Well-equipped Swordsman to Conquer Temple of Molecular Oncology? Chem. Biol. Drug Des. 2016, 87, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Blower, P.E.; Chung, J.-H.; Verducci, J.S.; Lin, S.; Park, J.-K.; Dai, Z.; Liu, C.-G.; Schmittgen, T.D.; Reinhold, W.C.; Croce, C.M.; et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol. Cancer Ther. 2008, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Sarmiento, C.; Tan, T.; Zhu, H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm. Sin. B 2017, 7, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, F.; Minakuchi, Y.; Nagahara, S.; Honma, K.; Sasaki, H.; Hirai, K.; Teratani, T.; Namatame, N.; Yamamoto, Y.; Hanai, K.; et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 12177–12182. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, D.; Campbell, N.R.; Karikari, C.; Chivukula, R.; Kent, O.A.; Mendell, J.T.; Maitra, A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol. Cancer Ther. 2011, 10, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, S.; Yu, B.; Golan, S.; Koh, C.-G.; Yang, J.; Huynh, L.; Yang, X.; Pang, J.; Muthusamy, N.; et al. Targeted Delivery of Antisense Oligodeoxynucleotide by Transferrin Conjugated pH-Sensitive Lipopolyplex Nanoparticles: A Novel Oligonucleotide-Based Therapeutic Strategy in Acute Myeloid Leukemia. Mol. Pharm. 2010, 7, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Velu, C.S.; Chaubey, A.; Phelan, J.D.; Horman, S.R.; Wunderlich, M.; Guzman, M.L.; Jegga, A.G.; Zeleznik-Le, N.J.; Chen, J.; Mulloy, J.C.; et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Investig. 2014, 124, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Schwind, S.; Santhanam, R.; Eisfeld, A.K; Chiang, C.L.; Lankenau, M.; Yu, B.; Hoellerbauer, P.; Jin, Y.; Tarighat, S.S.; et al. Targeting the RAS/MAPK pathway with miR-181a in acute myeloid leukemia. Oncotarget 2016, 7, 59273–59286. [Google Scholar] [CrossRef] [PubMed]

| Cell Stage | microRNA | Target | Function | References | |
|---|---|---|---|---|---|
| HSC | Let-7 | Hmg2a |  | self-renewal | [61] |
| miR-12 | Tip110 |  | differentiation | [62] | |
| miR-125a | BAK1 |  | apoptosis | [30,31,58] | |
| miR-125b | ABTB1/CDC25C/PPP1CA |  | proliferation | [63,64] | |
| Bmf/KLF13/p53 |  | apoptosis | [56,58,65] | ||
| STAT3/c-JUN/JUND/LIN28A/CBFB |  | differentiation | [63,66,67,68,69,70] | ||
| miR-126 | HOXA9/PI3K/AKT2/CRKII |  | self-renewal | [71,72] | |
| miR-132 | FOXO3 |  | proliferation | [73] | |
| miR-146a | TRAF6/IRAK1/STAT1 |  | self-renewal | [74,75,76] | |
| miR-17-92 cluster | E2F1/E2F2 |  | proliferation and block differentiation | [77,78] | |
| PTEN/Bim |  | apoptosis | [79,80] | ||
| miR-196b | HOXA9/MEIS1/FAS/HOXB8 |  | differentiation | [81,82] | |
| miR-24 | Bim/CASP9 |  | apoptosis | [83] | |
| miR-29a | Dnmt3a |  | self-renewal | [57] | |
| miR-33 | p53 | self-renewal | [84] | ||
| miR-22 | Tet2 |  | self-renewal | [85] | |
| MPP | miR-17/20/93/106 | SQSTM1 |  | differentiation towards myeloid progenitors | [54,86] |
| miR-24 | Unknown |  | differentiation towards myeloid progenitors | [83] | |
| miR-29a | HBP1, FZD5, TPM1 |  | differentiation towards myeloid progenitors | [55] | |
| miR-520h | ABCG2 | differentiation towards myeloid progenitors | [49] | ||
| miR-181a | Bcl2, CD69 |  | differentiation towards lymphoid progenitors | [87,88,89] | |
| CMP | miR-142-3p | CCNT2/TAB2 |  | granulocytic-macrophage differentiation | [90] |
| miR-155 | PU.1 |  | granulocytic-macrophage differentiation | [91] | |
| miR-29a | CCNT2/CDK6 | granulocytic-macrophage differentiation | [90] | ||
| miR-130a | C/EBPɛ |  | granulocytic differentiation | [92] | |
| GMP | miR-17-5p/20a/106a | RUNX1 |  | monocytic differentiation and maturation | [93] |
| miR-223 | MEF2C |  | progenitor proliferation and granulocyte differentiation | [94] | |
| miR-223 miR-27 | NFI-A/E2F1 |  | granulocytic differentiation | [95,96,97] | |
| RUNX1 |  | granulocytic differentiation | [98] | ||
| miR-30c | NOTCH1 |  | granulocytic differentiation | [99] | |
| miR-34a | E2F3 |  | granulocytic differentiation | [100] | |
| miR-424 | NFI-A |  | monocytic differentiation | [101] | |
| miR-486-3p | MAF | Skews from monocytopoiesis towards granulopoiesis | [102] | ||
| miR-105 | MYB | megakaryopoiesis | [103] | ||
| miR-22 | PU.1, MECOM |  | monocytic differentiation | [104,105] | |
| miR-181a | Unknown |  | megakaryocytic differentiation | [106] | |
| Lin28, let7 | megakaryocytic differentiation | [107] | |||
| MEP | miR-125b | Unknown |  | proliferation and self-renewal | [108] |
| miR-126 | MYB | Skews from erythropoiesis towards megakaryopoiesis | [109] | ||
| miR-145 | Fli-1 | Skews from megakaryopoiesis towards erythropoiesis | [110] | ||
| miR-146a | CXCR4 | Impairs megakaryocytic proliferation, differentiation and maturation | [111] | ||
| miR-15 | MYB |  | erythropoiesis | [112] | |
| miR-150 | MYB | Skews from erythropoiesis towards megakaryopoiesis | [109,113,114] | ||
| miR-155 | ETS-1/MEIS1 |  | megakaryocytic proliferation and differentiation | [115] | |
| miR-199b-5p | c-Kit |  | erythroid differentiation | [116] | |
| miR-221/222 | c-Kit | Impairs proliferation and accelerates differentiation of erythroid cells | [32] | ||
| miR-223 | LMO2 | Skews from erythroid towards megakaryocytic differentiation | [117] | ||
| miR-23 | SHP2 |  | erythroid differentiation | [118] | |
| miR-27a/24 | GATA2 |  | erythroid differentiation | [119] | |
| miR-299-5p | Unknown | Skews from erythroid-monocytic towards megakaryocytic-granulocytic differentiation | [120] | ||
| miR-34a | MYB/CDK4/CDK6 |  | megakaryocytic differentiation and inhibit cell cycle | [121] | |
| miR-376a | CDK2 |  | erythroid differentiation | [122] | |
| miR-451/144 | GATA2 |  | erythroid differentiation | [123,124,125] | |
| miR-486-3p | MAF/BCL11A | Skews from megakaryopoiesis towards erythropoiesis | [102,126] | ||
| miR-146b | PDGFRA |  | erythrocytic-megakaryocytic differentiation | [127] | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martiáñez Canales, T.; De Leeuw, D.C.; Vermue, E.; Ossenkoppele, G.J.; Smit, L. Specific Depletion of Leukemic Stem Cells: Can MicroRNAs Make the Difference? Cancers 2017, 9, 74. https://doi.org/10.3390/cancers9070074
Martiáñez Canales T, De Leeuw DC, Vermue E, Ossenkoppele GJ, Smit L. Specific Depletion of Leukemic Stem Cells: Can MicroRNAs Make the Difference? Cancers. 2017; 9(7):74. https://doi.org/10.3390/cancers9070074
Chicago/Turabian StyleMartiáñez Canales, Tania, David C. De Leeuw, Eline Vermue, Gert J. Ossenkoppele, and Linda Smit. 2017. "Specific Depletion of Leukemic Stem Cells: Can MicroRNAs Make the Difference?" Cancers 9, no. 7: 74. https://doi.org/10.3390/cancers9070074





