Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer
Abstract
:1. Introduction
2. Assessing CIN In Vitro and in Patient Tumor Samples
3. Current and Emerging CIN-Exploiting Therapies
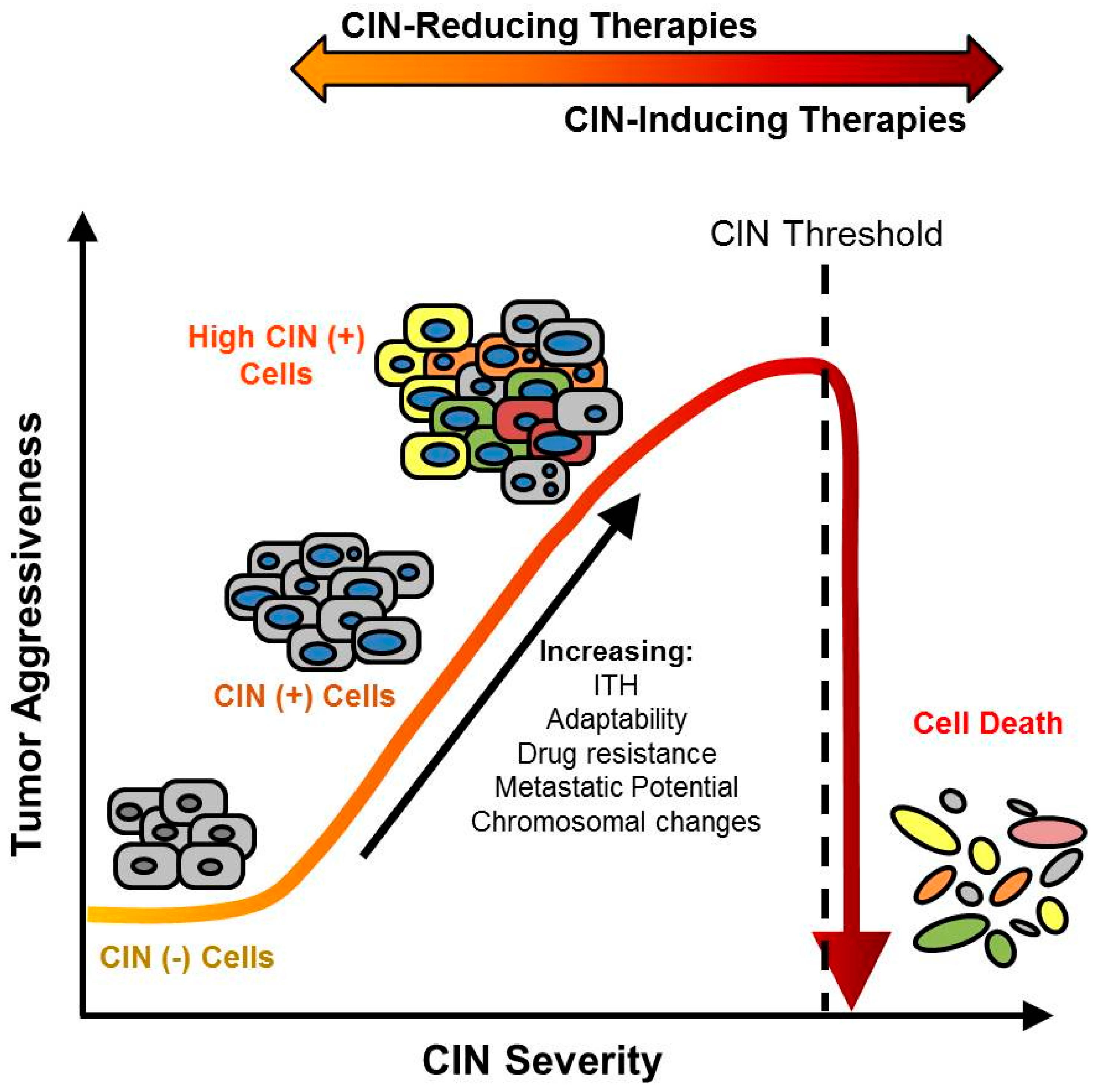
4. CIN Increases ITH and Drives Multidrug Resistance
5. Potential Risks Associated with CIN-Exploiting Therapies
6. Characterizing ITH to Identify Optimal Targets for CIN-Exploiting Therapies
7. Harnessing Synthetic Lethality to Develop Effective and Specific CIN Therapies
8. Combinatorial Chemotherapeutic Strategies May Circumvent ITH and Drug Resistance Stemming from CIN
9. Future Directions and Considerations for Targeting and Exploiting CIN in Cancer
10. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in Globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Galloro, G.; Ruggiero, S.; Russo, T.; Saunders, B. Recent advances to improve the endoscopic detection and differentiation of early colorectal neoplasia. Color. Dis. 2015, 17 (Suppl. 1), 25–30. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Ahn, C.; Symanski, E.; Lai, D.; Du, X.L. Relative impact of earlier diagnosis and improved treatment on survival for colorectal cancer: A us database study among elderly. Cancer Epidemiol. 2014, 38, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Colorectal Cancer Facts & Figures 2017–2019; American Cancer Society: Atlanta, GA, USA, 2017. [Google Scholar]
- Bruin, S.C.; He, Y.; Mikolajewska-Hanclich, I.; Liefers, G.J.; Klijn, C.; Vincent, A.; Verwaal, V.J.; de Groot, K.A.; Morreau, H.; van Velthuysen, M.L.; et al. Molecular alterations associated with liver metastases development in colorectal cancer patients. Br. J. Cancer 2011, 105, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, S.A.; Heflin, M.T. Cancer Screening in Older Adults. Clin. Geriatr. Med. 2016, 32, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Cisyk, A.L.; Penner-Goeke, S.; Lichtensztejn, Z.; Nugent, Z.; Wightman, R.H.; Singh, H.; McManus, K.J. Characterizing the prevalence of chromosome instability in interval colorectal cancer. Neoplasia 2015, 17, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instability in colorectal cancers. Nature 1997, 386, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Compton, D.A. Chromosomal instability and cancer: A complex relationship with therapeutic potential. J. Clin. Investig. 2012, 122, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.L.; Bakhoum, S.F.; Compton, D.A. Mechanisms of chromosomal instability. Curr. Biol. 2010, 20, R285–R295. [Google Scholar] [CrossRef] [PubMed]
- McClelland, S.E. Role of chromosomal instability in cancer progression. Endocr-Relat. Cancer 2017, 24, T23–T31. [Google Scholar] [CrossRef] [PubMed]
- Pierantoni, G.M.; Conte, A.; Rinaldo, C.; Tornincasa, M.; Gerlini, R.; Federico, A.; Valente, D.; Medico, E.; Fusco, A. Deregulation of HMGA1 expression induces chromosome instability through regulation of spindle assembly checkpoint genes. Oncotarget 2015, 6, 17342–17353. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Genovese, G.; Compton, D.A. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 2009, 19, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.; Wang, M.; Liu, X.; Wang, Z.; Xia, P.; Chu, Y.; Wang, X.; Liu, L.; Jiang, K.; Yu, H.; et al. Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic p300/CBP-associated factor (PCAF) orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J. Biol. Chem. 2013, 288, 15771–15785. [Google Scholar] [CrossRef] [PubMed]
- McManus, K.J.; Biron, V.L.; Heit, R.; Underhill, D.A.; Hendzel, M.J. Dynamic changes in histone H3 lysine 9 methylations: Identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J. Biol. Chem. 2006, 281, 8888–8897. [Google Scholar] [CrossRef] [PubMed]
- Barber, T.D.; McManus, K.; Yuen, K.W.; Reis, M.; Parmigiani, G.; Shen, D.; Barrett, I.; Nouhi, Y.; Spencer, F.; Markowitz, S.; et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 3443–3448. [Google Scholar] [CrossRef] [PubMed]
- Asbaghi, Y.; Thompson, L.L.; Lichtensztejn, Z.; McManus, K.J. KIF11 silencing and inhibition induces chromosome instability that may contribute to cancer. Genes Chromosom. Cancer 2017, 56, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Sajesh, B.V.; Lichtensztejn, Z.; McManus, K.J. Sister chromatid cohesion defects are associated with chromosome instability in Hodgkin lymphoma cells. BMC Cancer 2013, 13, 391. [Google Scholar] [CrossRef] [PubMed]
- Ganem, N.J.; Godinho, S.A.; Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009, 460, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Chen, L.; Wu, W.; Garribba, L.; Tian, H.; Liu, Z.; Vogel, I.; Li, C.; Hickson, I.D.; Liu, Y. Potential biomarkers of DNA replication stress in cancer. Oncotarget 2017, 8, 36996–37008. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; van der Burg, M.; Szuhai, K.; Kops, G.J.; Medema, R.H. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011, 333, 1895–1898. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, W.K.; Carson, C.C.; Omolo, B.; Filgo, A.J.; Sambade, M.J.; Simpson, D.A.; Shields, J.M.; Ibrahim, J.G.; Thomas, N.E. Mechanisms of chromosomal instability in melanoma. Environ. Mol. Mutagen. 2014, 55, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.A.; Komarova, N.L.; Sengupta, A.; Jallepalli, P.V.; Shih Ie, M.; Vogelstein, B.; Lengauer, C. The role of chromosomal instability in tumor initiation. Proc. Natl. Acad. Sci. USA 2002, 99, 16226–16231. [Google Scholar] [CrossRef] [PubMed]
- Miura, M.; Miura, Y.; Padilla-Nash, H.M.; Molinolo, A.A.; Fu, B.; Patel, V.; Seo, B.M.; Sonoyama, W.; Zheng, J.J.; Baker, C.C.; et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells 2006, 24, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Su, Y.; Koeman, J.; Haak, E.; Dykema, K.; Essenberg, C.; Hudson, E.; Petillo, D.; Khoo, S.K.; Vande Woude, G.F. Chromosome instability drives phenotypic switching to metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 14793–14798. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; Juul, N.; Johnston, S.R.; Reis-Filho, J.S.; Szallasi, Z.; Swanton, C. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J. Cell. Biochem. 2010, 111, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Endesfelder, D.; Rowan, A.J.; Walther, A.; Birkbak, N.J.; Futreal, P.A.; Downward, J.; Szallasi, Z.; Tomlinson, I.P.; Howell, M.; et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011, 71, 1858–1870. [Google Scholar] [CrossRef] [PubMed]
- Sotillo, R.; Schvartzman, J.M.; Socci, N.D.; Benezra, R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 2010, 464, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.L.; Eklund, A.C.; Kohane, I.S.; Harris, L.N.; Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006, 38, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Jallepalli, P.V.; Lengauer, C. Chromosome segregation and cancer: Cutting through the mystery. Nat. Rev. Cancer 2001, 1, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Roschke, A.V.; Kirsch, I.R. Targeting cancer cells by exploiting karyotypic complexity and chromosomal instability. Cell Cycle 2005, 4, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Geigl, J.B.; Obenauf, A.C.; Schwarzbraun, T.; Speicher, M.R. Defining ‘chromosomal instability’. Trends Genet. 2008, 24, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Gutenberg, A.; Gerdes, J.S.; Jung, K.; Sander, B.; Gunawan, B.; Bock, H.C.; Liersch, T.; Bruck, W.; Rohde, V.; Fuzesi, L. High chromosomal instability in brain metastases of colorectal carcinoma. Cancer Genet. Cytogenet. 2010, 198, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Giam, M.; Rancati, G. Aneuploidy and chromosomal instability in cancer: A jackpot to chaos. Cell Div. 2015, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Eklund, A.C.; Li, Q.; McClelland, S.E.; Endesfelder, D.; Tan, P.; Tan, I.B.; Richardson, A.L.; Szallasi, Z.; Swanton, C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011, 71, 3447–3452. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, A.; Kumar, Y. Relevance of microscopic indicators of chromosomal instability in routine reporting of malignancies. Diagn. Cytopathol. 2014, 42, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.L.; McManus, K.J. A novel multiplexed, image-based approach to detect phenotypes that underlie chromosome instability in human cells. PLoS ONE 2015, 10, e0123200. [Google Scholar] [CrossRef] [PubMed]
- Kronenwett, U.; Huwendiek, S.; Ostring, C.; Portwood, N.; Roblick, U.J.; Pawitan, Y.; Alaiya, A.; Sennerstam, R.; Zetterberg, A.; Auer, G. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004, 64, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Swanton, C.; Nicke, B.; Schuett, M.; Eklund, A.C.; Ng, C.; Li, Q.; Hardcastle, T.; Lee, A.; Roy, R.; East, P.; et al. Chromosomal instability determines taxane response. Proc. Natl. Acad. Sci. USA 2009, 106, 8671–8676. [Google Scholar] [CrossRef] [PubMed]
- Penner-Goeke, S.; Lichtensztejn, Z.; Neufeld, M.; Ali, J.L.; Altman, A.D.; Nachtigal, M.W.; McManus, K.J. The temporal dynamics of chromosome instability in ovarian cancer cell lines and primary patient samples. PLoS Genet. 2017, 13, e1006707. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, H.; Jallepalli, P.V.; Rago, C.; Velculescu, V.E.; Kinzler, K.W.; Vogelstein, B.; Lengauer, C. Inactivation of hCDC4 can cause chromosomal instability. Nature 2004, 428, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.M.; Seo, K.W.; Jang, S.J.; Oh, Y.M.; Shim, T.S.; Kim, W.S.; Lee, D.S.; Lee, S.D. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer 2009, 64, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Hossain, D.; Qian, J.; Adupe, J.; Drewnowska, K.; Bostwick, D.G. Differentiation of melanoma and benign nevi by fluorescence in-situ hybridization. Melanoma Res. 2011, 21, 426–430. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Burrell, R.A.; Endesfelder, D.; Novelli, M.R.; Swanton, C. Cancer chromosomal instability: Therapeutic and diagnostic challenges. EMBO Rep. 2012, 13, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Saji, H.; Idiris, A.; Kawasaki, N.; Hosaka, M.; Ogata, A.; Saijo, T.; Kato, H. Chromosomal instability detected by fluorescence in situ hybridization in surgical specimens of non-small cell lung cancer is associated with poor survival. Clin. Cancer Res. 2003, 9, 2294–2299. [Google Scholar] [CrossRef]
- Stirling, P.C.; Bloom, M.S.; Solanki-Patil, T.; Smith, S.; Sipahimalani, P.; Li, Z.; Kofoed, M.; Ben-Aroya, S.; Myung, K.; Hieter, P. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 2011, 7, e1002057. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Bester, A.C.; Roniger, M.; Oren, Y.S.; Im, M.M.; Sarni, D.; Chaoat, M.; Bensimon, A.; Zamir, G.; Shewach, D.S.; Kerem, B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011, 145, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.L.; Yazinski, S.A.; Nicolay, B.; Bryll, A.; Zou, L.; Dyson, N.J. Suppression of genome instability in pRB-deficient cells by enhancement of chromosome cohesion. Mol. Cell 2014, 53, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Orr, B.; Talje, L.; Liu, Z.; Kwok, B.H.; Compton, D.A. Adaptive Resistance to an Inhibitor of Chromosomal Instability in Human Cancer Cells. Cell Rep. 2016, 17, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Ertych, N.; Stolz, A.; Stenzinger, A.; Weichert, W.; Kaulfuss, S.; Burfeind, P.; Aigner, A.; Wordeman, L.; Bastians, H. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat. Cell Biol. 2014, 16, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Zasadil, L.M.; Andersen, K.A.; Yeum, D.; Rocque, G.B.; Wilke, L.G.; Tevaarwerk, A.J.; Raines, R.T.; Burkard, M.E.; Weaver, B.A. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl. Med. 2014, 6, 229ra243. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A. Taxanes for breast cancer: An evidence-based review of randomized phase II and phase III trials. Clin. Breast Cancer 2000, 1, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Wan, X.; Hirel, C.B.; Salmon, E.D. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 2006, 16, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; A’Hern, R.; Birkbak, N.J.; Gorman, P.; Gronroos, E.; Ngang, S.; Nicola, P.; Rahman, L.; Thanopoulou, E.; Kelly, G.; et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: A prospective validation cohort study from the TACT trial. Ann. Oncol. 2015, 26, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Roylance, R.; Endesfelder, D.; Gorman, P.; Burrell, R.A.; Sander, J.; Tomlinson, I.; Hanby, A.M.; Speirs, V.; Richardson, A.L.; Birkbak, N.J.; et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol. Biomark. Prev. 2011, 20, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Fanale, D.; Bronte, G.; Passiglia, F.; Calo, V.; Castiglia, M.; Di Piazza, F.; Barraco, N.; Cangemi, A.; Catarella, M.T.; Insalaco, L.; et al. Stabilizing versus destabilizing the microtubules: A double-edge sword for an effective cancer treatment option? Anal. Cell. Pathol. 2015, 2015, 690916. [Google Scholar] [CrossRef] [PubMed]
- U.S. National Institutes of Health. Clinicaltrials.Gov. Available online: https://prsinfo.clinicaltrials.gov (accessed on 31 October 2017).
- Jordan, M.A.; Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Colombo, R.; Caldarelli, M.; Mennecozzi, M.; Giorgini, M.L.; Sola, F.; Cappella, P.; Perrera, C.; Depaolini, S.R.; Rusconi, L.; Cucchi, U.; et al. Targeting the mitotic checkpoint for cancer therapy with NMS-P715, an inhibitor of MPS1 kinase. Cancer Res. 2010, 70, 10255–10264. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.H.; Hu, L.Y.; Xu, D.Q.; Li, X. A cell-based assay for screening spindle checkpoint inhibitors. Assay Drug Dev. Technol. 2012, 10, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Wengner, A.M.; Siemeister, G.; Koppitz, M.; Schulze, V.; Kosemund, D.; Klar, U.; Stoeckigt, D.; Neuhaus, R.; Lienau, P.; Bader, B.; et al. Novel Mps1 Kinase Inhibitors with Potent Antitumor Activity. Mol. Cancer Ther. 2016, 15, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Wei, X.; Fletcher, G.C.; Kiarash, R.; Brokx, R.; Hodgson, R.; Beletskaya, I.; Bray, M.R.; Mak, T.W. Functional characterization of CFI-402257, a potent and selective Mps1/TTK kinase inhibitor, for the treatment of cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3127–3132. [Google Scholar] [CrossRef] [PubMed]
- Sansregret, L.; Patterson, J.O.; Dewhurst, S.; Lopez-Garcia, C.; Koch, A.; McGranahan, N.; Chao, W.C.H.; Barry, D.J.; Rowan, A.; Instrell, R.; et al. APC/C Dysfunction Limits Excessive Cancer Chromosomal Instability. Cancer Discov. 2017, 7, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chang, L.; Alfieri, C.; Zhang, Z.; Yang, J.; Maslen, S.; Skehel, M.; Barford, D. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 2016, 533, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Sackton, K.L.; Dimova, N.; Zeng, X.; Tian, W.; Zhang, M.; Sackton, T.B.; Meaders, J.; Pfaff, K.L.; Sigoillot, F.; Yu, H.; et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 2014, 514, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Sigoillot, F.; Gaur, S.; Choi, S.; Pfaff, K.L.; Oh, D.-C.; Hathaway, N.; Dimova, N.; Cuny, G.D.; King, R.W. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 2010, 18, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Sen, S. Aurora Kinase inhibitors as Anticancer Molecules. Biochim. Biophys. Acta 2010, 1799, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Gollapudi, P.; Hasegawa, L.S.; Eastmond, D.A. A Comparative Study of the Aneugenic and Polyploidy-inducing Effects of Fisetin and Two Model Aurora Kinase Inhibitors. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2014, 767, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kollareddy, M.; Zheleva, D.; Dzubak, P.; Brahmkshatriya, P.S.; Lepsik, M.; Hajduch, M. Aurora kinase inhibitors: Progress towards the clinic. Investig. New Drugs 2012, 30, 2411–2432. [Google Scholar] [CrossRef] [PubMed]
- Diamond, J.R.; Bastos, B.R.; Hansen, R.J.; Gustafson, D.L.; Eckhardt, S.G.; Kwak, E.L.; Pandya, S.S.; Fletcher, G.C.; Pitts, T.M.; Kulikowski, G.N.; et al. Phase I Safety, Pharmacokinetic, and Pharmacodynamic Study of ENMD-2076, a Novel Angiogenic and Aurora Kinase Inhibitor, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2011, 17, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Bavetsias, V.; Linardopoulos, S. Aurora Kinase Inhibitors: Current Status and Outlook. Front. Oncol. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Woolery, J.E.; Mahadevan, D. Update on Aurora Kinase Targeted Therapeutics in Oncology. Expert Opin. Drug Discov. 2011, 6, 291–307. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.-Y.; Yu, X. The balance of Polo-like kinase 1 in tumorigenesis. Cell Div. 2009, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Burke, T.R.; Park, J.-E.; Bang, J.K.; Lee, E. Recent advances and new strategies in targeting Plk1 for anticancer therapy. Trends Pharmacol. Sci. 2015, 36, 858–877. [Google Scholar] [CrossRef] [PubMed]
- Schoffski, P. Polo-like kinase (PLK) inhibitors in preclinical and early clinical development in oncology. Oncologist 2009, 14, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; South, V.J.; Zhang, Y.; Davide, J.P.; Farrell, L.; Kohl, N.E.; Sepp-Lorenzino, L.; Lobell, R.B. Induction of apoptosis by an inhibitor of the mitotic kinesin KSP requires both activation of the spindle assembly checkpoint and mitotic slippage. Cancer Cell 2005, 8, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Woessner, R.; Tunquist, B.; Lemieux, C.; Chlipala, E.; Jackinsky, S.; Dewolf, W.J.; Voegtli, W.; Cox, A.; Rana, S.; Lee, P.; et al. ARRY-520, a novel KSP inhibitor with potent activity in hematological and taxane-resistant tumor models. Anticancer Res. 2009, 29, 4373–4380. [Google Scholar] [PubMed]
- Holen, K.; DiPaola, R.; Liu, G.; Tan, A.R.; Wilding, G.; Hsu, K.; Agrawal, N.; Chen, C.; Xue, L.; Rosenberg, E.; et al. A phase I trial of MK-0731, a Kinesin Spindle Protein (KSP) inhibitor, in patients with solid tumors. Investig. New Drugs 2012, 30, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.-X.; Yang, W.-X. KIFC1: A promising chemotherapy target for cancer treatment? Oncotarget 2016, 7, 48656–48670. [Google Scholar] [CrossRef] [PubMed]
- Bhakta-Guha, D.; Saeed, M.E.M.; Greten, H.J.; Efferth, T. Dis-organizing centrosomal clusters: Specific cancer therapy for a generic spread? Curr. Med. Chem. 2015, 22, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Watts, C.A.; Richards, F.M.; Bender, A.; Bond, P.J.; Korb, O.; Kern, O.; Riddick, M.; Owen, P.; Myers, R.M.; Raff, J.; et al. Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chem. Biol. 2013, 20, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, W.; Chen, D.; Boohaker, R.J.; Zhai, L.; Padmalayam, I.; Wennerberg, K.; Xu, B.; Zhang, W. KIFC1 is a novel potential therapeutic target for breast cancer. Cancer Biol. Ther. 2015, 16, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.; Bechi, B.; Tighe, A.; Thompson, S.; Procter, D.J.; Taylor, S.S. Cenp-E inhibitor GSK923295: Novel synthetic route and use as a tool to generate aneuploidy. Oncotarget 2015, 6, 20921–20932. [Google Scholar] [CrossRef] [PubMed]
- Kung, P.-P.; Martinez, R.; Zhu, Z.; Zager, M.; Blasina, A.; Rymer, I.; Hallin, J.; Xu, M.; Carroll, C.; Chionis, J.; et al. Chemogenetic evaluation of the mitotic kinesin CENP-E reveals a critical role in triple-negative breast cancer. Mol. Cancer Ther. 2014, 13, 2104–2115. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, E.; Fielding, A.B.; Kannan, N.; Balgi, A.; Eaves, C.J.; Roberge, M.; Dedhar, S. Identification of novel small molecule inhibitors of centrosome clustering in cancer cells. Oncotarget 2013, 4, 1763–1776. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.-S.; Perez, G.; Ngo, L.; Gui, C.-Y.; Marks, P.A. Induction of polyploidy by histone deacetylase inhibitor: A pathway for antitumor effects. Cancer Res. 2005, 65, 7832–7839. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-J.; Baek, K.-H.; Jeon, A.-H.; Kim, S.-J.; Jang, K.-L.; Sung, Y.-C.; Kim, C.-M.; Lee, C.-W. Inhibition of histone deacetylase activity increases chromosomal instability by the aberrant regulation of mitotic checkpoint activation. Oncogene 2003, 22, 3853–3858. [Google Scholar] [CrossRef] [PubMed]
- Bieliauskas, A.V.; Pflum, M.K.H. Isoform-selective histone deacetylase inhibitors. Chem. Soc. Rev. 2008, 37, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Mottamal, M.; Zheng, S.; Huang, T.L.; Wang, G. Histone Deacetylase Inhibitors in Clinical Studies as Templates for New Anticancer Agents. Molecules 2015, 20, 3898–3941. [Google Scholar] [CrossRef] [PubMed]
- Richon, V.M. Cancer biology: Mechanism of antitumour action of vorinostat (suberoylanilide hydroxamic acid), a novel histone deacetylase inhibitor. Br. J. Cancer 2006, 95, S2–S6. [Google Scholar] [CrossRef]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Boss, D.S.; Witteveen, P.O.; van der Sar, J.; Lolkema, M.P.; Voest, E.E.; Stockman, P.K.; Ataman, O.; Wilson, D.; Das, S.; Schellens, J.H. Clinical evaluation of AZD1152, an i.v. inhibitor of Aurora B kinase, in patients with solid malignant tumors. Ann. Oncol. 2011, 22, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Fox, D.T.; Duronio, R.J. Endoreplication and polyploidy: Insights into development and disease. Development 2013, 140, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, R.W.; Odedra, R.; Heaton, S.P.; Wedge, S.R.; Keen, N.J.; Crafter, C.; Foster, J.R.; Brady, M.C.; Bigley, A.; Brown, E.; et al. AZD1152, a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis. Clin. Cancer Res. 2007, 13, 3682–3688. [Google Scholar] [CrossRef] [PubMed]
- Falchook, G.S.; Bastida, C.C.; Kurzrock, R. Aurora Kinase Inhibitors in Oncology Clinical Trials: Current State of the Progress. Semin. Oncol. 2015, 42, 832–848. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, L.C.; Peterman, E.J.; Kwok, B.H.; Kim, J.H.; Kapoor, T.M.; Schmidt, C.F. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 2005, 435, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, G.; Tatrai, P.; Gergely, F. Hitting the brakes: Targeting microtubule motors in cancer. Br. J. Cancer 2015, 113, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Burris, H.A., III; Jones, S.F.; Williams, D.D.; Kathman, S.J.; Hodge, J.P.; Pandite, L.; Ho, P.T.; Boerner, S.A.; Lorusso, P. A phase I study of ispinesib, a kinesin spindle protein inhibitor, administered weekly for three consecutive weeks of a 28-day cycle in patients with solid tumors. Investig. New Drugs 2011, 29, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Drosopoulos, K.; Tang, C.; Chao, W.C.H.; Linardopoulos, S. APC/C is an essential regulator of centrosome clustering. Nat. Commun. 2014, 5, 3686. [Google Scholar] [CrossRef] [PubMed]
- Pannu, V.; Rida, P.C.G.; Celik, B.; Turaga, R.C.; Ogden, A.; Cantuaria, G.; Gopalakrishnan, J.; Aneja, R. Centrosome-declustering drugs mediate a two-pronged attack on interphase and mitosis in supercentrosomal cancer cells. Cell Death Dis. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Chavali, P.L.; Chandrasekaran, G.; Barr, A.R.; Tatrai, P.; Taylor, C.; Papachristou, E.K.; Woods, C.G.; Chavali, S.; Gergely, F. A CEP215-HSET complex links centrosomes with spindle poles and drives centrosome clustering in cancer. Nat. Commun. 2016, 7, 11005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef] [PubMed]
- McCabe, N.; Turner, N.C.; Lord, C.J.; Kluzek, K.; Bialkowska, A.; Swift, S.; Giavara, S.; O’Connor, M.J.; Tutt, A.N.; Zdzienicka, M.Z.; et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006, 66, 8109–8115. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Ison, G.; McKee, A.E.; Zhang, H.; Tang, S.; Gwise, T.; Sridhara, R.; Lee, E.; Tzou, A.; Philip, R.; et al. FDA Approval Summary: Olaparib Monotherapy in Patients with Deleterious Germline BRCA-Mutated Advanced Ovarian Cancer Treated with Three or More Lines of Chemotherapy. Clin. Cancer Res. 2015, 21, 4257–4261. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, B.; Shapira-Frommer, R.; Schmutzler, R.K.; Audeh, M.W.; Friedlander, M.; Balmana, J.; Mitchell, G.; Fried, G.; Stemmer, S.M.; Hubert, A.; et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015, 33, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrell, R.A.; Swanton, C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol. Oncol. 2014, 8, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Bozic, I.; Reiter, J.G.; Allen, B.; Antal, T.; Chatterjee, K.; Shah, P.; Moon, Y.S.; Yaqubie, A.; Kelly, N.; Le, D.T.; et al. Evolutionary dynamics of cancer in response to targeted combination therapy. eLife 2013, 2, e00747. [Google Scholar] [CrossRef] [PubMed]
- Waclaw, B.; Bozic, I.; Pittman, M.E.; Hruban, R.H.; Vogelstein, B.; Nowak, M.A. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 2015, 525, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.L.; Brough, R.; Lord, C.J.; Natrajan, R.; Vatcheva, R.; Levine, D.A.; Boyd, J.; Reis-Filho, J.S.; Ashworth, A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 2008, 451, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Carlino, M.S.; Todd, J.R.; Gowrishankar, K.; Mijatov, B.; Pupo, G.M.; Fung, C.; Snoyman, S.; Hersey, P.; Long, G.V.; Kefford, R.F.; et al. Differential activity of MEK and ERK inhibitors in BRAF inhibitor resistant melanoma. Mol. Oncol. 2014, 8, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Harismendy, O.; Schwab, R.B.; Alakus, H.; Yost, S.E.; Matsui, H.; Hasteh, F.; Wallace, A.M.; Park, H.L.; Madlensky, L.; Parker, B.; et al. Evaluation of ultra-deep targeted sequencing for personalized breast cancer care. Breast Cancer Res. 2013, 15, R115. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Horswell, S.; Larkin, J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N.; Santos, C.R.; et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014, 46, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Voss, M.H.; Hakimi, A.A.; Pham, C.G.; Brannon, A.R.; Chen, Y.B.; Cunha, L.F.; Akin, O.; Liu, H.; Takeda, S.; Scott, S.N.; et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin. Cancer Res. 2014, 20, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Bourgo, R.J.; Siddiqui, H.; Fox, S.; Solomon, D.; Sansam, C.G.; Yaniv, M.; Muchardt, C.; Metzger, D.; Chambon, P.; Roberts, C.W.; et al. SWI/SNF deficiency results in aberrant chromatin organization, mitotic failure, and diminished proliferative capacity. Mol. Biol. Cell 2009, 20, 3192–3199. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, P.M.; Chambers, A.L.; Cloney, R.; Bianchi, A.; Downs, J.A. BAF180 promotes cohesion and prevents genome instability and aneuploidy. Cell Rep. 2014, 6, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, S.R.; McGregor, G.A.; Murray, J.M.; Downs, J.A.; Savic, V. Novel synthetic lethality screening method identifies TIP60-dependent radiation sensitivity in the absence of BAF180. DNA Repair 2016, 46, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 2005, 5, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Sajesh, B.V.; Cisyk, A.L.; McManus, K.J. Synthetic Genetic Approaches in Colorectal Cancer: Exploiting and Targeting Genome Instability. In Genomic Instability and Cancer Metastasis: Mechanisms, Emerging Themes, and Novel Therapeutic Strategies; Maxwell, C., Roskelley, C., Eds.; Springer International Publishing: Cham, Germany, 2015; pp. 179–204. [Google Scholar]
- Tong, A.H.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Page, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.; Bussey, H.; et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.M.; Templeton, S.D.; Baharani, A.; Freywald, A.; Vizeacoumar, F.J. Building high-resolution synthetic lethal networks: A ‘Google map’ of the cancer cell. Trends Mol. Med. 2014, 20, 704–715. [Google Scholar] [CrossRef] [PubMed]
- McManus, K.J.; Barrett, I.J.; Nouhi, Y.; Hieter, P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc. Natl. Acad. Sci. USA 2009, 106, 3276–3281. [Google Scholar] [CrossRef] [PubMed]
- Sajesh, B.V.; Bailey, M.; Lichtensztejn, Z.; Hieter, P.; McManus, K.J. Synthetic lethal targeting of superoxide dismutase 1 selectively kills RAD54B-deficient colorectal cancer cells. Genetics 2013, 195, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Van Pel, D.M.; Barrett, I.J.; Shimizu, Y.; Sajesh, B.V.; Guppy, B.J.; Pfeifer, T.; McManus, K.J.; Hieter, P. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013, 9, e1003254. [Google Scholar] [CrossRef]
- Guppy, B.J.; McManus, K.J. Synthetic lethal targeting of RNF20 through PARP1 silencing and inhibition. Cell. Oncol. 2017, 40, 281–292. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, E.N.; Lepage, C.C.; McManus, K.J. The synthetic lethal killing of RAD54B-deficient colorectal cancer cells by PARP1 inhibition is enhanced with SOD1 inhibition. Oncotarget 2016, 7, 87417–87430. [Google Scholar] [CrossRef] [PubMed]
- Bajrami, I.; Frankum, J.R.; Konde, A.; Miller, R.E.; Rehman, F.L.; Brough, R.; Campbell, J.; Sims, D.; Rafiq, R.; Hooper, S.; et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014, 74, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; McDonald, S.; Swift, S.; Turner, N.C.; Ashworth, A. A high-throughput RNA interference screen for DNA repair determinants of PARP inhibitor sensitivity. DNA Repair 2008, 7, 2010–2019. [Google Scholar] [CrossRef] [PubMed]
- Min, A.; Im, S.A.; Yoon, Y.K.; Song, S.H.; Nam, H.J.; Hur, H.S.; Kim, H.P.; Lee, K.H.; Han, S.W.; Oh, D.Y.; et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol. Cancer Ther. 2013, 12, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Koppensteiner, R.; Samartzis, E.P.; Noske, A.; von Teichman, A.; Dedes, I.; Gwerder, M.; Imesch, P.; Ikenberg, K.; Moch, H.; Fink, D.; et al. Effect of MRE11 loss on PARP-inhibitor sensitivity in endometrial cancer in vitro. PLoS ONE 2014, 9, e100041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, N.C.; Lord, C.J.; Iorns, E.; Brough, R.; Swift, S.; Elliott, R.; Rayter, S.; Tutt, A.N.; Ashworth, A. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 2008, 27, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.E.; Li, S.; Vizeacoumar, F.S.; Bhanumathy, K.K.; Lee, J.S.; Parameswaran, S.; Furber, L.; Abuhussein, O.; Paul, J.M.; McDonald, M.; et al. Therapeutic relevance of the protein phosphatase 2A in cancer. Oncotarget 2016, 7, 61544–61561. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Fam, H.K.; Wang, Y.K.; Styles, E.B.; Kim, J.H.; Ang, J.S.; Singh, T.; Larionov, V.; Shah, S.P.; Andrews, B.; et al. Overexpression screens identify conserved dosage chromosome instability genes in yeast and human cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 9967–9976. [Google Scholar] [CrossRef] [PubMed]
- Sajesh, B.V.; McManus, K.J. Targeting SOD1 induces synthetic lethal killing in BLM- and CHEK2-deficient colorectal cancer cells. Oncotarget 2015, 6, 27907–27922. [Google Scholar] [CrossRef] [PubMed]
- Al-Lazikani, B.; Banerji, U.; Workman, P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012, 30, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.G.; Makohon-Moore, A.P.; Gerold, J.M.; Bozic, I.; Chatterjee, K.; Iacobuzio-Donahue, C.A.; Vogelstein, B.; Nowak, M.A. Reconstructing metastatic seeding patterns of human cancers. Nat. Commun. 2017, 8, 14114. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Wagle, N.; Emery, C.; Berger, M.F.; Davis, M.J.; Sawyer, A.; Pochanard, P.; Kehoe, S.M.; Johannessen, C.M.; Macconaill, L.E.; Hahn, W.C.; et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011, 29, 3085–3096. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Ascierto, P.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandala, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
- Moriceau, G.; Hugo, W.; Hong, A.; Shi, H.; Kong, X.; Yu, C.C.; Koya, R.C.; Samatar, A.A.; Khanlou, N.; Braun, J.; et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015, 27, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Fung, C.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Hyman, J.; Shahheydari, H.; Tembe, V.; Thompson, J.F.; Saw, R.P.; et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014, 5, 5694. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; Kops, G.J.P.L.; Medema, R.H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. USA 2009, 106, 19108–19113. [Google Scholar] [CrossRef] [PubMed]
- Jemaa, M.; Galluzzi, L.; Kepp, O.; Senovilla, L.; Brands, M.; Boemer, U.; Koppitz, M.; Lienau, P.; Prechtl, S.; Schulze, V.; et al. Characterization of novel MPS1 inhibitors with preclinical anticancer activity. Cell Death Differ. 2013, 20, 1532–1545. [Google Scholar] [CrossRef] [PubMed]
- Faisal, A.; Mak, G.W.Y.; Gurden, M.D.; Xavier, C.P.R.; Anderhub, S.J.; Innocenti, P.; Westwood, I.M.; Naud, S.; Hayes, A.; Box, G.; et al. Characterisation of CCT271850, a selective, oral and potent MPS1 inhibitor, used to directly measure in vivo MPS1 inhibition vs therapeutic efficacy. Br. J. Cancer 2017, 116, 1166–1176. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.T.; Erdal, S.; Huang, S.; Poon, R.Y.C. Synergism between inhibitors of Aurora A and KIF11 overcomes KIF15-dependent drug resistance. Mol. Oncol. 2014, 8, 1404–1418. [Google Scholar] [CrossRef] [PubMed]
- Cokol, M.; Chua, H.N.; Tasan, M.; Mutlu, B.; Weinstein, Z.B.; Suzuki, Y.; Nergiz, M.E.; Costanzo, M.; Baryshnikova, A.; Giaever, G.; et al. Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 2011, 7, 544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dy, G.K.; Adjei, A.A. Understanding, recognizing, and managing toxicities of targeted anticancer therapies. CA Cancer J. Clin. 2013, 63, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Karanam, V.; Marslin, G.; Krishnamoorthy, B.; Chellan, V.; Siram, K.; Natarajan, T.; Bhaskar, B.; Franklin, G. Poly (varepsilon-caprolactone) nanoparticles of carboplatin: Preparation, characterization and in vitro cytotoxicity evaluation in U-87 MG cell lines. Colloids Surf. B Biointerfaces 2015, 130, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Du Bois, A.; Luck, H.-J.; Meier, W.; Adams, H.-P.; Mobus, V.; Costa, S.; Bauknecht, T.; Richter, B.; Warm, M.; Schroder, W.; et al. A randomized clinical trial of cisplatin/paclitaxel versus carboplatin/paclitaxel as first-line treatment of ovarian cancer. J. Natl. Cancer Inst. 2003, 95, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- McGuire, W.P.; Hoskins, W.J.; Brady, M.F.; Kucera, P.R.; Partridge, E.E.; Look, K.Y.; Clarke-Pearson, D.L.; Davidson, M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N. Engl. J. Med. 1996, 334, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Monk, B.J.; Sood, A.K.; Herzog, T.J. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat. Rev. Clin. Oncol. 2013, 10, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, Z.; Liu, D.; Choo, A.; Hussain, R.; O’Keefe, L.; Richards, R.; Saint, R.; Gregory, S.L. Chromosomal instability causes sensitivity to metabolic stress. Oncogene 2015, 34, 4044–4055. [Google Scholar] [CrossRef] [PubMed]
| Principle | Advantage | Limitation | |
|---|---|---|---|
| Indirect Indicators of Chromosome Instability (CIN) | |||
| Gene expression signatures | The genes of which the expression is most highly correlated with high levels of aneuploidy were selected to define the CIN70 expression signatures [31]. The signature was confirmed to correlate with structural and numerical CIN [37]. | Applicable to published gene expression datasets for research purposes. Could be used in routine clinical practice with fresh or fixed patient samples. | No direct measurement of the level of CIN. |
| Array comparative genome hybridization (CGH) | The genome to be tested and a reference genome are labeled with distinct fluorescent probes and competitively hybridized to arrayed DNA sequences. The fluorescent signal indicates whether the test genome harbors a gain or loss of material at a given locus. | Detailed resolution of the recurrent copy number alterations present in the tumor. | No direct measurement of the level of CIN. Cannot distinguish between CIN and stable aneuploidy. |
| Nuclear area heterogeneity | Nuclear area is correlated with DNA content. Nuclear area heterogeneity is suggestive of underlying DNA content differences [38,39]. | Observable in routine pathology reports. | Follow-up assessment with single-cell method is necessary to confirm nuclear area heterogeneity is due to CIN. |
| Micronucleus formation | Micronuclei contain missegregated chromosomes or large chromosomal fragments. An increase in micronucleus formation is indicative of DNA content changes/structural DNA damage [34,38,39]. | Observable in routine pathology reports. | Follow-up assessment with single-cell method is necessary to confirm increased micronucleus formation is due to CIN. |
| Direct Measurements of CIN | |||
| DNA image cytometry | Nuclei are extracted from the test samples and DNA is stained with the Feulgen method. Nuclei are microscopically imaged and optical density is recorded for each nucleus to calculate DNA content. Cell-to-cell variability is assessed with the Stemline Scatter Index (SSI), which is equal to the sum of the percentage of cells in the S-phase region, the percentage of cell with DNA content exceeding G2 and the coefficient of variation of the DNA content stemline [40,41]. | Higher throughput than FISH. Applicable to formalin fixed, parafin embedded (FFPE) tumor samples. | No distinction between numerical and structural CIN. |
| Fluorescence in situ Hybridization (FISH) | Fluorescent probes detect centromere copy numbers to assess gain or loss of chromosomes in individual cells [8,10,42,43]. Alternatively, probes binding to chromosomal arms can be employed to assess segmental aneuploidy and structural CIN [34]. | Accurate measurement of cell-to-cell heterogeneity. Hundreds of cells evaluated at a time. Applicable to FFPE tumor samples. | Labor intensive. Microscope capacity generally limits the analysis to 3–4 probes at a time. Cannot measure structural and numerical CIN at the same time. |
| Single-cell CGH | CGH is performed to analyze the DNA of individual tumor cells after amplification of their genome. CIN level in the tumor samples is inferred from degree of cell-to-cell heterogeneity. | High resolution of copy number alterations present in individual cells. | Lower resolution of copy number alterations than single-cell sequencing. Technology still under development, not yet reliably applicable in clinical setting. |
| Single-cell sequencing | New generation sequencing technology is applied to isolated single tumor cells after amplification of their genome. Copy number variations can be assessed across the whole genome. CIN level is inferred from degree of cell-to-cell heterogeneity. | Detailed resolution of copy number alterations Base-pair resolution of mutations. | Technology still under development, not yet reliably applicable in clinical setting. |
| Drug Family | Mechanism of Action | Effect on CIN | Drug Examples | Molecular Target | Clinical Trials | Clinicaltrials.gov Identifier [60] |
|---|---|---|---|---|---|---|
| Microtubule Dynamics | ||||||
| Microtubule stabilizers (taxanes, epothilones) | Bind tubulin subunits to inhibit microtubule depolymerization | I [54,61] | Paclitaxel | β-tubulin | FDA approved (breast, ovarian, non-small cell lung cancer, Kaposi sarcoma) | FDA approved |
| Docetaxel | β-tubulin | FDA approved (breast, prostate, gastric, head & neck, non-small cell lung cancer) | FDA approved | |||
| Ixabepilone | β-tubulin | FDA approved (breast) | FDA approved | |||
| Microtubule destabilizers (vinca alkaloids, colchicine analogs) | Inhibit microtubule polymerization and induce mitotic arrest | U | Vincristine | β-tubulin | FDA approved (leukemia) | FDA approved |
| Vinblastine | β-tubulin | FDA approved (breast, testicular, Hodgkin lymphoma, non-Hodgkin lymphoma, Kaposi sarcoma) | FDA approved | |||
| Vinorelbine | β-tubulin | FDA approved (non-small cell lung cancer) | FDA approved | |||
| Mitotic Checkpoints | ||||||
| Spindle assembly checkpoint inhibitors | Induce premature mitotic exit and chromosome missegregation | I [62,63] | BAY1217389 | MPS1 [64] | Phase I | NCT02366949 |
| BAY1161909 | MPS1 [64] | Phase I | NCT02138812 | |||
| CFI-402257 | MPS1 [65] | Phase I | NCT02792465 | |||
| Anaphase- promoting complex/cyclosome (APC/C) inhibitors | Inhibit mitotic exit and induce metaphase arrest | R [66] | Tosyl-L-arginine methyl ester (TAME) | APC/C [67] | Preclinical [68,69] | Preclinical |
| Mitotic Kinases | ||||||
| Aurora kinase inhibitors | Interfere with mitotic chromosome alignment, spindle assembly, and cytokinesis | I [70,71] | ENMD-2076 | Aurora Kinase A [72,73] | Phase II | NCT01104675 NCT01639248 |
| Alisertib (MLN8237) | Aurora Kinase A [74] | Phase I/II | NCT02187991 NCT01923337 | |||
| Barasertib (AZD1152) | Aurora Kinase B [74] | Phase II/III | NCT00952588 | |||
| GSK1070916 | Aurora Kinase B/C [75] | Phase I | NCT01118611 | |||
| Polo-like kinase inhibitors | Inhibit bipolar spindle formation, sister chromatid separation, and cytokinesis | I [76,77,78] | Volasertib (BI 6727) | PLK1 [78] | Phase I/II | NCT02273388 NCT01121406 |
| BI 2536 | PLK1 [78] | Phase II | NCT00706498 NCT00710710 | |||
| Rigosertib (ON 01910.Na) | PLK1 [78] | Phase I/II | NCT01168011 NCT01807546 | |||
| Microtubule-associated Motor Proteins | ||||||
| KIF11 (Eg5) inhibitors | Interfere with centrosome separation and cause monopolar spindle formation | I [19] or R [79] | Filanesib (ARRY-520) | KIF11 [80] | Phase I/II | NCT00821249 |
| MK0731 | KIF11 [81] | Phase I | NCT00104364 | |||
| KIFC1 (HSET) inhibitors | Inhibit centrosomal clustering activity of KIFC1, resulting in multipolar spindle formation | I [82] | CW069 | KIFC1 [82] | Preclinical [83,84] | Preclinical |
| PJ34 | KIFC1 [82] | Preclinical [85] | Preclinical | |||
| CENP-E inhibitors | Inhibit CENP-E mediated chromosomal alignment in metaphase | I [86] | GSK923295 | CENP-E [86] | Phase I | NCT00504790 |
| PF-2771 | CENP-E [87] | Preclinical | Preclinical | |||
| KIF2C potentiators | Enhance KIF2C activity and destabilize kinetochore-microtubule attachments, leading to a reduction in erroneous attachments | R [52] | UMK57 | Uncharacterized | Preclinical | Preclinical |
| Centrosome Dynamics | ||||||
| Centrosomal clustering inhibitors | Inhibit supernumerary centrosomal clustering, leading to multipolar spindle formation | I [88] | CCCI01 | Uncharacterized | Preclinical [88] | Preclinical |
| Chromatin Modification | ||||||
| Histone deacetylase (HDAC) inhibitors | Accumulation of acetylated histones disrupts centromere function and causes mitotic abnormalities | I [89,90] | Romidepsin | HDAC1/2 [91] | FDA approved (cutaneous T-cell lymphoma) | FDA approved |
| Entinostat | Class I HDACs [92] | Phase I/II | NCT01105377 NCT00020579 | |||
| Vorinostat | Class I/II HDACs [93] | Phase I/II | NCT01045538 NCT00365599 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, L.L.; Jeusset, L.M.-P.; Lepage, C.C.; McManus, K.J. Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer. Cancers 2017, 9, 151. https://doi.org/10.3390/cancers9110151
Thompson LL, Jeusset LM-P, Lepage CC, McManus KJ. Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer. Cancers. 2017; 9(11):151. https://doi.org/10.3390/cancers9110151
Chicago/Turabian StyleThompson, Laura L., Lucile M-P. Jeusset, Chloe C. Lepage, and Kirk J. McManus. 2017. "Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer" Cancers 9, no. 11: 151. https://doi.org/10.3390/cancers9110151





