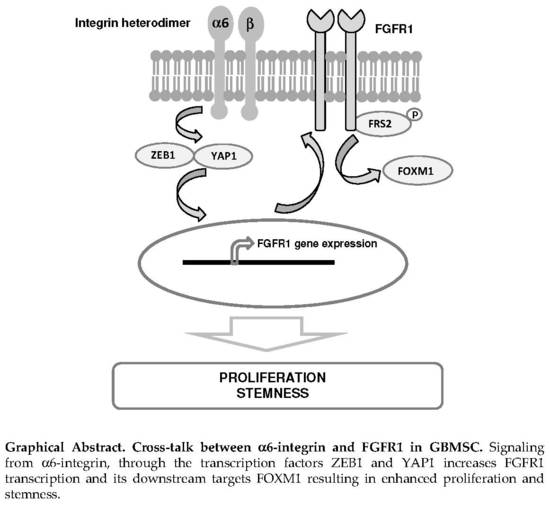
Alpha6-Integrin Regulates FGFR1 Expression through the ZEB1/YAP1 Transcription Complex in Glioblastoma Stem Cells Resulting in Enhanced Proliferation and Stemness
Abstract
:1. Introduction
2. Results
2.1. α6-Integrin Regulates FGFR1 Signaling
2.2. α6-Integrin and FGFR1 Cooperate in GBMSC to Regulate Cell Cycle and Proliferation
2.3. α6-Integrin Regulates FGFR1 Expression Via the Transcription Factors ZEB1 and YAP1
2.4. α6-Integrin and FGFR1 Cooperate in GBMSC to Regulate Neurospheres Formation and Stemness
2.5. α6-Integrin Expression Correlates with the Target Genes ZEB1, YAP1, FGFR1, and FOXM1
2.6. High Expression of the Five Genes Signature: α6-Integrin/ZEB1/YAP1/FGFR1/FOXM1 Is Prognostic of the Overall Survival of GBM Patients
3. Discussion
4. Materials and Methods
4.1. GBM Patient-Derived Cells
4.2. SiRNA Transfection, RNA Extraction, Reverse Transcription, and Real-Time PCR
4.3. D Spheroid Formation
4.4. Western-Blot Analysis
4.5. Cell Cycle Analysis
4.6. Statistical Analysis for In Vitro Studies
4.7. Genes Correlations in TGCA and Rembrandt Databases
4.8. Association Between Five Genes: Integrin α6/ZEB1/YAP1/FGFR1/FOXM1 and Overall Survival in GBM Patients
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weller, M.; Butowski, N.; Tran, D.D.; Recht, L.D.; Lim, M.; Hirte, H.; Ashby, L.; Mechtler, L.; Goldlust, S.A.; Iwamoto, F.; et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): A randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017, 18, 1373–1385. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Liu, G.; Yuan, X.; Zeng, Z.; Tunici, P.; Ng, H.; Abdulkadir, I.R.; Lu, L.; Irvin, D.; Black, K.L.; Yu, J.S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. [Google Scholar] [CrossRef]
- Bianconi, D.; Unseld, M.; Prager, G.W. Integrins in the Spotlight of Cancer. Int. J. Mol. Sci. 2016, 17, 2037. [Google Scholar] [CrossRef]
- Meldolesi, J. Pharmacology of the cell/matrix form of adhesion. Pharmacol. Res. 2016, 107, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Gingras, M.C.; Roussel, E.; Bruner, J.M.; Branch, C.D.; Moser, R.P. Comparison of cell adhesion molecule expression between glioblastoma multiforme and autologous normal brain tissue. J. Neuroimmunol. 1995, 57, 143–153. [Google Scholar] [CrossRef]
- Previtali, S.; Quattrini, A.; Nemni, R.; Truci, G.; Ducati, A.; Wrabetz, L.; Canal, N. Alpha6 beta4 and alpha6 beta1 integrins in astrocytomas and other CNS tumors. J. Neuropathol. Exp. Neurol. 1996, 55, 456–465. [Google Scholar] [CrossRef]
- Hall, P.E.; Lathia, J.D.; Miller, N.G.; Caldwell, M.A.; ffrench-Constant, C. Integrins are markers of human neural stem cells. Stem Cells 2006, 24, 2078–2084. [Google Scholar] [CrossRef]
- Krebsbach, P.H.; Villa-Diaz, L.G. The Role of Integrin alpha6 (CD49f) in Stem Cells: More than a Conserved Biomarker. Stem Cells Dev. 2017, 26, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Gallagher, J.; Heddleston, J.M.; Wang, J.; Eyler, C.E.; Macswords, J.; Wu, Q.; Vasanji, A.; McLendon, R.E.; Hjelmeland, A.B.; et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell 2010, 6, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Katoh, M.; Nakagama, H. FGF receptors: Cancer biology and therapeutics. Med. Res. Rev. 2014, 34, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Morrison, R.S.; Yamaguchi, F.; Bruner, J.M.; Tang, M.; McKeehan, W.; Berger, M.S. Fibroblast growth factor receptor gene expression and immunoreactivity are elevated in human glioblastoma multiforme. Cancer Res. 1994, 54, 2794–2799. [Google Scholar] [PubMed]
- Morrison, R.S.; Yamaguchi, F.; Saya, H.; Bruner, J.M.; Yahanda, A.M.; Donehower, L.A.; Berger, M. Basic fibroblast growth factor and fibroblast growth factor receptor I are implicated in the growth of human astrocytomas. J. Neurooncol. 1994, 18, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Auguste, P.; Gursel, D.B.; Lemiere, S.; Reimers, D.; Cuevas, P.; Carceller, F.; Di Santo, J.P.; Bikfalvi, A. Inhibition of fibroblast growth factor/fibroblast growth factor receptor activity in glioma cells impedes tumor growth by both angiogenesis-dependent and -independent mechanisms. Cancer Res. 2001, 61, 1717–1726. [Google Scholar]
- Murphy, P.R.; Sato, Y.; Knee, R.S. Phosphorothioate antisense oligonucleotides against basic fibroblast growth factor inhibit anchorage-dependent and anchorage-independent growth of a malignant glioblastoma cell line. Mol. Endocrinol. 1992, 6, 877–884. [Google Scholar] [CrossRef]
- Yamada, S.M.; Yamaguchi, F.; Brown, R.; Berger, M.S.; Morrison, R.S. Suppression of glioblastoma cell growth following antisense oligonucleotide-mediated inhibition of fibroblast growth factor receptor expression. Glia 1999, 28, 66–76. [Google Scholar] [CrossRef]
- Haley, E.M.; Kim, Y. The role of basic fibroblast growth factor in glioblastoma multiforme and glioblastoma stem cells and in their in vitro culture. Cancer Lett. 2014, 346, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Loilome, W.; Joshi, A.D.; ap Rhys, C.M.; Piccirillo, S.; Vescovi, A.L.; Gallia, G.L.; Riggins, G.J. Glioblastoma cell growth is suppressed by disruption of Fibroblast Growth Factor pathway signaling. J. Neurooncol. 2009, 94, 359–366. [Google Scholar] [CrossRef]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef]
- Chang, K.W.; Huang, Y.L.; Wong, Z.R.; Su, P.H.; Huang, B.M.; Ju, T.K.; Yang, H.Y. Fibroblast growth factor-2 up-regulates the expression of nestin through the Ras-Raf-ERK-Sp1 signaling axis in C6 glioma cells. Biochem. Biophys. Res. Commun. 2013, 434, 854–860. [Google Scholar] [CrossRef]
- Kowalski-Chauvel, A.; Modesto, A.; Gouaze-Andersson, V.; Baricault, L.; Gilhodes, J.; Delmas, C.; Lemarie, A.; Toulas, C.; Cohen-Jonathan-Moyal, E.; Seva, C. Alpha-6 integrin promotes radioresistance of glioblastoma by modulating DNA damage response and the transcription factor Zeb1. Cell Death Dis. 2018, 9, 872. [Google Scholar] [CrossRef]
- Gouaze-Andersson, V.; Gherardi, M.J.; Lemarie, A.; Gilhodes, J.; Lubrano, V.; Arnauduc, F.; Cohen-Jonathan Moyal, E.; Toulas, C. FGFR1/FOXM1 pathway: A key regulator of glioblastoma stem cells radioresistance and a prognosis biomarker. Oncotarget 2018, 9, 31637–31649. [Google Scholar] [CrossRef]
- Liao, G.B.; Li, X.Z.; Zeng, S.; Liu, C.; Yang, S.M.; Yang, L.; Hu, C.J.; Bai, J.Y. Regulation of the master regulator FOXM1 in cancer. Cell Commun. Signal 2018, 16, 57. [Google Scholar] [CrossRef]
- Xiong, J.; Balcioglu, H.E.; Danen, E.H. Integrin signaling in control of tumor growth and progression. Int. J. Biochem. Cell Biol. 2013, 45, 1012–1015. [Google Scholar] [CrossRef]
- Lehmann, W.; Mossmann, D.; Kleemann, J.; Mock, K.; Meisinger, C.; Brummer, T.; Herr, R.; Brabletz, S.; Stemmler, M.P.; Brabletz, T. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat. Commun. 2016, 7, 10498. [Google Scholar] [CrossRef] [Green Version]
- Siebzehnrubl, F.A.; Silver, D.J.; Tugertimur, B.; Deleyrolle, L.P.; Siebzehnrubl, D.; Sarkisian, M.R.; Devers, K.G.; Yachnis, A.T.; Kupper, M.D.; Neal, D.; et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol. Med. 2013, 5, 1196–1212. [Google Scholar] [CrossRef] [Green Version]
- Tsou, R.; Isik, F.F. Integrin activation is required for VEGF and FGF receptor protein presence on human microvascular endothelial cells. Mol. Cell. Biochem. 2001, 224, 81–89. [Google Scholar] [CrossRef]
- Mori, S.; Wu, C.Y.; Yamaji, S.; Saegusa, J.; Shi, B.; Ma, Z.; Kuwabara, Y.; Lam, K.S.; Isseroff, R.R.; Takada, Y.K.; et al. Direct binding of integrin alphavbeta3 to FGF1 plays a role in FGF1 signaling. J. Biol. Chem. 2008, 283, 18066–18075. [Google Scholar] [CrossRef]
- Rusnati, M.; Tanghetti, E.; Dell’Era, P.; Gualandris, A.; Presta, M. alphavbeta3 integrin mediates the cell-adhesive capacity and biological activity of basic fibroblast growth factor (FGF-2) in cultured endothelial cells. Mol. Biol. Cell 1997, 8, 2449–2461. [Google Scholar] [CrossRef]
- Zou, L.; Cao, S.; Kang, N.; Huebert, R.C.; Shah, V.H. Fibronectin induces endothelial cell migration through beta1 integrin and Src-dependent phosphorylation of fibroblast growth factor receptor-1 at tyrosines 653/654 and 766. J. Biol. Chem. 2012, 287, 7190–7202. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.S.; Tan, L.; Smith, A.; Gray, N.S.; Wendt, M.K. Covalent Targeting of Fibroblast Growth Factor Receptor Inhibits Metastatic Breast Cancer. Mol. Cancer Ther. 2016, 15, 2096–2106. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Roth, B.; Choi, W.; Black, P.C.; Dinney, C.; McConkey, D.J. Fibroblast growth factor receptors-1 and -3 play distinct roles in the regulation of bladder cancer growth and metastasis: Implications for therapeutic targeting. PLoS ONE 2013, 8, e57284. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, D.; Saito, T.; Ito, S.; Masuda, T.; Kurashige, J.; Kuroda, Y.; Eguchi, H.; Kodera, Y.; Mimori, K. Overexpression of FGFR1 Promotes Peritoneal Dissemination Via Epithelial-to-Mesenchymal Transition in Gastric Cancer. Cancer Genom. Proteom. 2018, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Cosset, E.; Ilmjarv, S.; Dutoit, V.; Elliott, K.; von Schalscha, T.; Camargo, M.F.; Reiss, A.; Moroishi, T.; Seguin, L.; Gomez, G.; et al. Glut3 Addiction Is a Druggable Vulnerability for a Molecularly Defined Subpopulation of Glioblastoma. Cancer Cell 2017, 32, 856–868.e855. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.F.; Liu, A.M.; Hong, W.; Xu, Z.; Luk, J.M. Integrin alpha2beta1 inhibits MST1 kinase phosphorylation and activates Yes-associated protein oncogenic signaling in hepatocellular carcinoma. Oncotarget 2016, 7, 77683–77695. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.; Yamada, D.; Hirsova, P.; Bronk, S.F.; Werneburg, N.W.; Krishnan, A.; Salim, W.; Zhang, L.; Trushina, E.; Truty, M.J.; et al. A Hippo and Fibroblast Growth Factor Receptor Autocrine Pathway in Cholangiocarcinoma. J. Biol. Chem. 2016, 291, 8031–8047. [Google Scholar] [CrossRef]
- Lu, T.; Li, Z.; Yang, Y.; Ji, W.; Yu, Y.; Niu, X.; Zeng, Q.; Xia, W.; Lu, S. The Hippo/YAP1 pathway interacts with FGFR1 signaling to maintain stemness in lung cancer. Cancer Lett. 2018, 423, 36–46. [Google Scholar] [CrossRef] [PubMed]







| Genes | ITGA6 | FOXM1 | FGFR1 | ZEB1 | YAP1 |
|---|---|---|---|---|---|
| ITGA6 | 1.0000 | ||||
| FOXM1 | 0.1215 ** p = 0.0047 | 1.0000 | |||
| FGFR1 | 0.2610 *** p < 0.0001 | 0.2543 *** p < 0.0001 | 1.0000 | ||
| ZEB1 | 0.2611 p < 0.0001 | 0.2810 *** p < 0.0001 | 0.3414 *** p < 0.0001 | 1.0000 | |
| YAP1 | 0.4111 *** p < 0.0001 | 0.0548 p = 2042 | 0.3145 *** p <0.0001 | 0.1579 ** p = 0.0002 | 1.0000 |
| Genes | ITGA6 | FOXM1 | FGFR1 | ZEB1 | YAP1 |
|---|---|---|---|---|---|
| ITGA6 | 1.0000 | ||||
| FOXM1 | 0.1307 p = 0.077 | 1.0000 | |||
| FGFR1 | 0.1662 * p = 0.0241 | 0.3943 *** p < 0.0001 | 1.0000 | ||
| ZEB1 | 0.2141 ** p = 0.0035 | 0.4381 *** p < 0.0001 | 0.2492 *** p = 0.0006 | 1.0000 | |
| YAP1 | 0.5133 *** p < 0.0001 | 0.2296 ** p = 0.0017 | 0.3958 *** p < 0.0001 | 0.1960 ** p = 0.0077 | 1.0000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
KOWALSKI-CHAUVEL, A.; GOUAZE-ANDERSSON, V.; BARICAULT, L.; MARTIN, E.; DELMAS, C.; TOULAS, C.; COHEN-JONATHAN-MOYAL, E.; SEVA, C. Alpha6-Integrin Regulates FGFR1 Expression through the ZEB1/YAP1 Transcription Complex in Glioblastoma Stem Cells Resulting in Enhanced Proliferation and Stemness. Cancers 2019, 11, 406. https://doi.org/10.3390/cancers11030406
KOWALSKI-CHAUVEL A, GOUAZE-ANDERSSON V, BARICAULT L, MARTIN E, DELMAS C, TOULAS C, COHEN-JONATHAN-MOYAL E, SEVA C. Alpha6-Integrin Regulates FGFR1 Expression through the ZEB1/YAP1 Transcription Complex in Glioblastoma Stem Cells Resulting in Enhanced Proliferation and Stemness. Cancers. 2019; 11(3):406. https://doi.org/10.3390/cancers11030406
Chicago/Turabian StyleKOWALSKI-CHAUVEL, Aline, Valerie GOUAZE-ANDERSSON, Laurent BARICAULT, Elodie MARTIN, Caroline DELMAS, Christine TOULAS, Elizabeth COHEN-JONATHAN-MOYAL, and Catherine SEVA. 2019. "Alpha6-Integrin Regulates FGFR1 Expression through the ZEB1/YAP1 Transcription Complex in Glioblastoma Stem Cells Resulting in Enhanced Proliferation and Stemness" Cancers 11, no. 3: 406. https://doi.org/10.3390/cancers11030406