A Microfluidic Chip for Cell Patterning Utilizing Paired Microwells and Protein Patterns †
Abstract
:1. Introduction
2. Materials and Methods
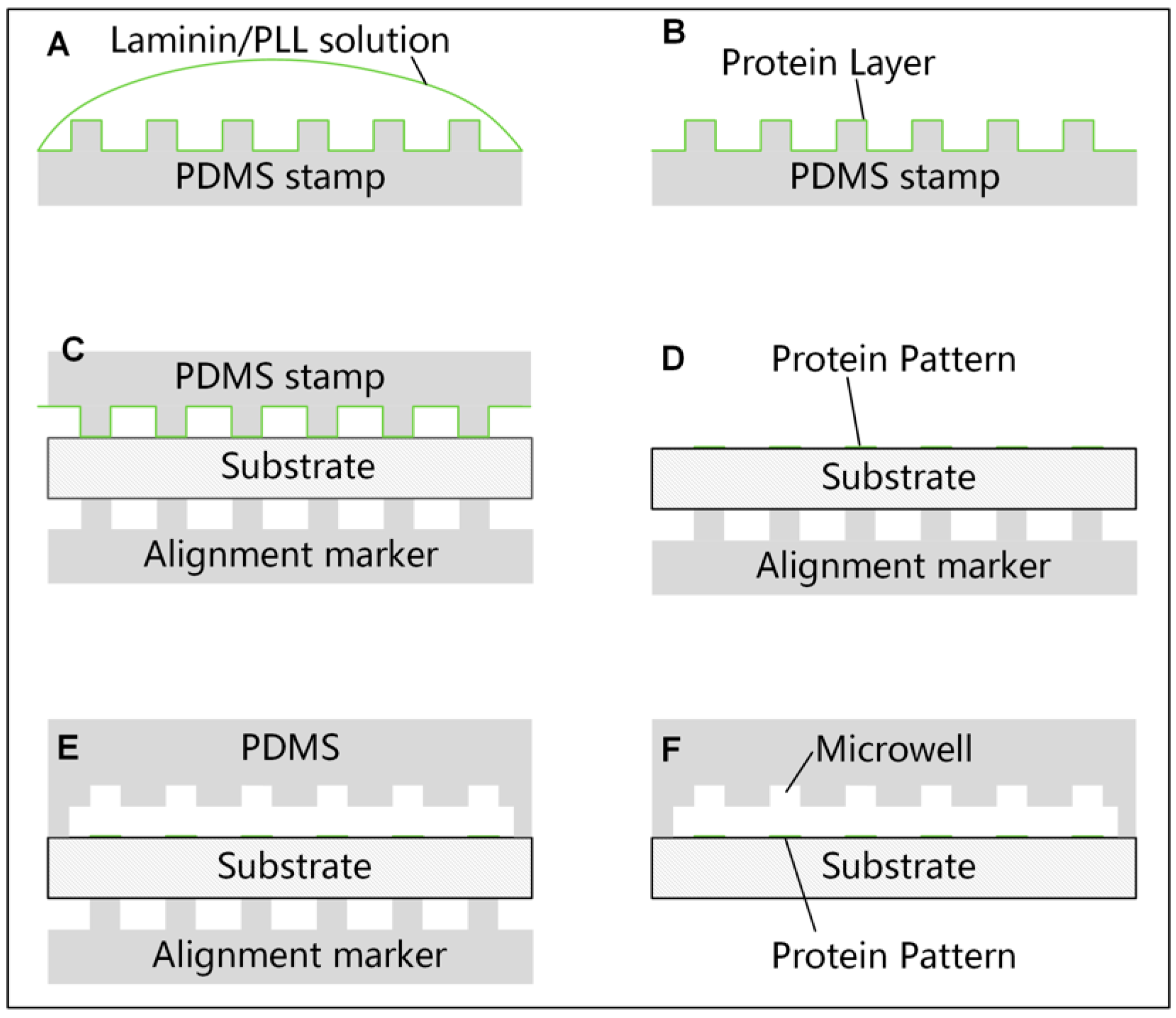
2.1. Micro Contact Printing of Protein on the Substrate
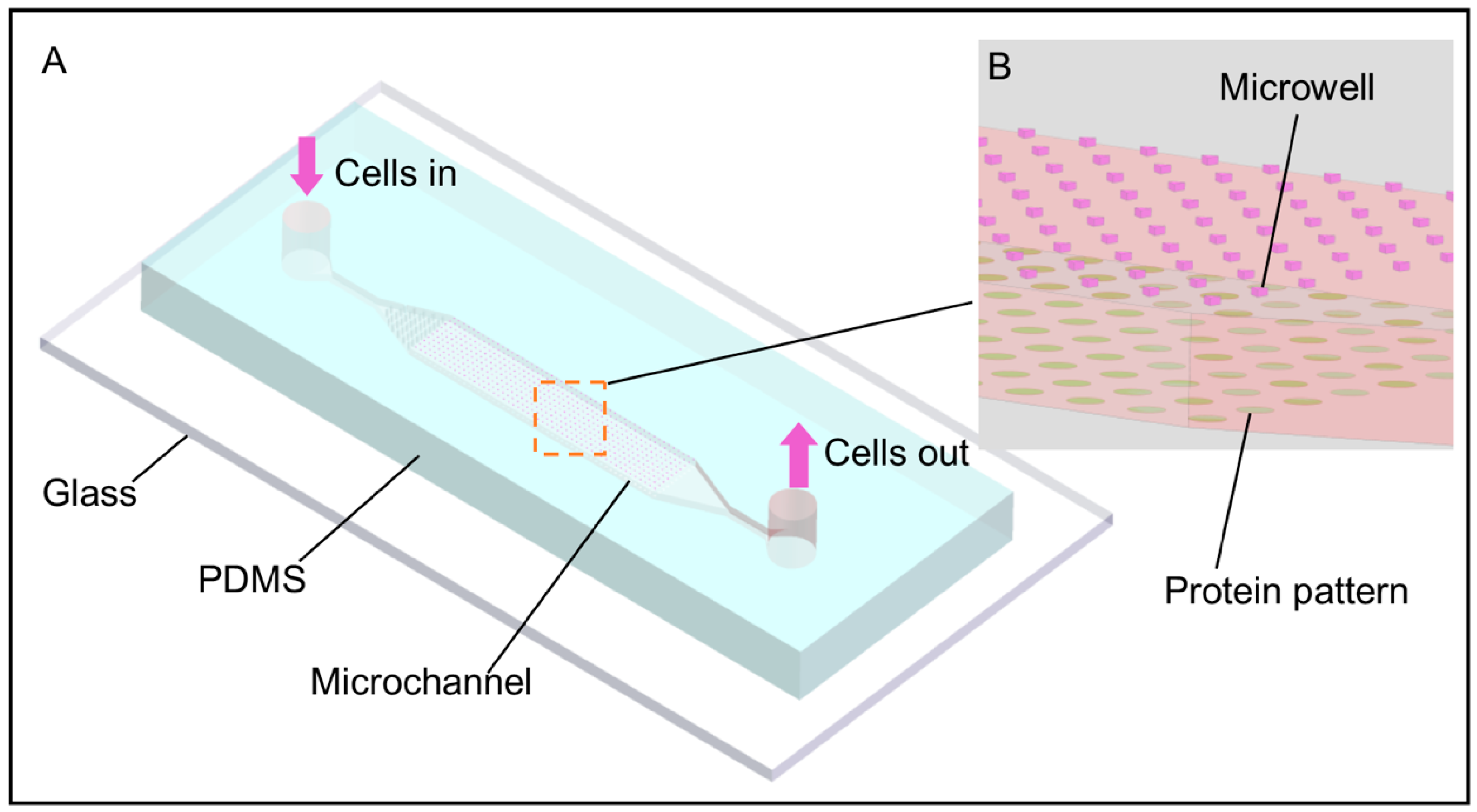
2.2. Fabrication and Preparation of the Microfluidic Chip
2.3. Cell Culture and Cell Suspension Preparation
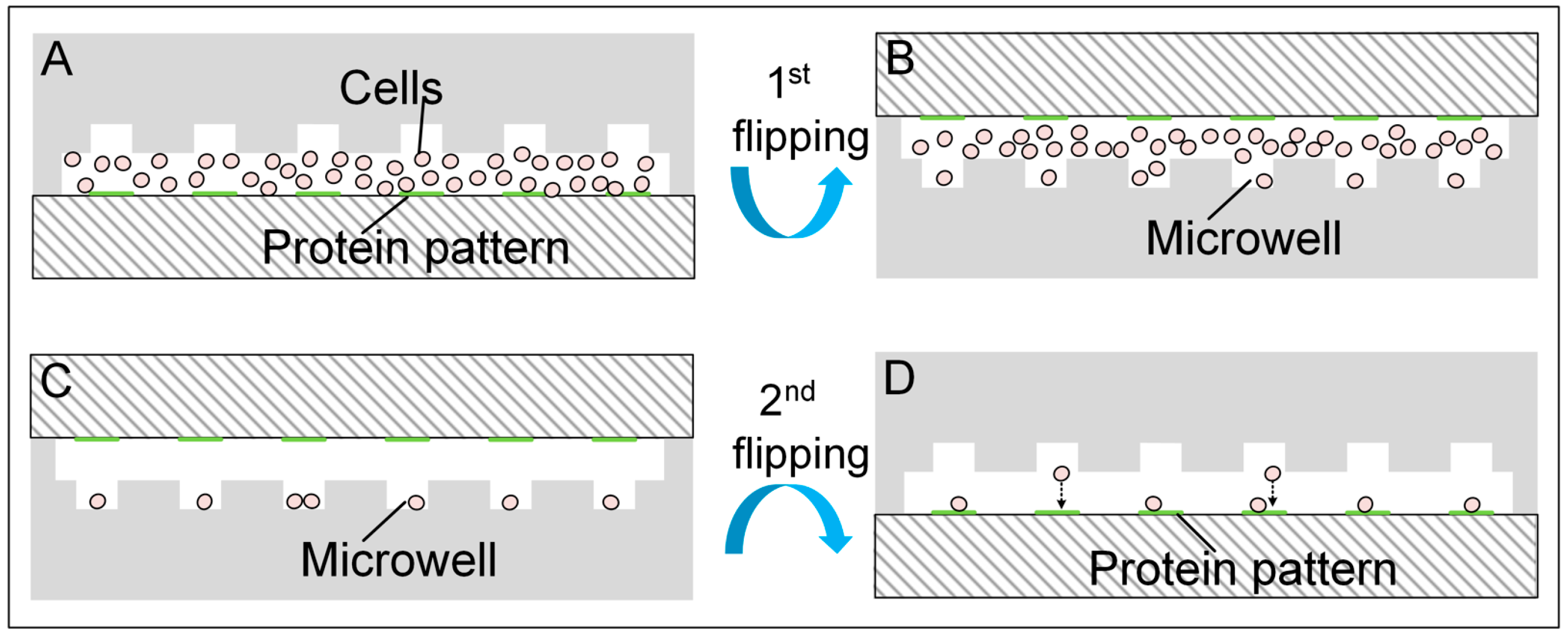
2.4. Cell Loading and Experiment Setup
2.5. Cell Culture in the Microfluidic Chip
2.6. Imaging and Cell Analysis
3. Results and Discussion
3.1. Cell Patterning Microfluidic Device with Paired Microwells and Protein Patterns
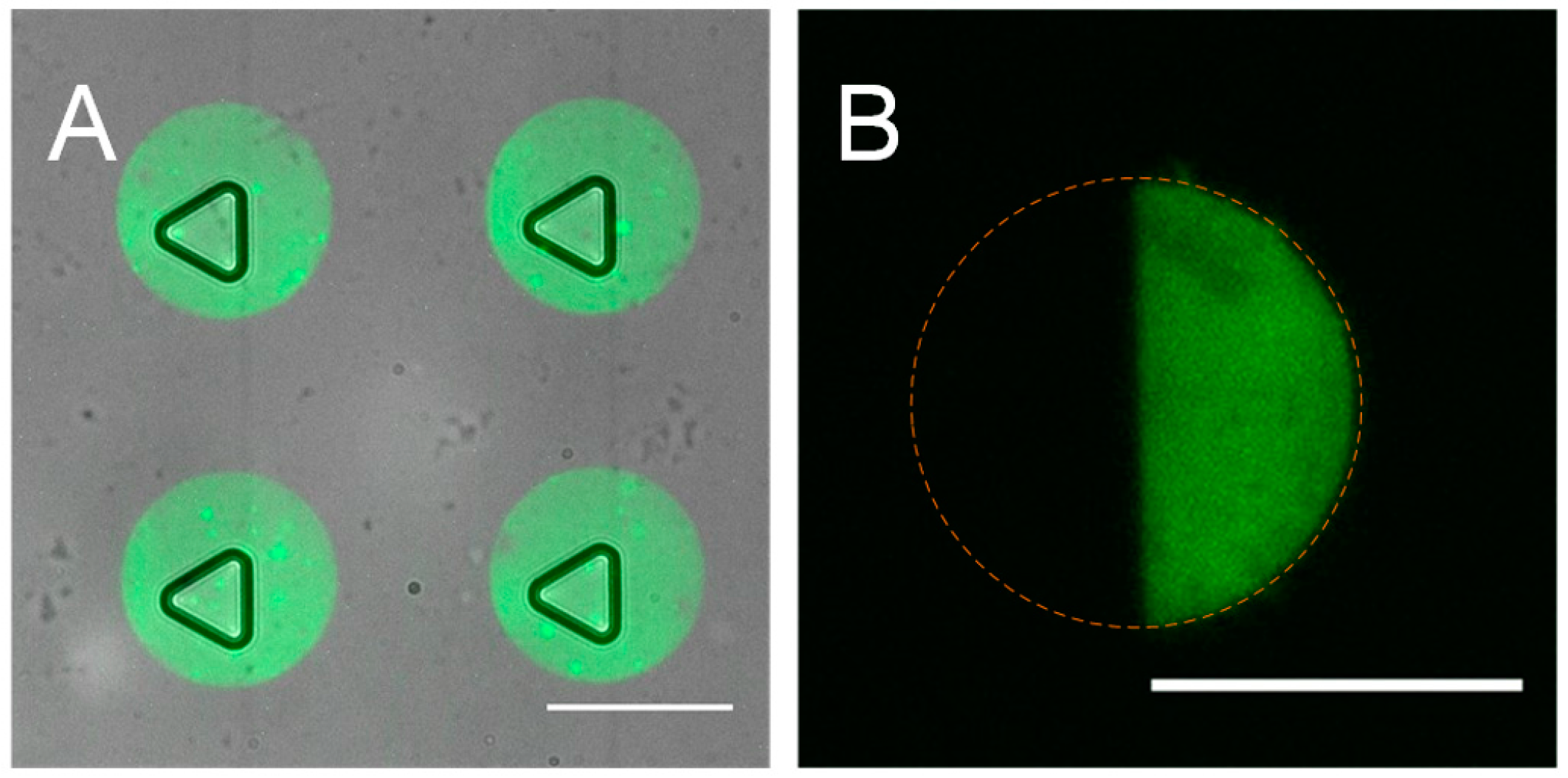
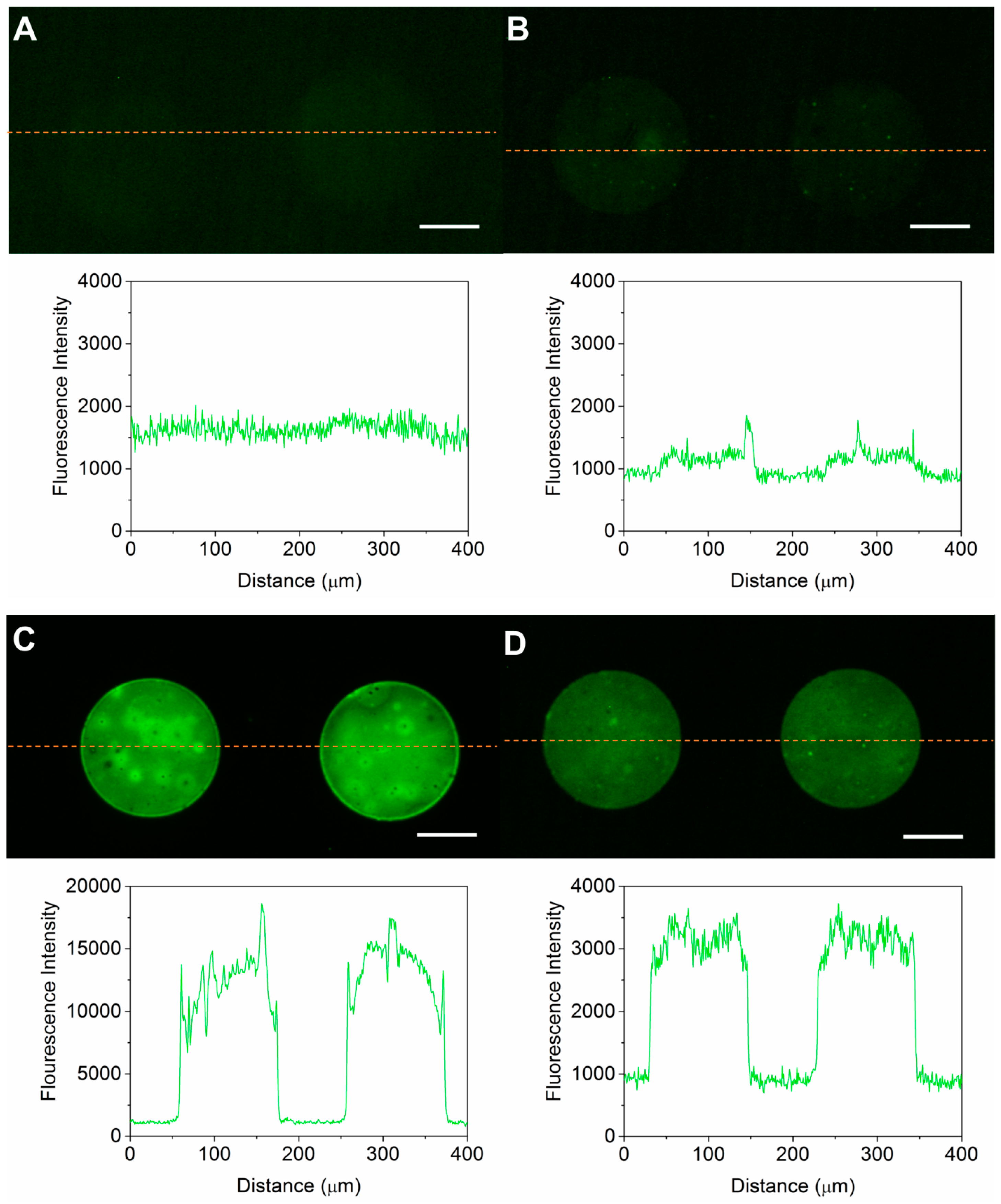
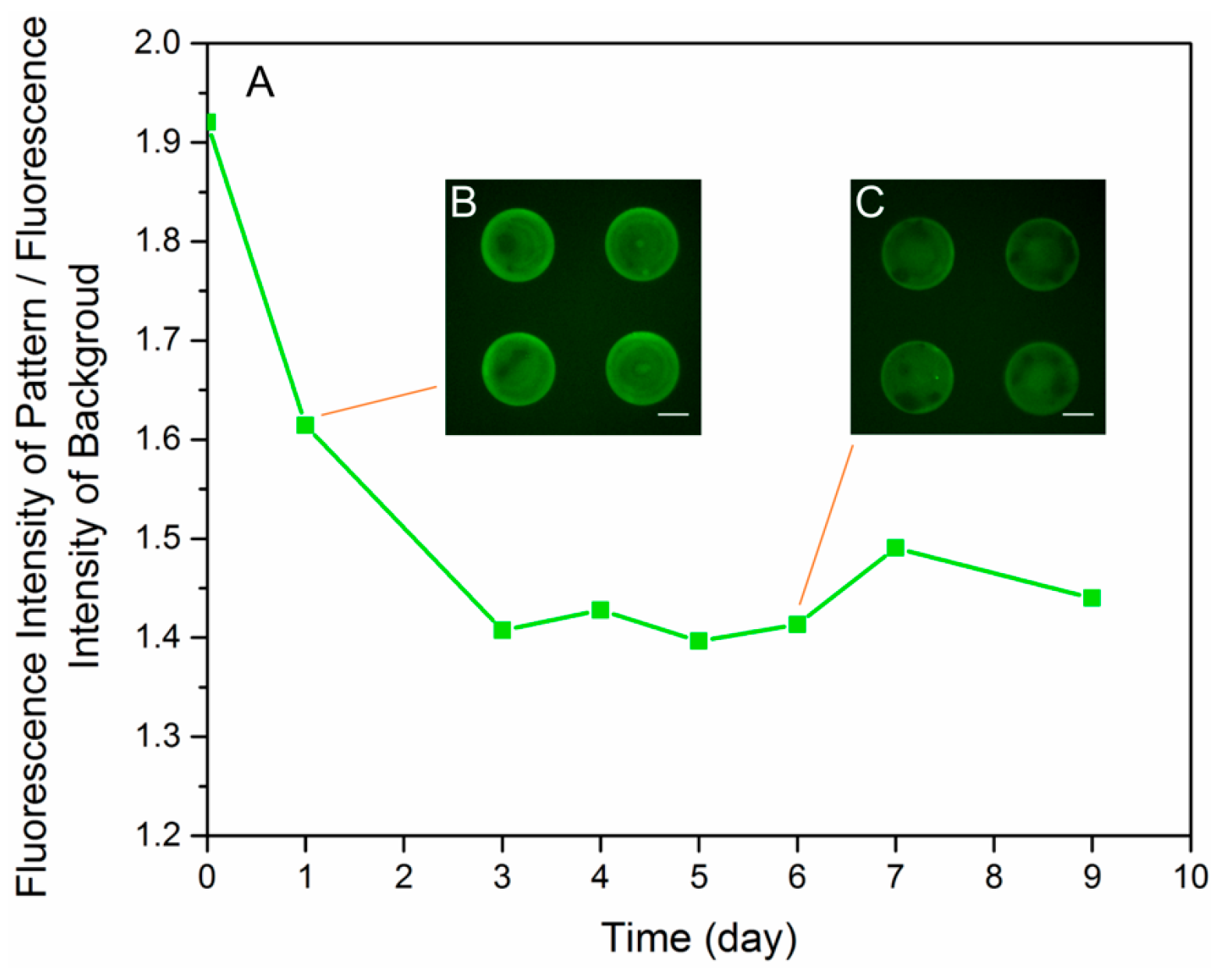
3.2. Optimization of the Micro Contact Printing
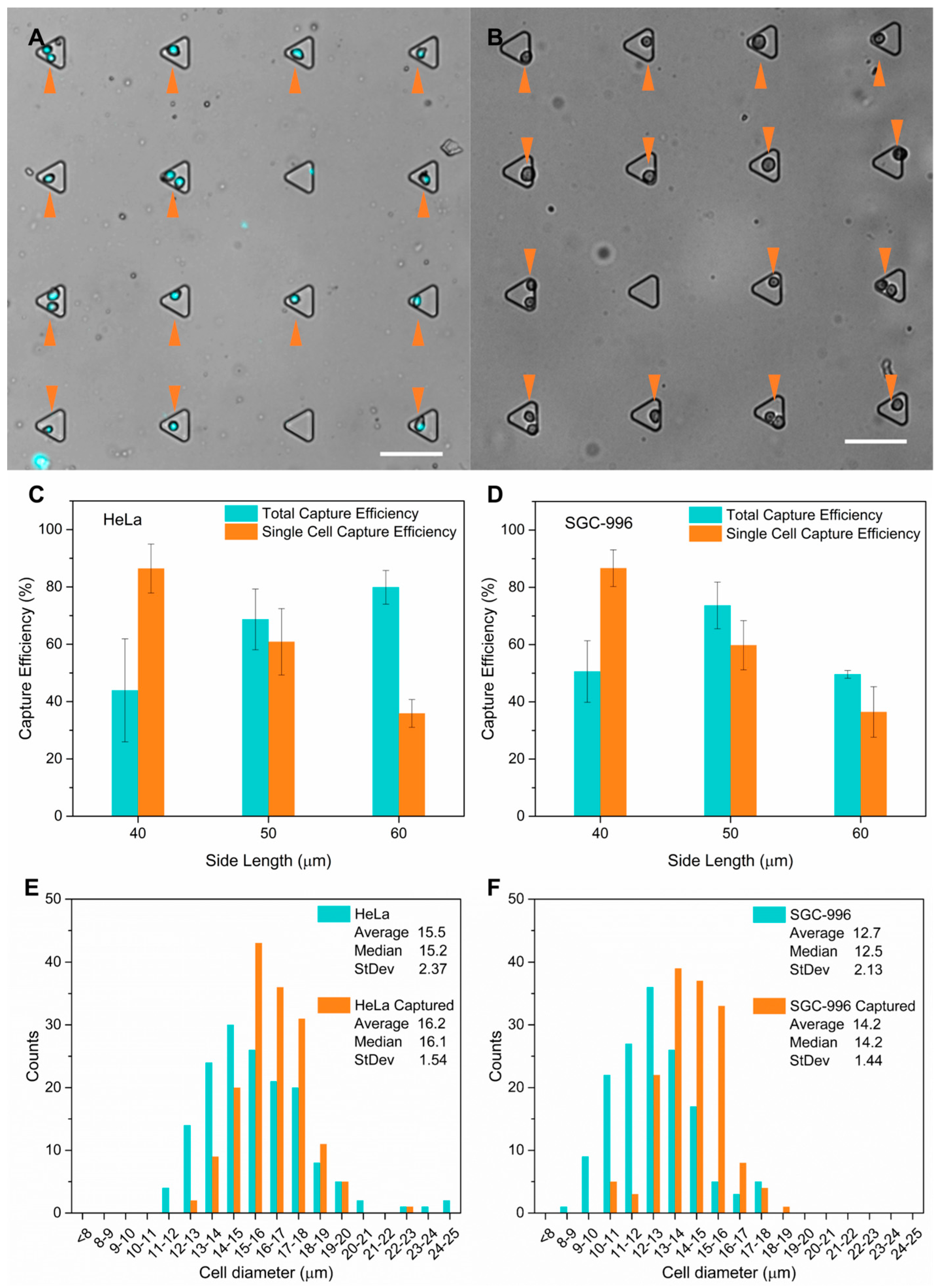
3.3. Cell Capture Performance Demonstrated with HeLa Cells and SGC-996 Cells
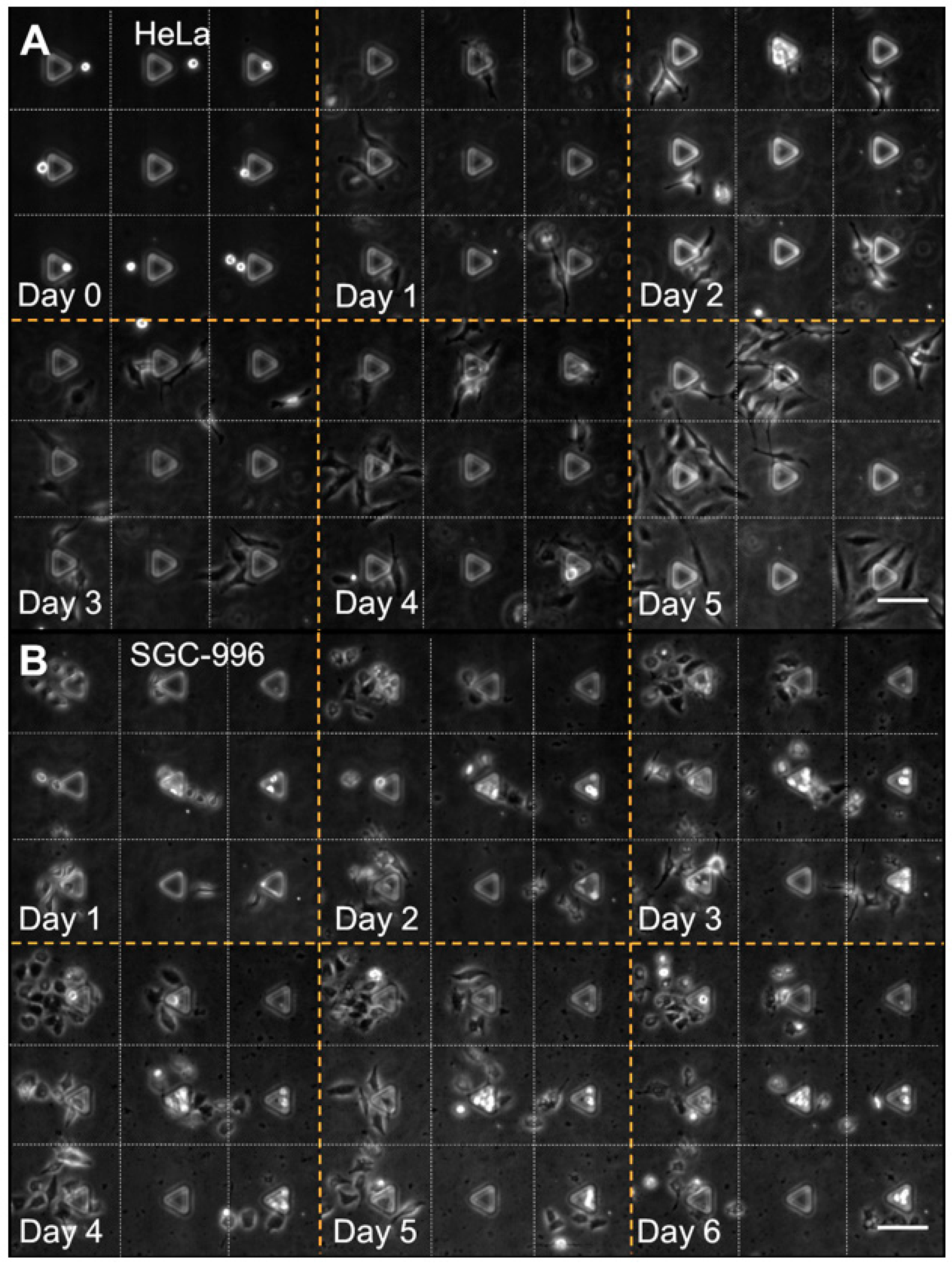
3.4. Cell Patterning Performance with HeLa and SGC-996 Cells
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Csucs, G.; Quirin, K.; Danuser, G. Locomotion of fish epidermal keratocytes on spatially selective adhesion patterns. Cell Motil. Cytoskelet. 2007, 64, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Xia, N.; Thodeti, C.K.; Hunt, T.P.; Xu, Q.B.; Ho, M.; Whitesides, G.M.; Westervelt, R.; Ingber, D.E. Directional control of cell motility through focal adhesion positioning and spatial control of rac activation. FASEB J. 2008, 22, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Thery, M.; Racine, V.; Pepin, A.; Piel, M.; Chen, Y.; Sibarita, J.B.; Bornens, M. The extracellular matrix guides the orientation of the cell division axis. Nat. Cell Biol. 2005, 7, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.A.; Wissner-Gross, Z.D.; Yanik, M.F. Ultra-Rapid laser protein micropatterning: Screening for directed polarization of single neurons. Lab Chip 2012, 12, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Desai, R.A.; Gao, L.; Raghavan, S.; Liu, W.F.; Chen, C.S. Cell polarity triggered by cell-cell adhesion via e-cadherin. J. Cell Sci. 2009, 122, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, A.; Macdonald, A.; Voldman, J. Cell patterning chip for controlling the stem cell microenvironment. Biomaterials 2007, 28, 3208–3216. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Geometric control of cell life and death. Science 1997, 276, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.T.; Lin, R.Z.; Chen, R.J.; Chin, C.K.; Gong, S.E.; Chang, H.Y.; Peng, H.L.; Hsu, L.; Yew, T.R.; Chang, S.F.; et al. Liver-Cell patterning lab chip: Mimicking the morphology of liver lobule tissue. Lab Chip 2013, 13, 3578–3587. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A.; Langer, R.; Borenstein, J.; Vacanti, J.P. Microscale technologies for tissue engineering and biology. Proc. Natl. Acad. Sci. USA 2006, 103, 2480–2487. [Google Scholar] [CrossRef] [PubMed]
- Hardelauf, H.; Waide, S.; Sisnaiske, J.; Jacob, P.; Hausherr, V.; Schobel, N.; Janasek, D.; van Thriel, C.; West, J. Micropatterning neuronal networks. Analyst 2014, 139, 3256–3264. [Google Scholar] [CrossRef] [PubMed]
- Takayama, Y.; Kotake, N.; Haga, T.; Suzuki, T.; Mabuchi, K. Formation of one-way-structured cultured neuronal networks in microfluidic devices combining with micropatterning techniques. J. Biosci. Bioeng. 2012, 114, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Mrksich, M.; Whitesides, G.M. Patterning self-assembled monolayers using microcontact printing: A new technology for biosensors? Trends Biotechnol. 1995, 13, 228–235. [Google Scholar] [CrossRef]
- Hynes, W.F.; Doty, N.J.; Zarembinski, T.I.; Schwartz, M.P.; Toepke, M.W.; Murphy, W.L.; Atzet, S.K.; Clark, R.; Melendez, J.A.; Cady, N.C. Micropatterning of 3D microenvironments for living biosensor applications. Biosensors 2014, 4, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.B.; Zhang, Y.; Yu, J.J.; Mak, A.F.T.; Li, Y.; Yang, M. A microfluidic chip with poly(ethylene glycol) hydrogel microarray on nanoporous alumina membrane for cell patterning and drug testing. Sens. Actuators B Chem. 2010, 143, 776–783. [Google Scholar] [CrossRef]
- Millet, L.J.; Gillette, M.U. New perspectives on neuronal development via microfluidic environments. Trends Neurosci. 2012, 35, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Weaver, W.M.; Tseng, P.; Kunze, A.; Masaeli, M.; Chung, A.J.; Dudani, J.S.; Kittur, H.; Kulkarni, R.P.; Di Carlo, D. Advances in high-throughput single-cell microtechnologies. Curr. Opin. Biotechnol. 2014, 25, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Yusof, A.; Keegan, H.; Spillane, C.D.; Sheils, O.M.; Martin, C.M.; O’Leary, J.J.; Zengerle, R.; Koltay, P. Inkjet-Like printing of single-cells. Lab Chip 2011, 11, 2447–2454. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, M.; Pisanic, T.; Scheel, J.; Barlow, C.; Esener, S.; Bhatia, S.N. Electro-Optical platform for the manipulation of live cells. Langmuir 2003, 19, 1532–1538. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, K.K. Optical tweezers for single cells. J. R. Soc. Interface 2008, 5, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, A.; Voldman, J. Dielectrophoretic traps for single-particle patterning. Biophys J. 2005, 88, 2193–2205. [Google Scholar] [CrossRef] [PubMed]
- Gray, D.S.; Tan, J.L.; Voldman, J.; Chen, C.S. Dielectrophoretic registration of living cells to a microelectrode array. Biosens. Bioelectron. 2004, 19, 1765–1774. [Google Scholar] [CrossRef]
- Odde, D.J.; Renn, M.J. Laser-Guided direct writing of living cells. Biotechnol. Bioeng. 2000, 67, 312–318. [Google Scholar] [CrossRef]
- Schiele, N.R.; Corr, D.T.; Huang, Y.; Raof, N.A.; Xie, Y.; Chrisey, D.B. Laser-based direct-write techniques for cell printing. Biofabrication 2010, 2, 032001. [Google Scholar] [CrossRef] [PubMed]
- Rettig, J.R.; Folch, A. Large-Scale single-cell trapping and imaging using microwell arrays. Anal. Chem. 2005, 77, 5628–5634. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-H.; Hsiao, Y.-H.; Chang, H.-C.; Yeh, C.-F.; He, C.-K.; Salm, E.M.; Chen, C.; Chiu, I.-M.; Hsu, C.-H. A microfluidic dual-well device for high-throughput single-cell capture and culture. Lab Chip 2015, 15, 2928–2938. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A.; Ferreira, L.; Blumling, J., 3rd; Yeh, J.; Karp, J.M.; Fukuda, J.; Langer, R. Co-Culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates. Biomaterials 2006, 27, 5968–5977. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Morgan, M.; Sachs, A.N.; Samorezov, J.; Teller, R.; Shen, Y.; Pienta, K.J.; Takayama, S. Single cell trapping in larger microwells capable of supporting cell spreading and proliferation. Microfluid. Nanofluid. 2010, 8, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D.; Wu, L.Y.; Lee, L.P. Dynamic single cell culture array. Lab Chip 2006, 6, 1445–1449. [Google Scholar] [CrossRef] [PubMed]
- Skelley, A.M.; Kirak, O.; Suh, H.; Jaenisch, R.; Voldman, J. Microfluidic control of cell pairing and fusion. Nat. Methods 2009, 6, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chou, C.K.; Xia, X.; Hung, M.C.; Qin, L. Block-Cell-Printing for live single-cell printing. Proc. Natl. Acad. Sci. USA 2014, 111, 2948–2953. [Google Scholar] [CrossRef] [PubMed]
- Kane, R.S.; Takayama, S.; Ostuni, E.; Ingber, D.E.; Whitesides, G.M. Patterning proteins and cells using soft lithography. Biomaterials 1999, 20, 2363–2376. [Google Scholar] [CrossRef]
- Théry, M.; Piel, M. Adhesive micropatterns for cells: A microcontact printing protocol. Cold Spring Harb. Protoc. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- Corbin, E.A.; Dorvel, B.R.; Millet, L.J.; King, W.P.; Bashir, R. Micro-Patterning of mammalian cells on suspended MEMS resonant sensors for long-term growth measurements. Lab Chip 2014, 14, 1401–1404. [Google Scholar] [CrossRef] [PubMed]
- Millet, L.J.; Collens, M.B.; Perry, G.L.; Bashir, R. Pattern analysis and spatial distribution of neurons in culture. Integr. Biol. Quant. Biosci. Nano Macro 2011, 3, 1167–1178. [Google Scholar] [CrossRef] [PubMed]
- Jing, G.; Perry, S.F.; Tatic-Lucic, S. Precise cell patterning using cytophobic self-assembled monolayer deposited on top of semi-transparent gold. Biomed. Microdevices 2010, 12, 935–948. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Nedelec, S.; Wichterle, H.; Kam, L.C. Combined microfluidics/protein patterning platform for pharmacological interrogation of axon pathfinding. Lab Chip 2010, 10, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Millet, L.J.; Stewart, M.E.; Nuzzo, R.G.; Gillette, M.U. Guiding neuron development with planar surface gradients of substrate cues deposited using microfluidic devices. Lab Chip 2010, 10, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Ostuni, E.; Chen, C.S.; Ingber, D.E.; Whitesides, G.M. Selective deposition of proteins and cells in arrays of microwells. Langmuir 2001, 17, 2828–2834. [Google Scholar] [CrossRef]
- Rhee, S.W.; Taylor, A.M.; Tu, C.H.; Cribbs, D.H.; Cotman, C.W.; Jeon, N.L. Patterned cell culture inside microfluidic devices. Lab Chip 2005, 5, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Chu, Y.-S.; Thiery, J.P.; Lim, C.T.; Rodriguez, I. Microfluidic cell trap array for controlled positioning of single cells on adhesive micropatterns. Lab Chip 2013, 13, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Torisawa, Y.S.; Mosadegh, B.; Luker, G.D.; Morell, M.; O’Shea, K.S.; Takayama, S. Microfluidic hydrodynamic cellular patterning for systematic formation of co-culture spheroids. Integr. Biol. Quant. Biosci. Nano Macro 2009, 1, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.T.; Jeon, N.L.; Huang, S.; Kane, R.S.; Wargo, C.J.; Choi, I.S.; Ingber, D.E.; Whitesides, G.M. Patterned deposition of cells and proteins onto surfaces by using three-dimensional microfluidic systems. Proc. Natl. Acad. Sci. USA 2000, 97, 2408–2413. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.R.; Cox, E.C.; Austin, R.H.; Sturm, J.C. Continuous particle separation through deterministic lateral displacement. Science 2004, 304, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.L.; Tien, J.; Chen, C.S. Microcontact printing of proteins on mixed self-assembled monolayers. Langmuir 2002, 18, 519–523. [Google Scholar] [CrossRef]
- Alom Ruiz, S.; Chen, C.S. Microcontact printing: A tool to pattern. Soft Matter 2007, 3, 168–177. [Google Scholar] [CrossRef]
- Walker, G.M.; Zeringue, H.C.; Beebe, D.J. Microenvironment design considerations for cellular scale studies. Lab Chip 2004, 4, 91–97. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, C.; Huang, B.; Zhou, J.; Liang, Y.; Tian, J.; Ji, L.; Liang, X.; Ye, X. A Microfluidic Chip for Cell Patterning Utilizing Paired Microwells and Protein Patterns. Micromachines 2017, 8, 1. https://doi.org/10.3390/mi8010001
Tu C, Huang B, Zhou J, Liang Y, Tian J, Ji L, Liang X, Ye X. A Microfluidic Chip for Cell Patterning Utilizing Paired Microwells and Protein Patterns. Micromachines. 2017; 8(1):1. https://doi.org/10.3390/mi8010001
Chicago/Turabian StyleTu, Chunlong, Bobo Huang, Jian Zhou, Yitao Liang, Jian Tian, Lin Ji, Xiao Liang, and Xuesong Ye. 2017. "A Microfluidic Chip for Cell Patterning Utilizing Paired Microwells and Protein Patterns" Micromachines 8, no. 1: 1. https://doi.org/10.3390/mi8010001





