Chiral Orientation of Skeletal Muscle Cells Requires Rigid Substrate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. Micropatterning on Substrates with Varied Stiffness
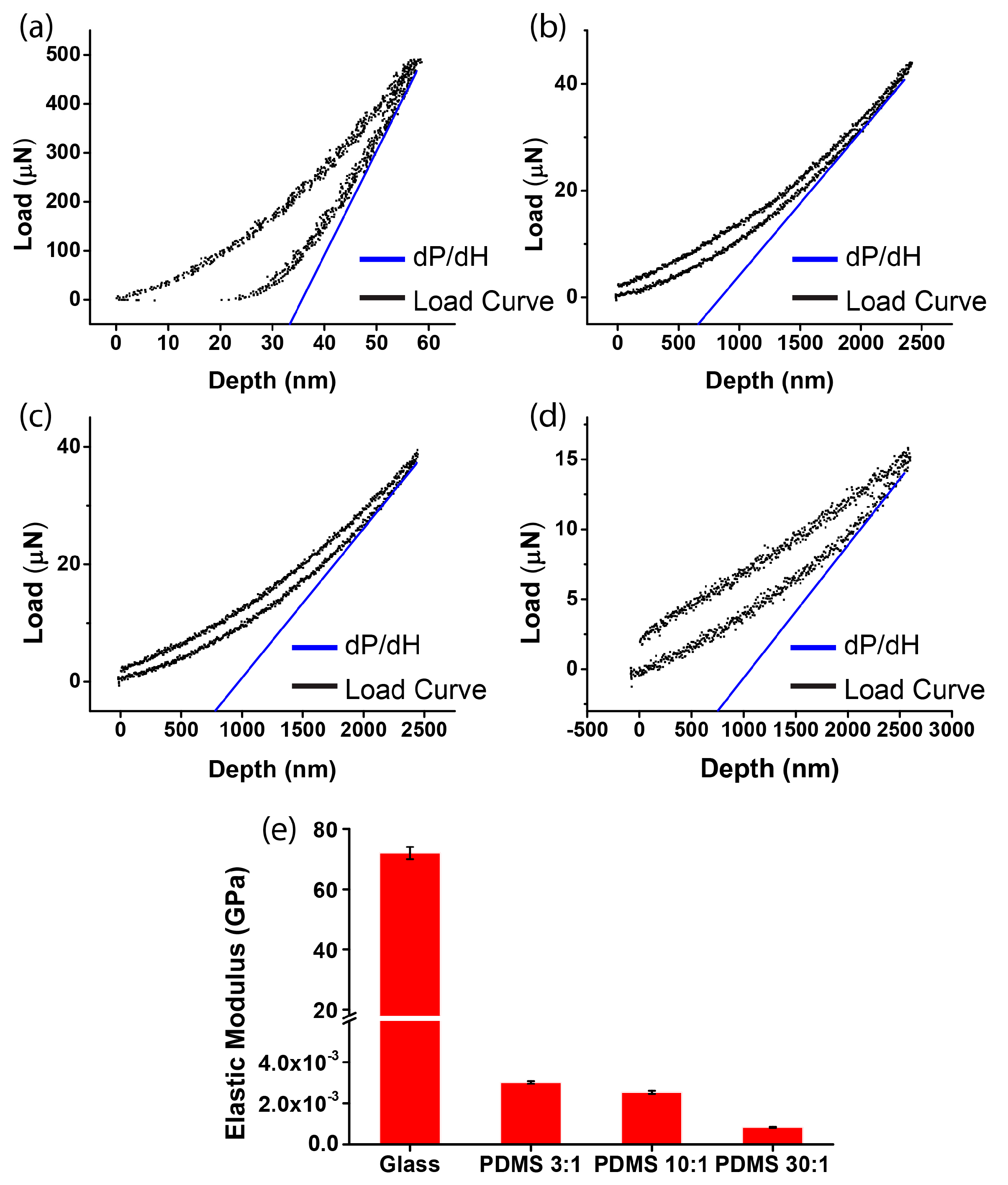
2.3. Nano-Indentation Measurement
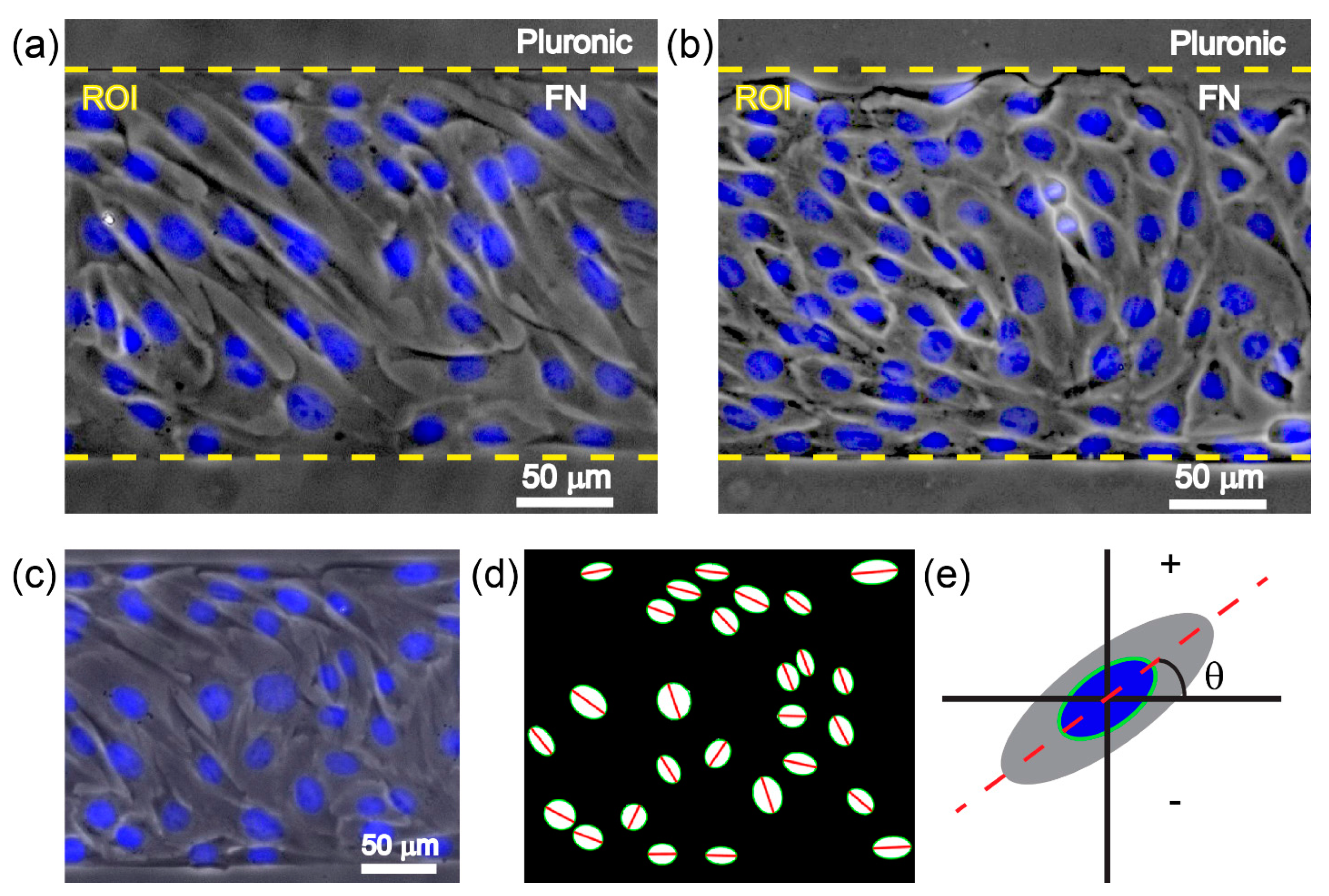
2.4. Fluorescence Staining and Imaging
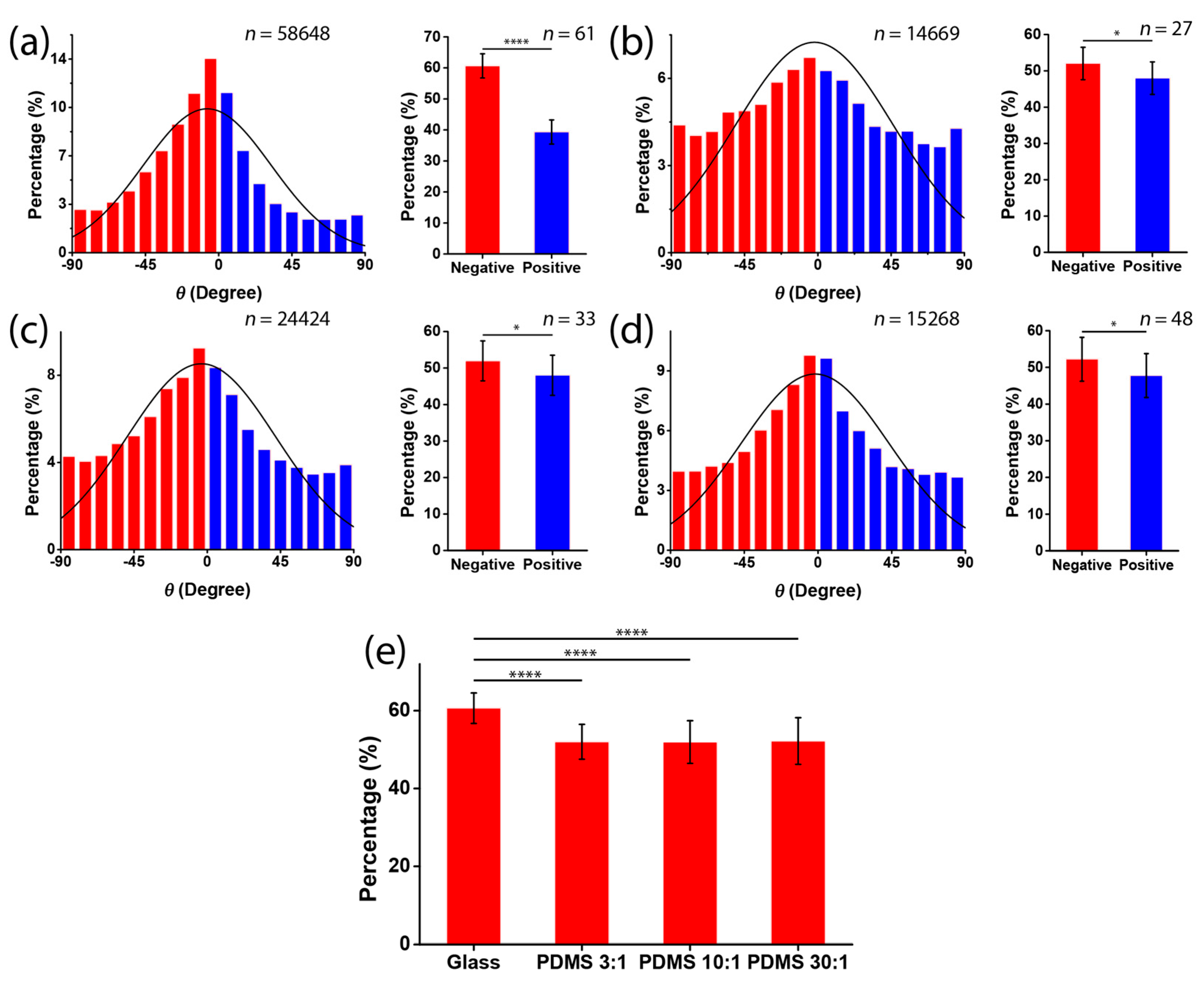
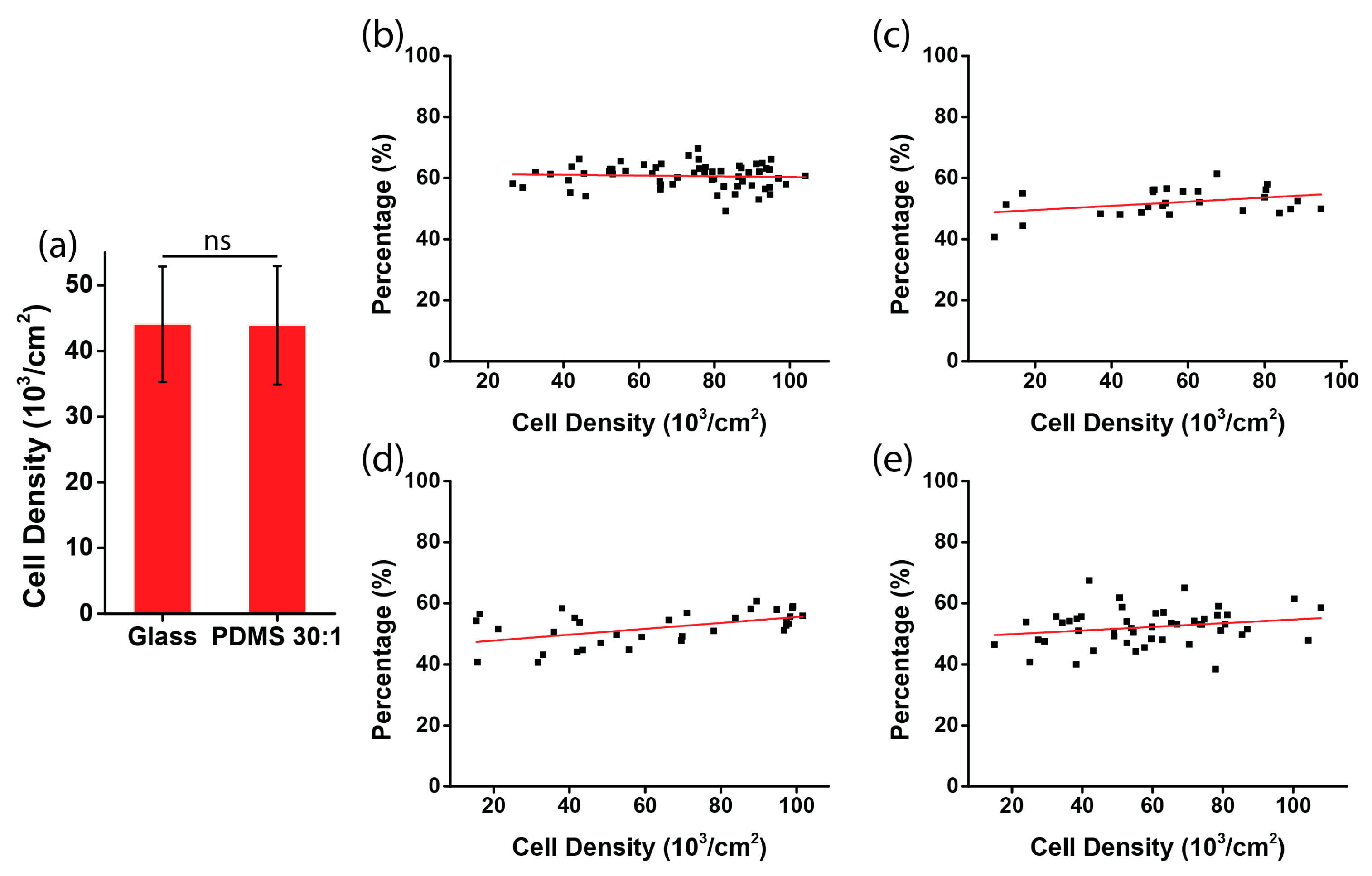
2.5. Analysis of Cell Orientation and Density
2.6. Statistical Analysis
3. Result
3.1. Measurement of Substrate Stiffness
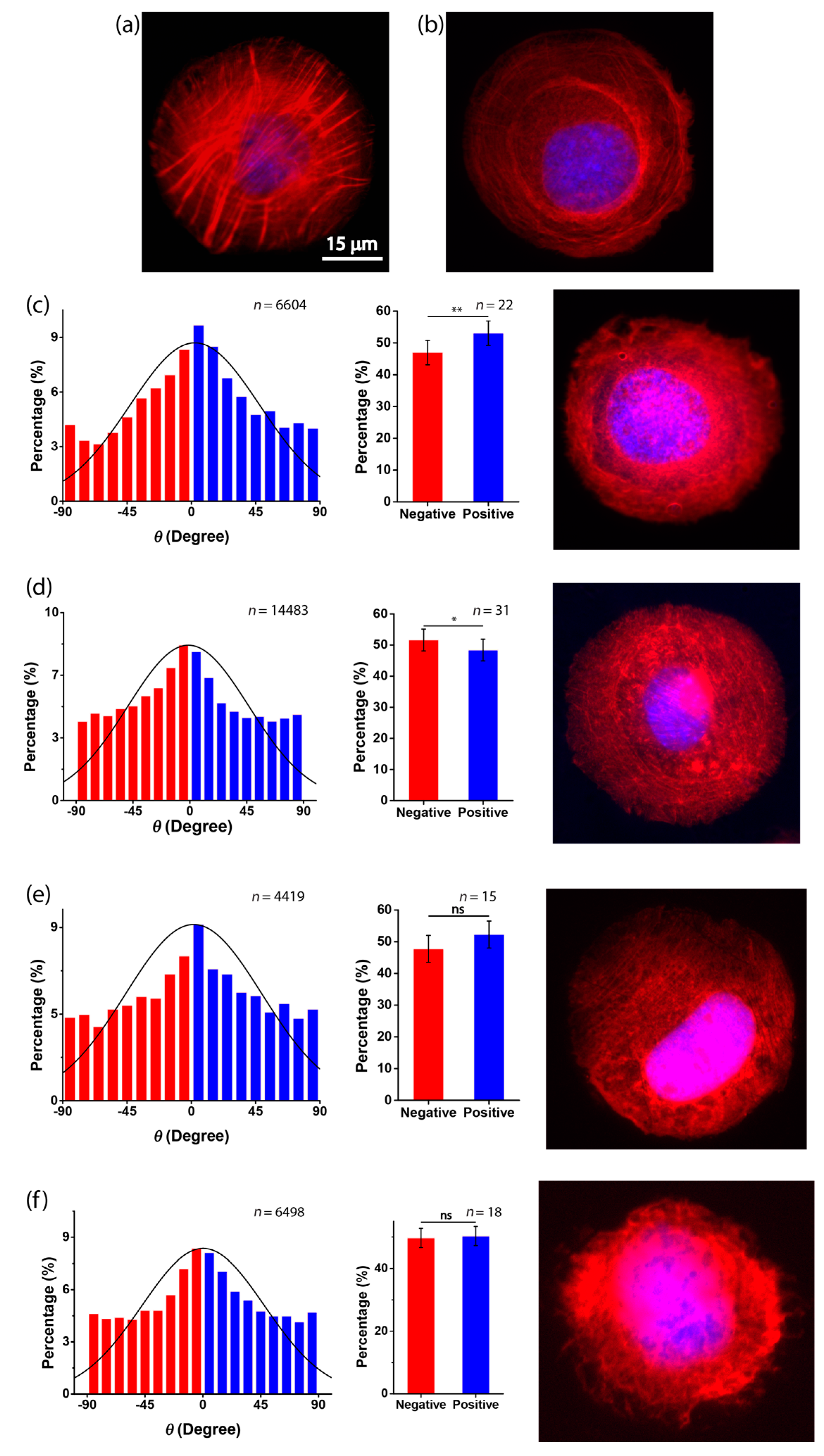
3.2. Chirality of Cell Orientation
3.3. Role of Actin
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Edwards, W.; Moles, A.T.; Franks, P. The global trend in plant twining direction. Glob. Ecol. Biogeogr. 2007, 16, 795–800. [Google Scholar] [CrossRef]
- Hozumi, S.; Maeda, R.; Taniguchi, K.; Kanai, M.; Shirakabe, S.; Sasamura, T.; Spéder, P.; Noselli, S.; Aigaki, T.; Murakami, R.; et al. An unconventional myosin in drosophila reverses the default handedness in visceral organs. Nature 2006, 440, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.M.; Gong, A.G.W.; Xu, M.L.; Lam, C.T.W.; Zhang, L.M.L.; Bi, C.W.C.; Cui, D.; Cheng, A.W.M.; Dong, T.T.X.; Tsim, K.W.K.; et al. Transcriptional activity of acetylcholinesterase gene is regulated by DNA methylation during C2C12 myogenesis. Brain Res. 2016, 1642, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; van Eeden, F.J.; Warren, K.S.; Chin, A.; Nüsslein-Volhard, C.; Haffter, P.; Fishman, M.C. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 1997, 124, 4373–4382. [Google Scholar] [PubMed]
- Chen, T.-H.; Hsu, J.J.; Zhao, X.; Guo, C.; Wong, M.N.; Huang, Y.; Li, Z.; Garfinkel, A.; Ho, C.-M.; Tintut, Y.; et al. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ. Res. 2012, 110, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, H.; Imamura, T.; Morino, K.; Shimosato, T.; Tawa, M.; Ugi, S.; Sakurai, H.; Maegawa, H.; Okamura, T. MicroRNA-494 plays a role in fiber type-specific skeletal myogenesis in human induced pluripotent stem cells. Biochem. Biophys. Res. Commun. 2015, 468, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Bao, Y.; Lam, M.L.; Xu, T.; Xie, K.; Man, H.S.; Chan, E.Y.; Zhu, N.; Lam, R.H.W.; Chen, T.-H. Nanowire magnetoscope reveals a cellular torque with left–right bias. ACS Nano 2016, 10, 7409–7417. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Duffy, R.; Lee, A.; Feinberg, A.W. Optimizing the structure and contractility of engineered skeletal muscle thin films. Acta Biomater. 2013, 9, 7885–7894. [Google Scholar] [CrossRef] [PubMed]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin stress fibers—Assembly, dynamics and biological roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.Q.; Ronaldson, K.; Park, M.; Taylor, G.; Zhang, Y.; Gimble, J.M.; Vunjak-Novakovic, G. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc. Natl. Acad. Sci. USA 2011, 108, 12295–12300. [Google Scholar] [CrossRef] [PubMed]
- Dokukina, I.V.; Gracheva, M.E. A model of fibroblast motility on substrates with different rigidities. Biophys. J. 2010, 98, 2794–2803. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Griffin, M.A.; Sen, S.; Bönnemann, C.G.; Sweeney, H.L.; Discher, D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004, 166, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Huang, Y.; Lam, M.L.; Xu, T.; Zhu, N.; Guo, Z.; Cui, X.; Lam, R.H.W.; Chen, T.H. Substrate stiffness regulates the development of left-right asymmetry in cell orientation. ACS Appl. Mater. Interfaces 2016, 8, 17976–17986. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Guo, Z.; Chen, T.H. Left-right asymmetry in cell orientation requires high substrate rigidity. In Proceedings of the 2015 9th IEEE International Conference on Nano/Molecular Medicine & Engineering (NANOMED), Honolulu, HI, USA, 15–18 November 2015; pp. 7–11. [Google Scholar]
- Yeung, T.; Georges, P.C.; Flanagan, L.A.; Marg, B.; Ortiz, M.; Funaki, M.; Zahir, N.; Ming, W.; Weaver, V.; Janmey, P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskelet. 2005, 60, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Tee, Y.H.; Shemesh, T.; Thiagarajan, V.; Hariadi, R.F.; Anderson, K.L.; Page, C.; Volkmann, N.; Hanein, D.; Sivaramakrishnan, S.; Kozlov, M.M.; et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat. Cell Biol. 2015, 17, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Tannenbaum, J.; Miranda, A.F. Stimulation of actin synthesis by cytochalasin D is specific for the isoactins normally expressed in muscle or non-muscle cells. J. Cell Sci. 1986, 84, 253–262. [Google Scholar] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, N.; Kwong, H.K.; Bao, Y.; Chen, T.-H. Chiral Orientation of Skeletal Muscle Cells Requires Rigid Substrate. Micromachines 2017, 8, 181. https://doi.org/10.3390/mi8060181
Zhu N, Kwong HK, Bao Y, Chen T-H. Chiral Orientation of Skeletal Muscle Cells Requires Rigid Substrate. Micromachines. 2017; 8(6):181. https://doi.org/10.3390/mi8060181
Chicago/Turabian StyleZhu, Ninghao, Hoi Kwan Kwong, Yuanye Bao, and Ting-Hsuan Chen. 2017. "Chiral Orientation of Skeletal Muscle Cells Requires Rigid Substrate" Micromachines 8, no. 6: 181. https://doi.org/10.3390/mi8060181





