Turing Instability-Driven Biofabrication of Branching Tissue Structures: A Dynamic Simulation and Analysis Based on the Reaction–Diffusion Mechanism † †
Abstract
:1. Introduction
2. Mathematical Model
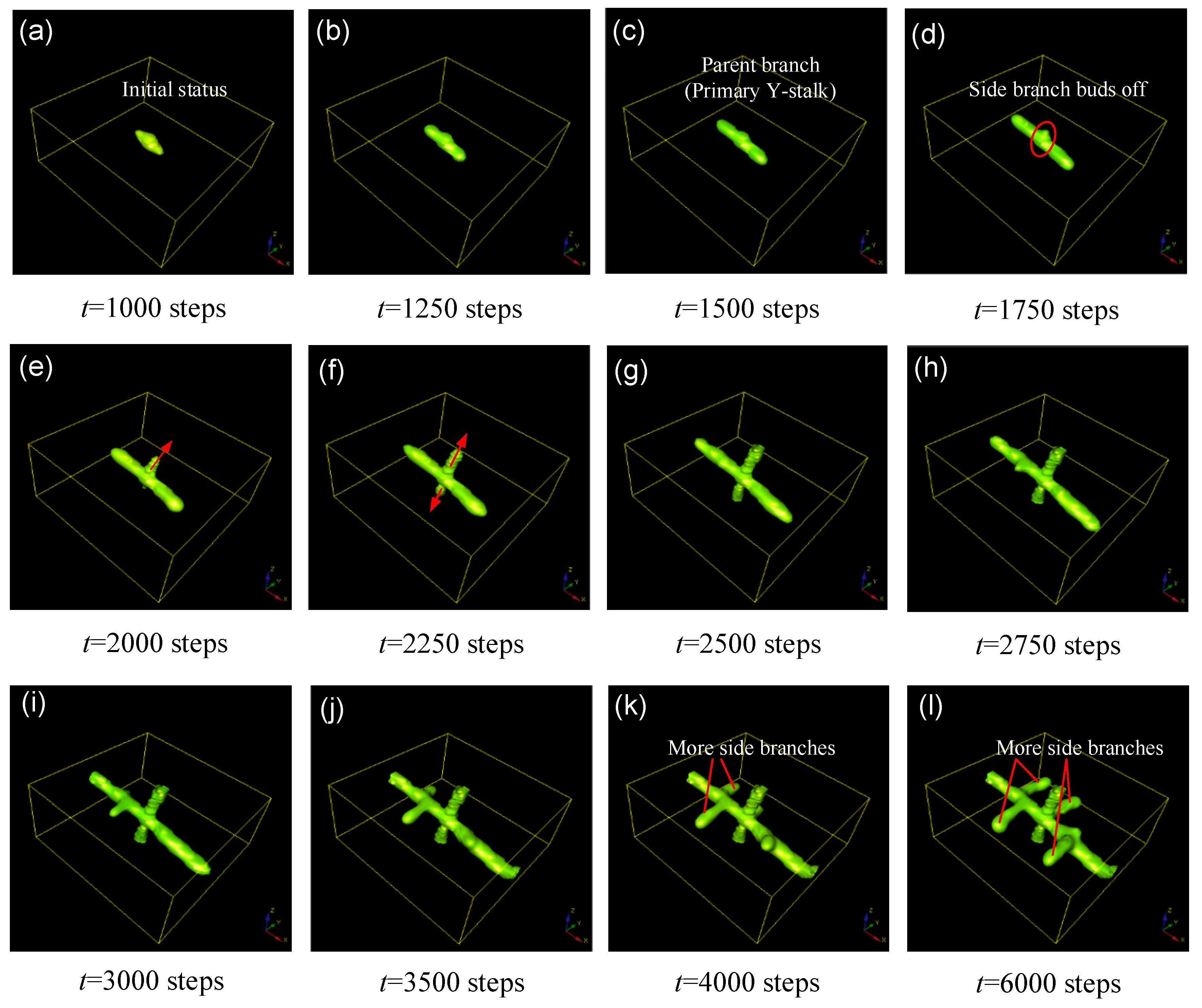
3. The Evolution of Side Branching
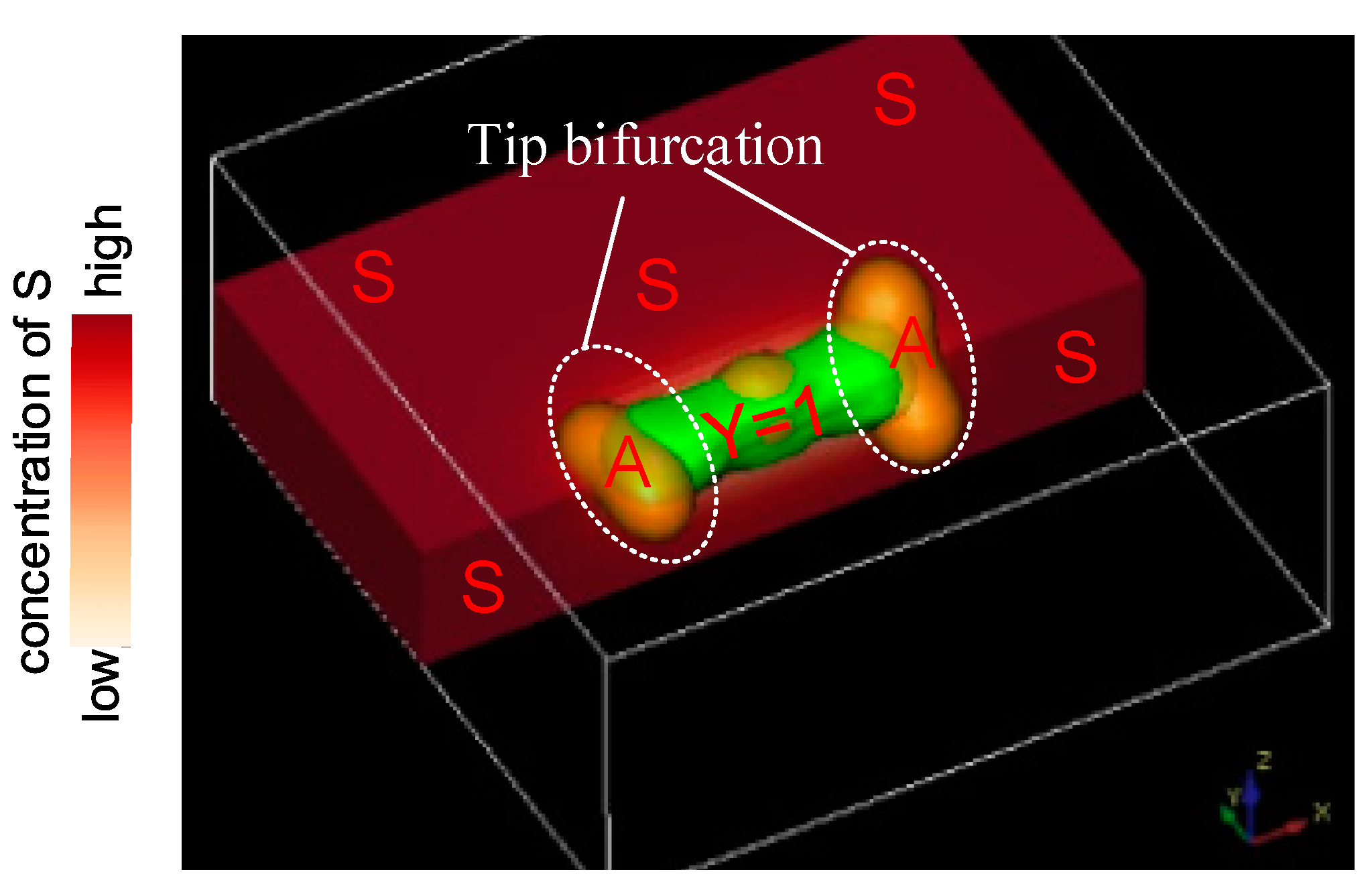
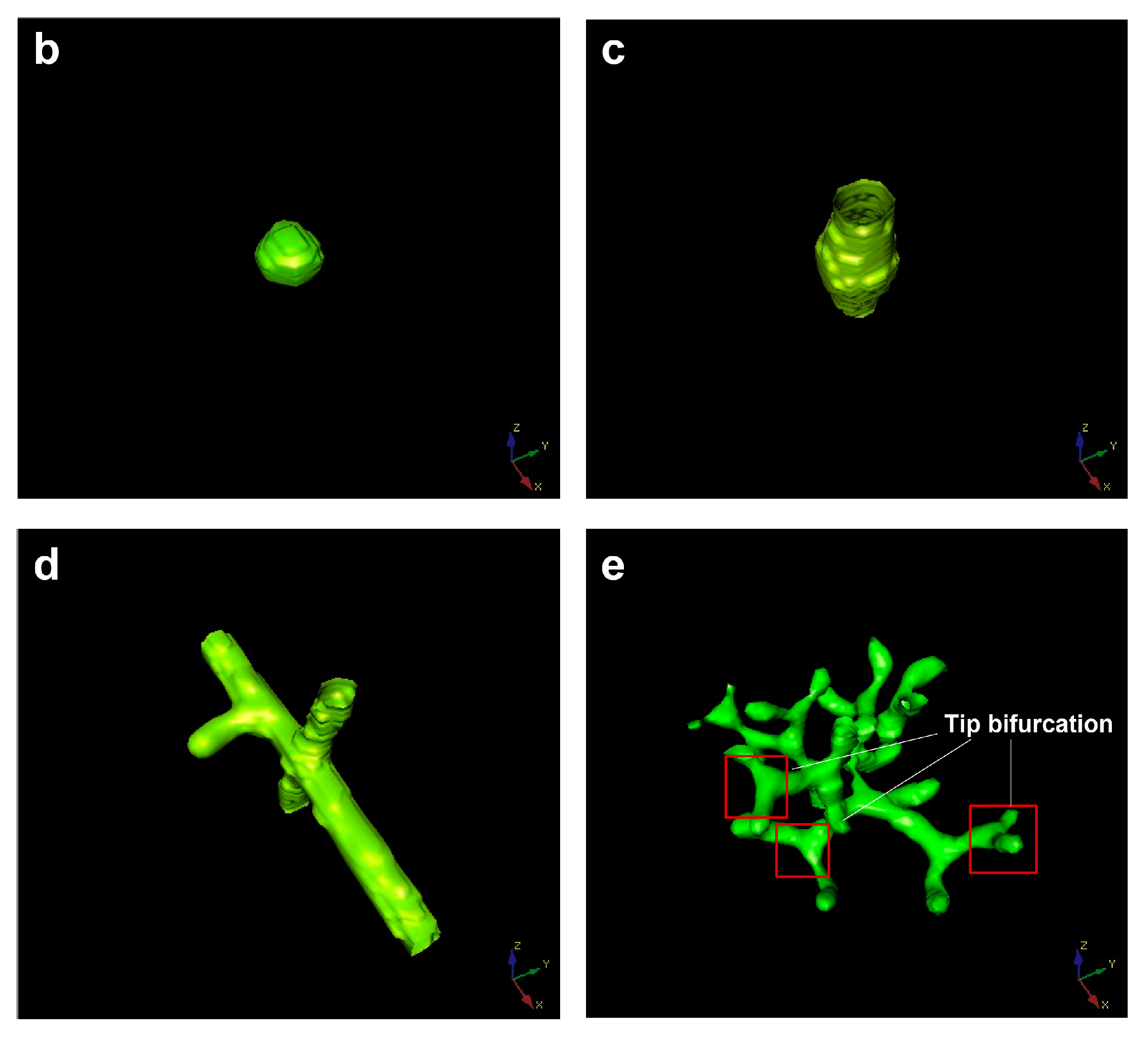
4. Tip Bifurcation
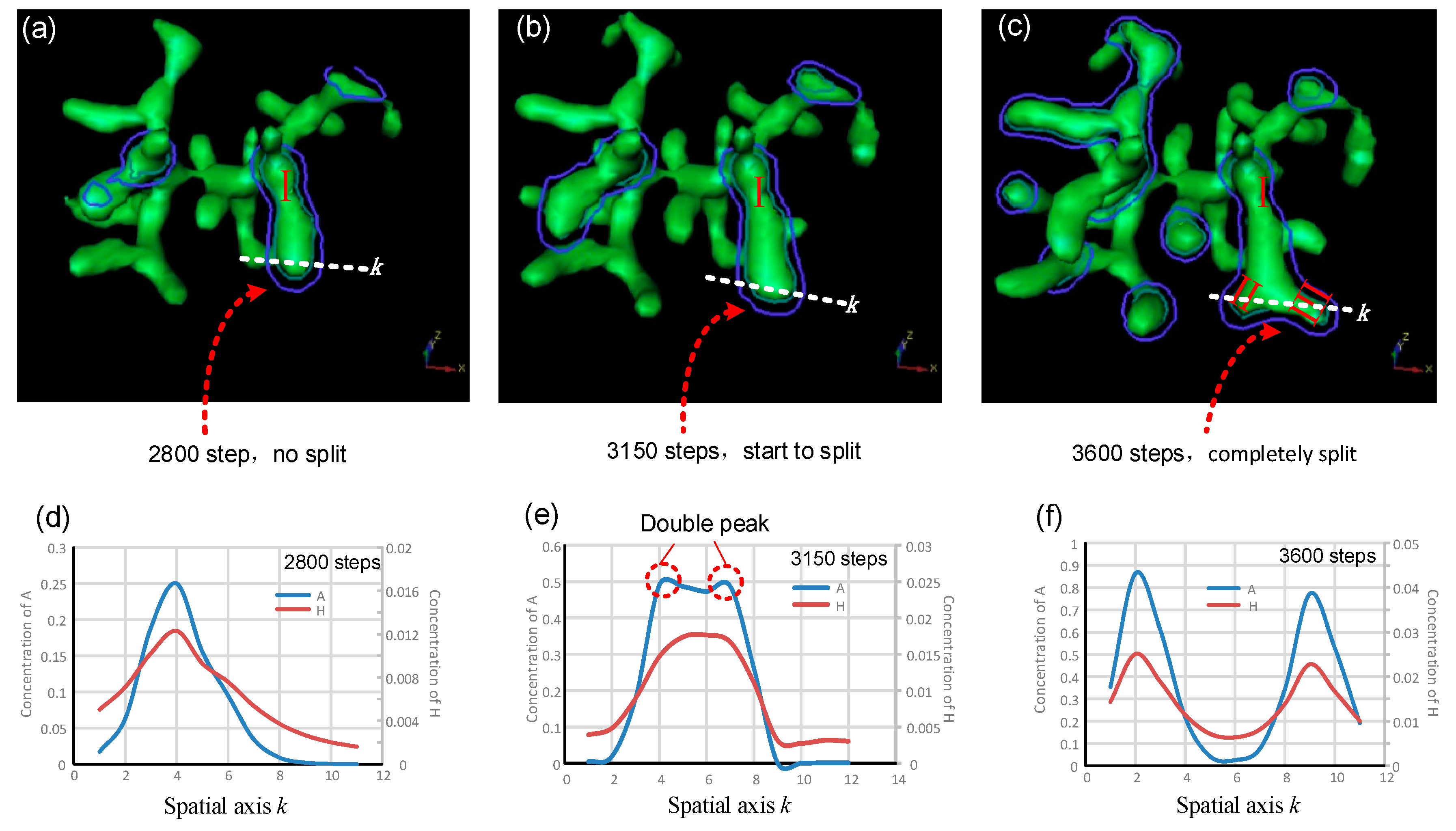
5. The Evolution of Tip Bifurcation
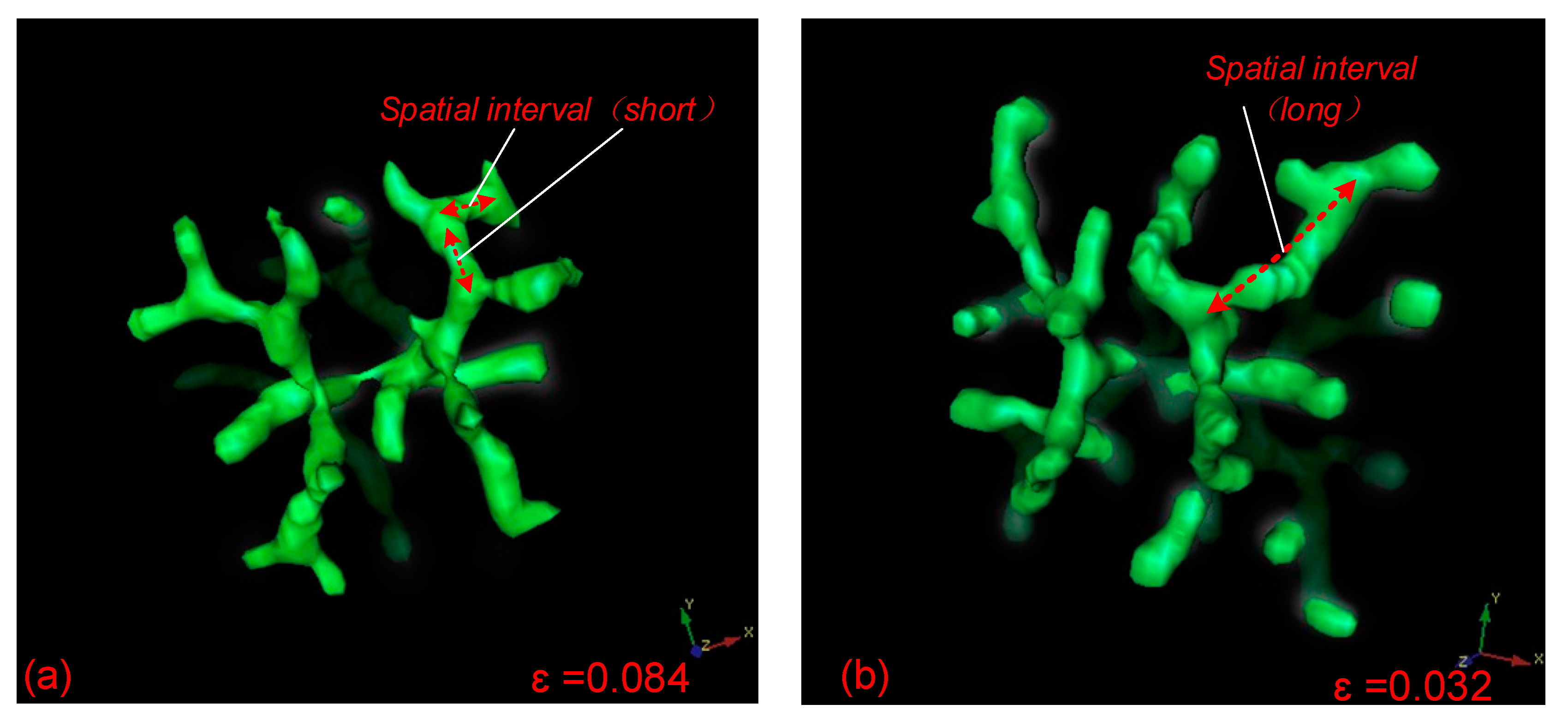
6. Distribution Law of Bifurcation Patterns in Parameter Domains
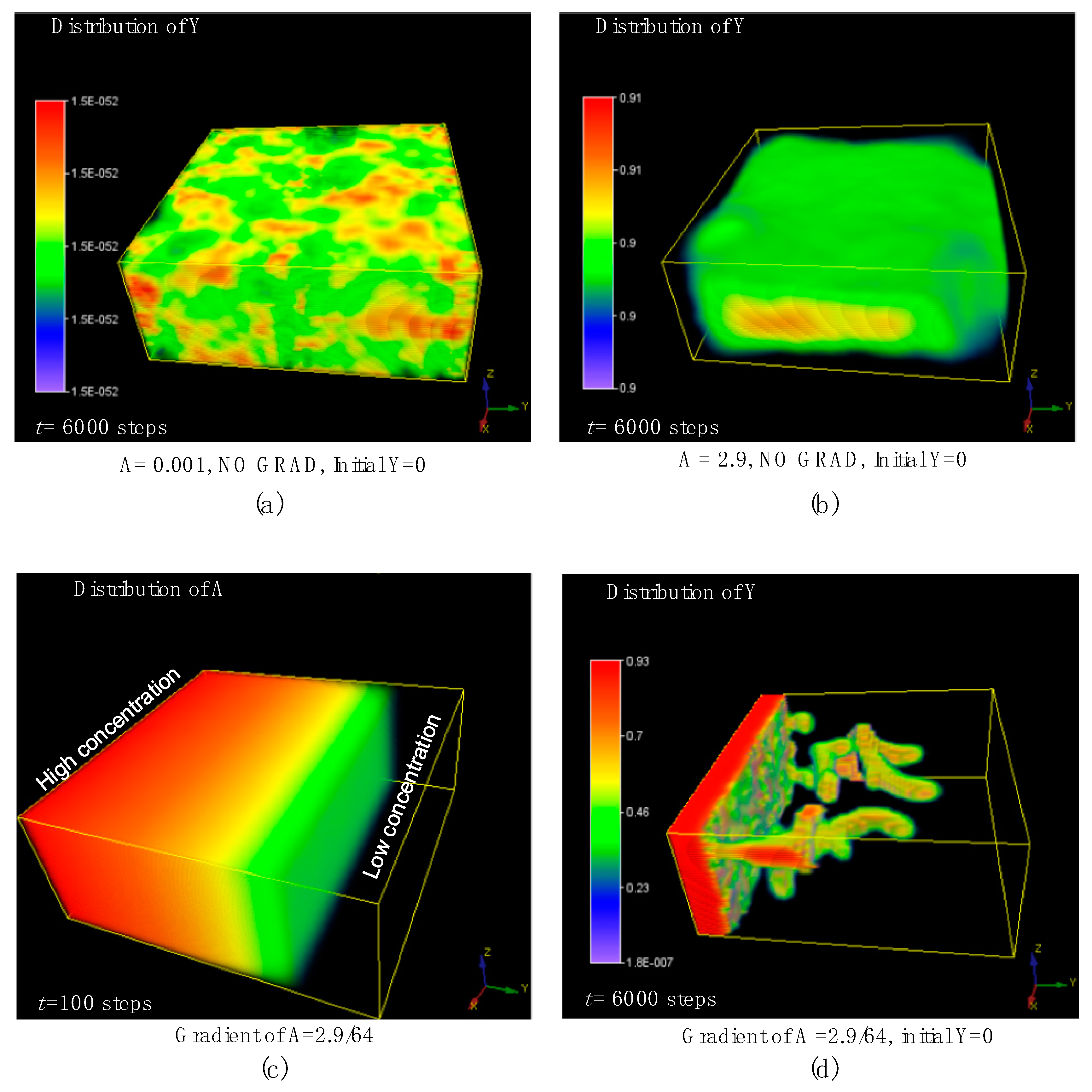
7. The 3D Morphological Change under an External Concentration Gradient of Activator
8. Selected Morphogens and Their Roles in Lung Development
9. The Limitations of this Model
10. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chia, H.N.; Wu, B.M. Recent advances in 3D printing of biomaterials. J. Biol. Eng. 2015, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gojgini, S.; Chen, T.H.; Teng, F.; Fei, P.; Dong, S.; Segura, T.; Ho, C.-M. Three dimensional tubular structure self-assembled by vascular mesenchymal cells at stiffness interfaces of hydrogels. Biomed. Pharmacother. 2016, 83, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gojgini, S.; Chen, T.H.; Fei, P.; Dong, S.; Ho, C.-M.; Segura, T. Directing three-dimensional multicellular morphogenesis by self-organization of vascular mesenchymal cells in hyaluronic acid hydrogels. J. Biol. Eng. 2017, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H. Tissue regeneration: From synthetic scaffolds to self-organizing morphogenesis. Curr. Stem Cell Res. Ther. 2014, 9, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Yang, Q.; Zhao, X.; Jin, G.; Ma, Y.; Xu, F. 4D bioprinting for biomedical applications. Trends Biotechnol. 2016, 34, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Datta, P.; Ayan, B.; Ozbolat, I.T. Bioprinting for vascular and vascularized tissue biofabrication. Acta Biomater. 2017. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Zhang, Y.S.; Akpek, A.; Shin, S.R.; Khademhosseini, A. 4D bioprinting: The next-generation technology for biofabrication enabled by stimuli-responsive materials. Biofabrication 2017, 9, 012001. [Google Scholar] [CrossRef] [PubMed]
- Morouço, P.; Lattanzi, W.; Alves, N.; Morouço, P.; Lattanzi, W.; Alves, N. 4D bioprinting as a new era for tissue engineering and regenerative medicine. Front. Bioeng. Biotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kirillova, A.; Maxson, R.; Stoychev, G.; Gomillion, C.T.; Ionov, L. 4D biofabrication using shape-morphing hydrogels. Adv. Mater. 2017, 29, 1703443. [Google Scholar] [CrossRef] [PubMed]
- Stroganov, V.; Pant, J.; Stoychev, G.; Janke, A.; Jehnichen, D.; Fery, A.; Handa, H.; Ionov, L. 4D biofabrication: 3D cell patterning using shape-changing films. Adv. Funct. Mater. 2018. [Google Scholar] [CrossRef]
- Metzger, R.J.; Klein, O.D.; Martin, G.R.; Krasnow, M.A. The branching programme of mouse lung development. Nature 2008, 453, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Tzou, D.; Spurlin, J.W.; Pavlovich, A.L.; Stewart, C.R.; Gleghorn, J.P.; Nelson, C.M. Morphogenesis and morphometric scaling of lung airway development follows phylogeny in chicken, quail, and duck embryos. EvoDevo 2016, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Sampogna, R.V.; Schneider, L.; Al-Awqati, Q. Developmental programming of branching morphogenesis in the kidney. J. Am. Soc. Nephrol. 2015, 26, 2414–2422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yokoyama, S.; Herriges, J.C.; Zhang, Z.; Young, R.E.; Verheyden, J.M.; Sun, X. E3 ubiquitin ligase RFWD2 controls lung branching through protein-level regulation of ETV transcription factors. Proc. Natl. Acad. Sci. USA 2016, 113, 7557–7562. [Google Scholar] [CrossRef] [PubMed]
- Morrisey, E.E.; Hogan, B.L.M. Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev. Cell 2010, 18, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Mandelbrot, B.B. The Fractal Geometry of Nature; Freeman: New York, NY, USA, 1983. [Google Scholar]
- Meinhardt, H. Morphogenesis of lines and nets. Differentiation 1976, 6, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, H.; Takaki, R.; Suki, B. A three-dimensional model of the human airway tree. J. Appl. Physiol. 1999, 87, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, H. Three-dimensional model of the human airway tree based on a fractal branching algorithm. In Fractals in Biology and Medicine; Losa, G.A., Merlini, D., Nonnenmacher, T.F., Weibel, E.R., Eds.; Birkhäuser: Basel, Switzerland, 2002; pp. 39–46. [Google Scholar]
- Tebockhorst, S.; Lee, D.; Wexler, A.S.; Oldham, M.J. Interaction of epithelium with mesenchyme affects global features of lung architecture: A computer model of development. J. Appl. Physiol. 2007, 102, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Turing, A.M. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1952, 237, 37–72. [Google Scholar] [CrossRef]
- Chen, T.H.; Hsu, J.J.; Zhao, X.; Guo, C.Y.; Wong, M.N.; Huang, Y.; Li, Z.W.; Garfinkel, A.; Ho, C.M.; Tintut, Y.; et al. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ. Res. 2012, 110, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Miura, T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 2010, 329, 1616–1620. [Google Scholar] [CrossRef] [PubMed]
- Menshykau, D.; Iber, D. Kidney branching morphogenesis under the control of a ligand-receptor-based turing mechanism. Phys. Biol. 2013, 10, 046003. [Google Scholar] [CrossRef] [PubMed]
- Menshykau, D.; Blanc, P.; Unal, E.; Sapin, V.; Iber, D. An interplay of geometry and signaling enables robust lung branching morphogenesis. Development 2014, 141, 4526–4536. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yang, Y. Hollow spheroid formation by cells at the mechanical interface of hyaluronic acid hydrogels in three dimensions. In Proceedings of the 2017 IEEE 17th International Conference on Nanotechnology (IEEE-NANO), Pittsburgh, PA, USA, 25–28 July 2017; pp. 218–222. [Google Scholar]
- Zhu, X.; Yang, Y. Simulation for tubular and spherical structure formation via self- organization of vascular mesenchymal cells in three dimensions. In Proceedings of the 2016 9th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Taiyuan, China, 15–17 October 2016; pp. 1654–1659. [Google Scholar]
- Xu, H.; Sun, M.; Zhao, X. Turing mechanism underlying a branching model for lung morphogenesis. PLoS ONE 2017, 12, e0174946. [Google Scholar] [CrossRef] [PubMed]
- Marcon, L.; Sharpe, J. Turing patterns in development: What about the horse part? Curr. Opin. Genet. Dev. 2012, 22, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Menshykau, D.; Kraemer, C.; Iber, D. Branch mode selection during early lung development. PLoS Comp. Biol. 2012, 8, e1002377. [Google Scholar] [CrossRef] [PubMed]
- Cellière, G.; Menshykau, D.; Iber, D. Simulations demonstrate a simple network to be sufficient to control branch point selection, smooth muscle and vasculature formation during lung branching morphogenesis. Biol. Open 2012, 1, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Miura, T. Models of lung branching morphogenesis. J. Biochem. 2015, 157, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Inoue, Y.; Adachi, T. Three-dimensional vertex model for simulating multicellular morphogenesis. Biophys. Physicobiol. 2015, 12, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Zhu, X.; Pan, L.; Zeng, X.; Garfinkel, A.; Tintut, Y.; Demer, L.L.; Zhao, X.; Ho, C.M. Directing tissue morphogenesis via self-assembly of vascular mesenchymal cells. Biomaterials 2012, 33, 9019–9026. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.N.; Sun, M.Z.; Garfinkel, A.; Zhao, X. Mechanisms of side branching and tip splitting in a model of branching morphogenesis. PLoS ONE 2014, 9, e102718. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, T.H.; Zeng, X.J.; Warburton, D.; Bostrom, K.I.; Ho, C.M.; Zhao, X.; Garfinkel, A. Branching patterns emerge in a mathematical model of the dynamics of lung development. J. Physiol. Lond. 2014, 592, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yang, H. 4D biofabrication of branching multicellular structures: A morphogenesis simulation based on turing’s reaction-diffusion dynamics. In IOP Conference Series: Materials Science and Engineering; IOP Publishing Ltd.: Bristol, UK, 2017; Volume 280, p. 012018. [Google Scholar]
- Yao, Y.; Nowak, S.; Yochelis, A.; Garfinkel, A.; Bostrom, K.I. Matrix gla protein, an inhibitory morphogen in pulmonary vascular development. J. Biol. Chem. 2007, 282, 30131–30142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Yang, H. In-Silico constructing three-dimensional hollow structure via self-organization of vascular mesenchymal cells. In Proceedings of the 16th International Conference on Nanotechnology (IEEE NANO 2016), Sendai, Japan, 22–25 August 2016; IEEE: Sendai, Japan, 2016; pp. 468–471. [Google Scholar]
- Zhu, X.L.; Ding, X.T. Study on a 3D hydrogel-based culture model for characterizing growth of fibroblasts under viral infection and drug treatment. SLAS Discov. 2017, 22, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Lorda-Diez, C.I.; Montero, J.A.; Rodriguez-Leon, J.; Garcia-Porrero, J.A.; Hurle, J.M. Expression and functional study of extracellular bmp antagonists during the morphogenesis of the digits and their associated connective tissues. PLoS ONE 2013, 8, e60423. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, I.; Camargo, L.; Pereira, M.M.; Fernandez-Martin, R.; Paz, D.A.; Salamone, D.F. Effects of bone morphogenic protein 4 (BMP4) and its inhibitor, Noggin, on in vitro maturation and culture of bovine preimplantation embryos. Reprod. Biol. Endocrinol. 2011, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, J.; Roderer, G.; Gunther, K.P.; Brenner, R.E. BMP-2, BMP-4, and PDGF-BB stimulate chemotactic migration of primary human mesenchymal progenitor cells. J. Cell. Biochem. 2002, 87, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Peluso, C.E.; Umulis, D.; Kim, Y.J.; O’Connor, M.B.; Serpe, M. Shaping BMP morphogen gradients through enzyme-substrate interactions. Dev. Cell 2011, 21, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Zebboudj, A.F.; Imura, M.; Boström, K. Matrix GLA protein, a regulatory protein for bone morphogenetic protein-2. J. Biol. Chem. 2002, 277, 4388–4394. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, A.; Del-Moral, P.M.; Ilovich, O.; Mishani, E.; Warburton, D.; Keshet, E. A perfusion-independent role of blood vessels in determining branching stereotypy of lung airways. Development 2011, 138, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Weaver, M.; Dunn, N.R.; Hogan, B.L. BMP4 and FGF10 play opposing roles during lung bud morphogenesis. Development 2000, 127, 2695–2704. [Google Scholar] [PubMed]
- Weaver, M.; Yingling, J.M.; Dunn, N.R.; Bellusci, S.; Hogan, B.L. BMP signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development 1999, 126, 4005–4015. [Google Scholar] [PubMed]
- Mailleux, A.A.; Tefft, D.; Ndiaye, D.; Itoh, N.; Thiery, J.P.; Warburton, D.; Bellusci, S. Evidence that SPROUTY2 functions as an inhibitor of mouse embryonic lung growth and morphogenesis. Mech. Dev. 2001, 102, 81–94. [Google Scholar] [CrossRef]
- Shu, W.; Guttentag, S.; Wang, Z.; Andl, T.; Ballard, P.; Lu, M.M.; Piccolo, S.; Birchmeier, W.; Whitsett, J.A.; Millar, S.E.; et al. Wnt/β-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal–distal patterning in the lung. Dev. Biol. 2005, 283, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Bellusci, S.; Henderson, R.; Winnier, G.; Oikawa, T.; Hogan, B.L. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (BMP-4) plays a role in mouse embryonic lung morphogenesis. Development 1996, 122, 1693–1702. [Google Scholar] [PubMed]
- Shi, W.; Zhao, J.; Anderson, K.D.; Warburton, D. Gremlin negatively modulates BMP-4 induction of embryonic mouse lung branching morphogenesis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 280, L1030–L1039. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.C.; Zebboudj, A.F.; Shao, E.; Perez, M.; Bostrom, K. Regulation of bone morphogenetic protein-4 by matrix GLA protein in vascular endothelial cells involves activin-like kinase receptor 1. J. Biol. Chem. 2006, 281, 33921–33930. [Google Scholar] [CrossRef] [PubMed]
- Garfinkel, A.; Tintut, Y.; Petrasek, D.; Bostrom, K.; Demer, L.L. Pattern formation by vascular mesenchymal cells. Proc. Natl. Acad. Sci. USA 2004, 101, 9247–9250. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Jumabay, M.; Wang, A.; Boström, K.I. Matrix GLA protein deficiency causes arteriovenous malformations in mice. J. Clin. Investig. 2011, 121, 2993–3004. [Google Scholar] [CrossRef] [PubMed]
- Lebeche, D.; Malpel, S.; Cardoso, W.V. Fibroblast growth factor interactions in the developing lung. Mech. Dev. 1999, 86, 125–136. [Google Scholar] [CrossRef]
- Warburton, D.; El-Hashash, A.; Carraro, G.; Tiozzo, C.; Sala, F.; Rogers, O.; De Langhe, S.; Kemp, P.J.; Riccardi, D.; Torday, J.; et al. Lung organogenesis. Curr. Top. Dev. Biol. 2010, 90, 73–158. [Google Scholar] [PubMed]
- Oster, G.F.; Murray, J.D.; Harris, A.K. Mechanical aspects of mesenchymal morphogenesis. J. Embryol. Exp. Morphol. 1984, 78, 83–125. [Google Scholar]
- Harris, A.K.; Warner, P.; Stopak, D. Generation of spatially periodic patterns by a mechanical instability: A mechanical alternative to the turing model. J. Embryol. Exp. Morphol. 1984, 80, 1–20. [Google Scholar] [PubMed]
- Oster, G.F. Lateral inhibition models of developmental processes. Math. Comput. Model. 1989, 12, 1179. [Google Scholar] [CrossRef]
- Lubarsky, B.; Krasnow, M.A. Tube morphogenesis: Making and shaping biological tubes. Cell 2003, 112, 19–28. [Google Scholar] [CrossRef]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Yang, H. Turing Instability-Driven Biofabrication of Branching Tissue Structures: A Dynamic Simulation and Analysis Based on the Reaction–Diffusion Mechanism †. Micromachines 2018, 9, 109. https://doi.org/10.3390/mi9030109
Zhu X, Yang H. Turing Instability-Driven Biofabrication of Branching Tissue Structures: A Dynamic Simulation and Analysis Based on the Reaction–Diffusion Mechanism †. Micromachines. 2018; 9(3):109. https://doi.org/10.3390/mi9030109
Chicago/Turabian StyleZhu, Xiaolu, and Hao Yang. 2018. "Turing Instability-Driven Biofabrication of Branching Tissue Structures: A Dynamic Simulation and Analysis Based on the Reaction–Diffusion Mechanism †" Micromachines 9, no. 3: 109. https://doi.org/10.3390/mi9030109



