Cytostretch, an Organ-on-Chip Platform
Abstract
:1. Introduction
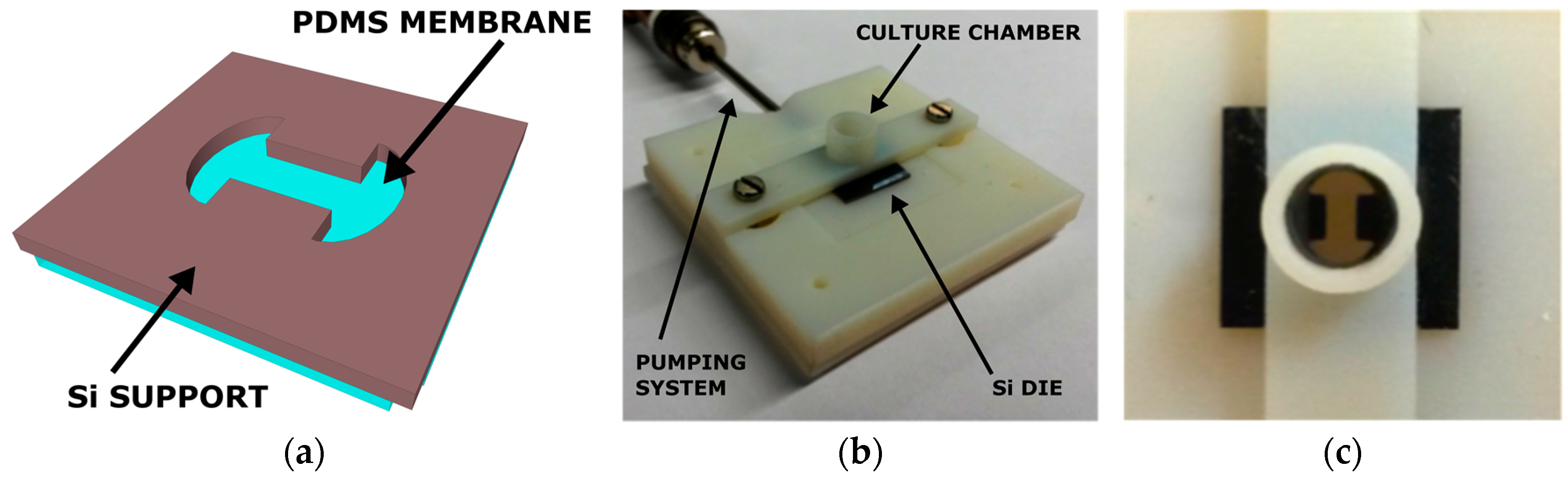
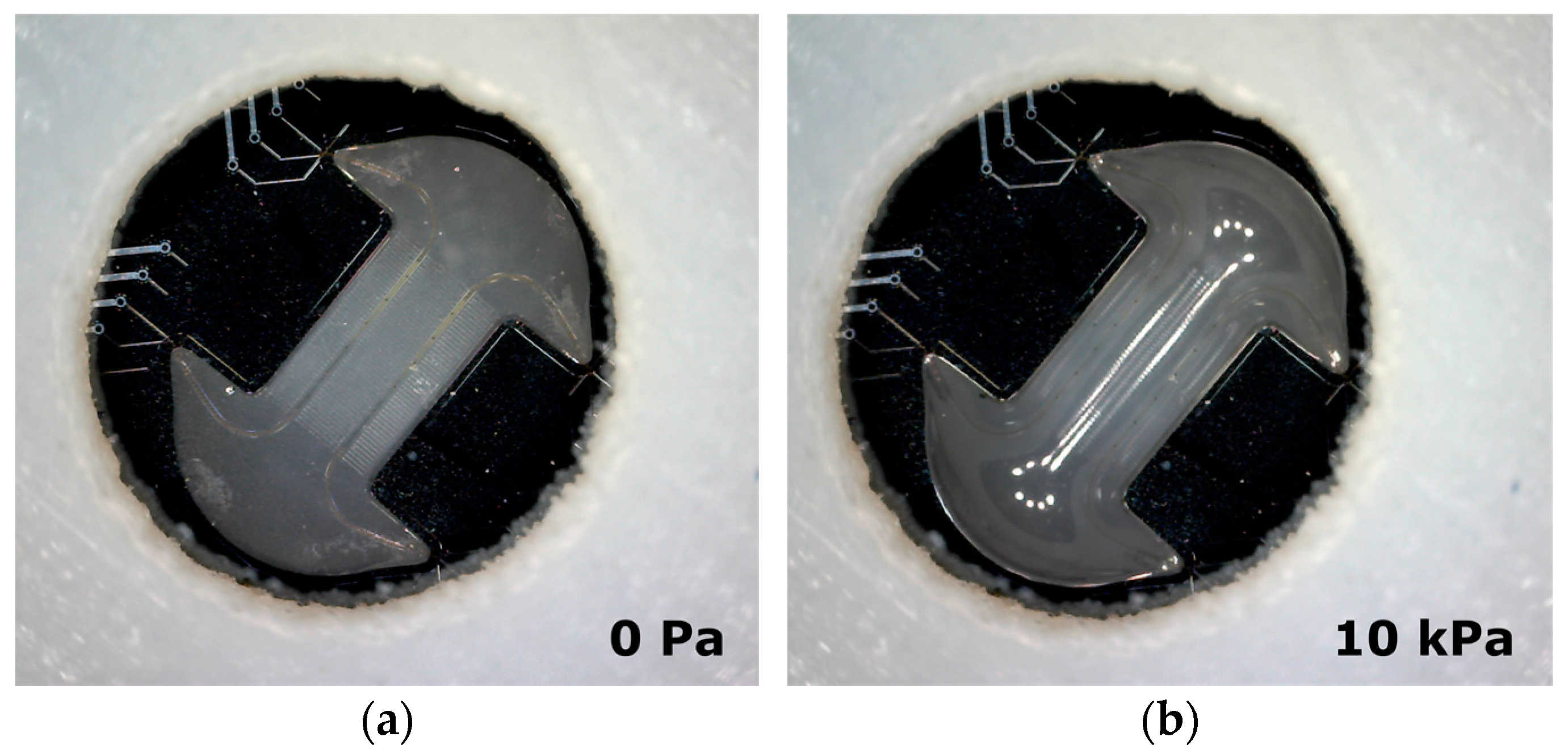
2. Cytostretch Membrane Product Platform
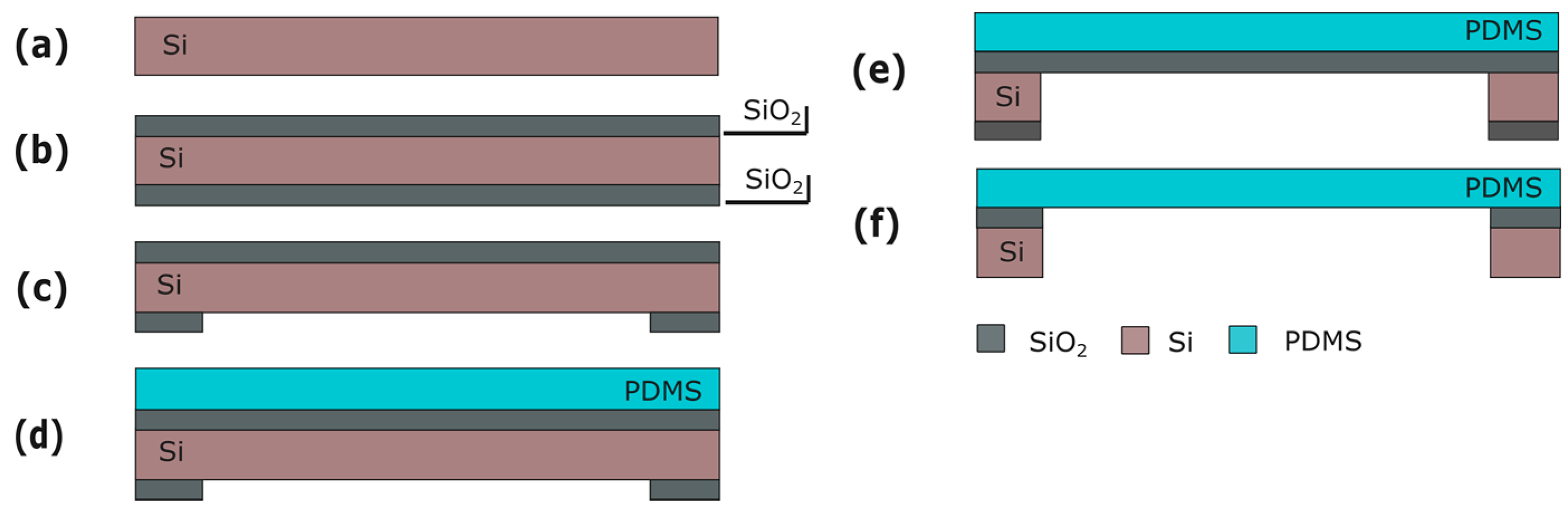
Fabrication
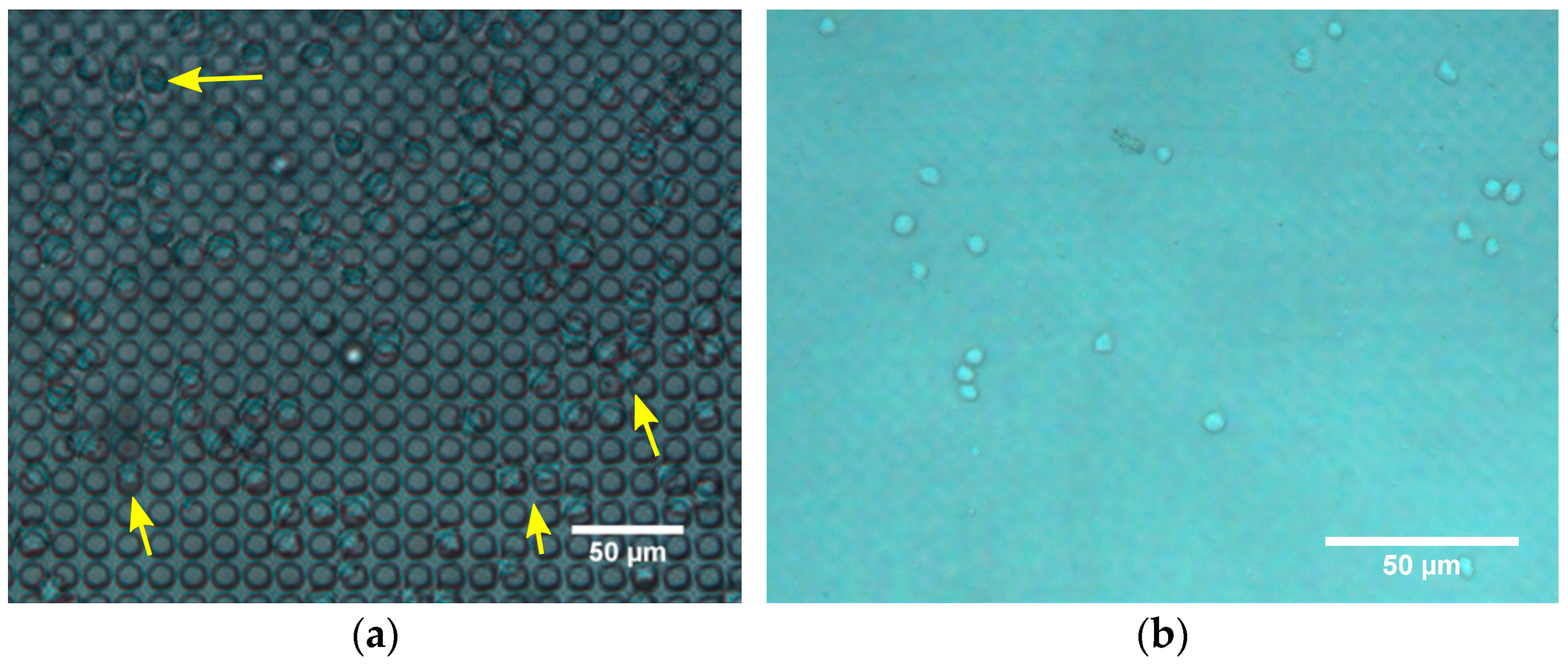
3. Through-Membrane Micro-Pores
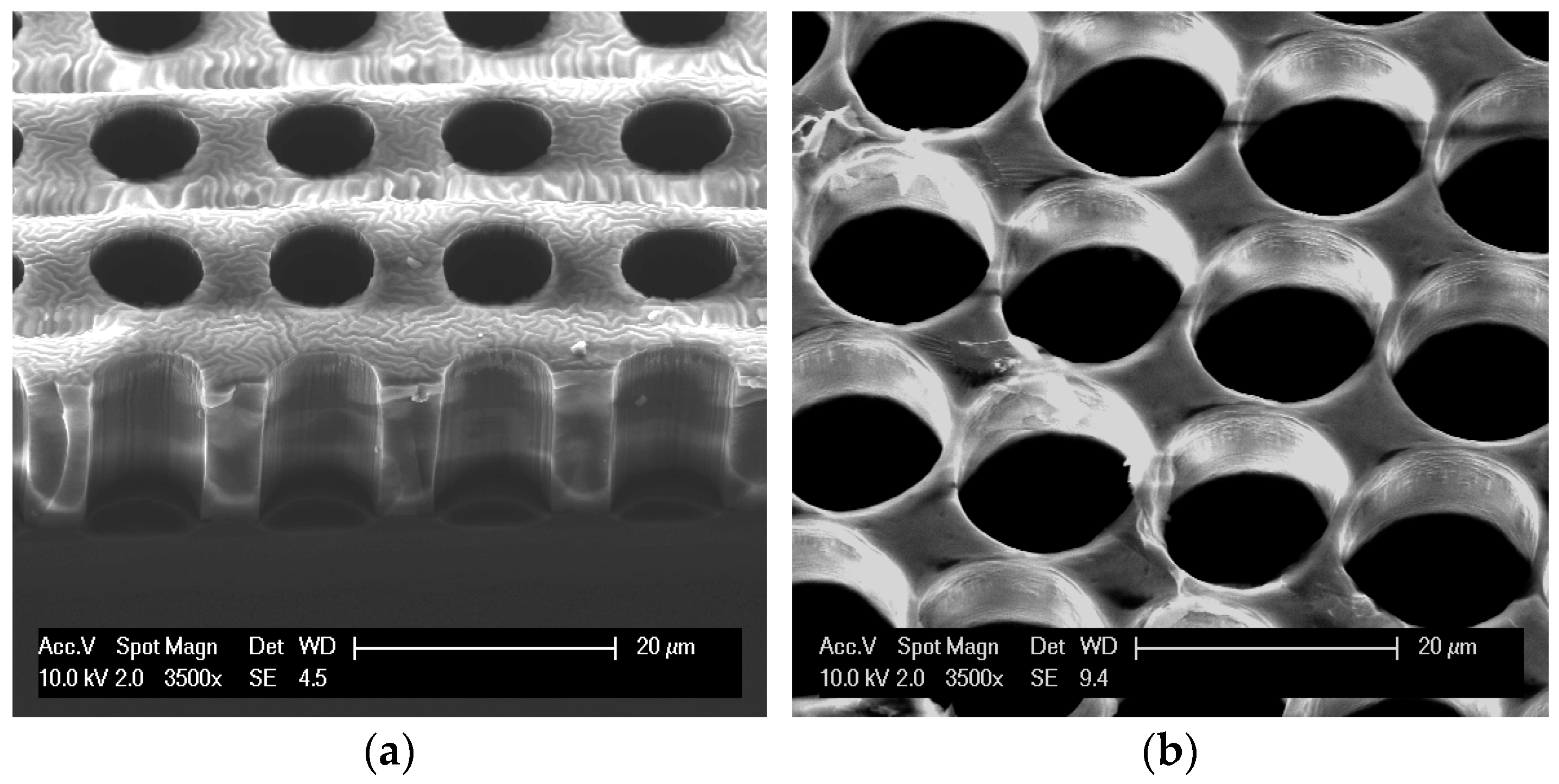
3.1. Fabrication
3.2. Characterization
3.3. Results and Discussion
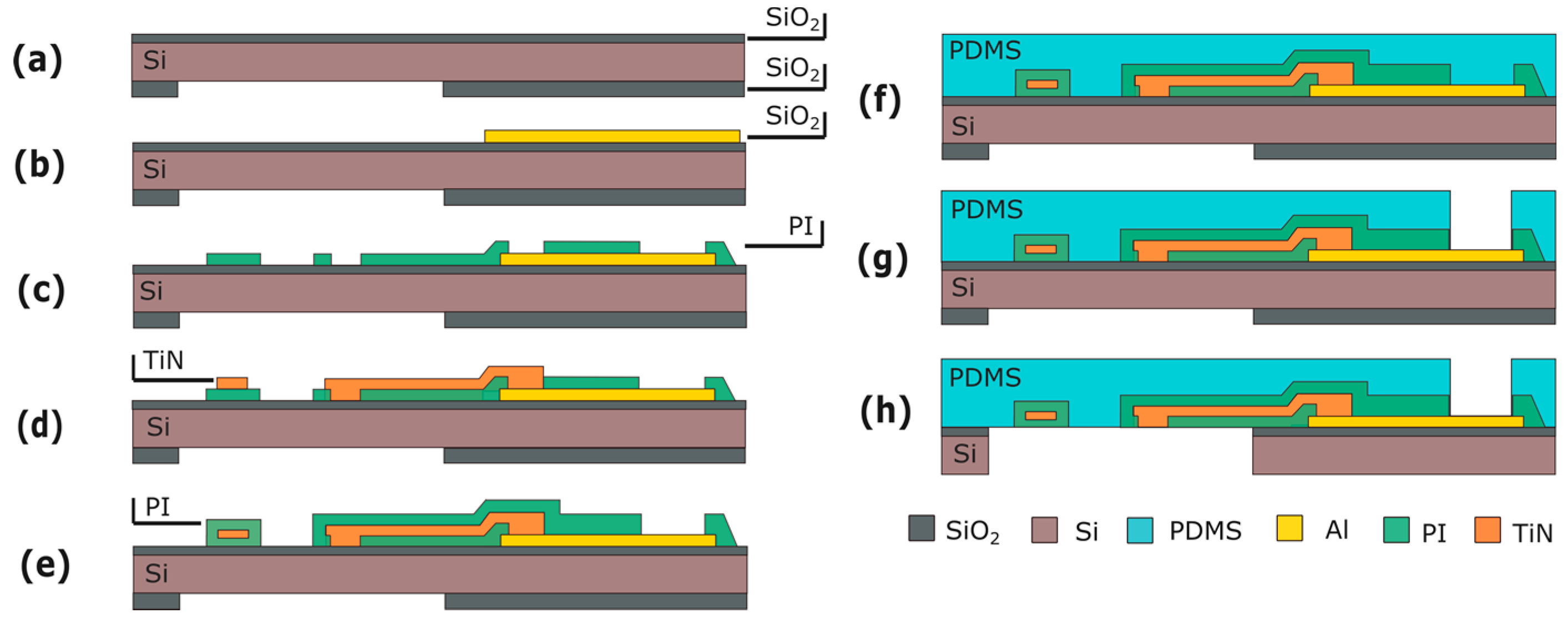
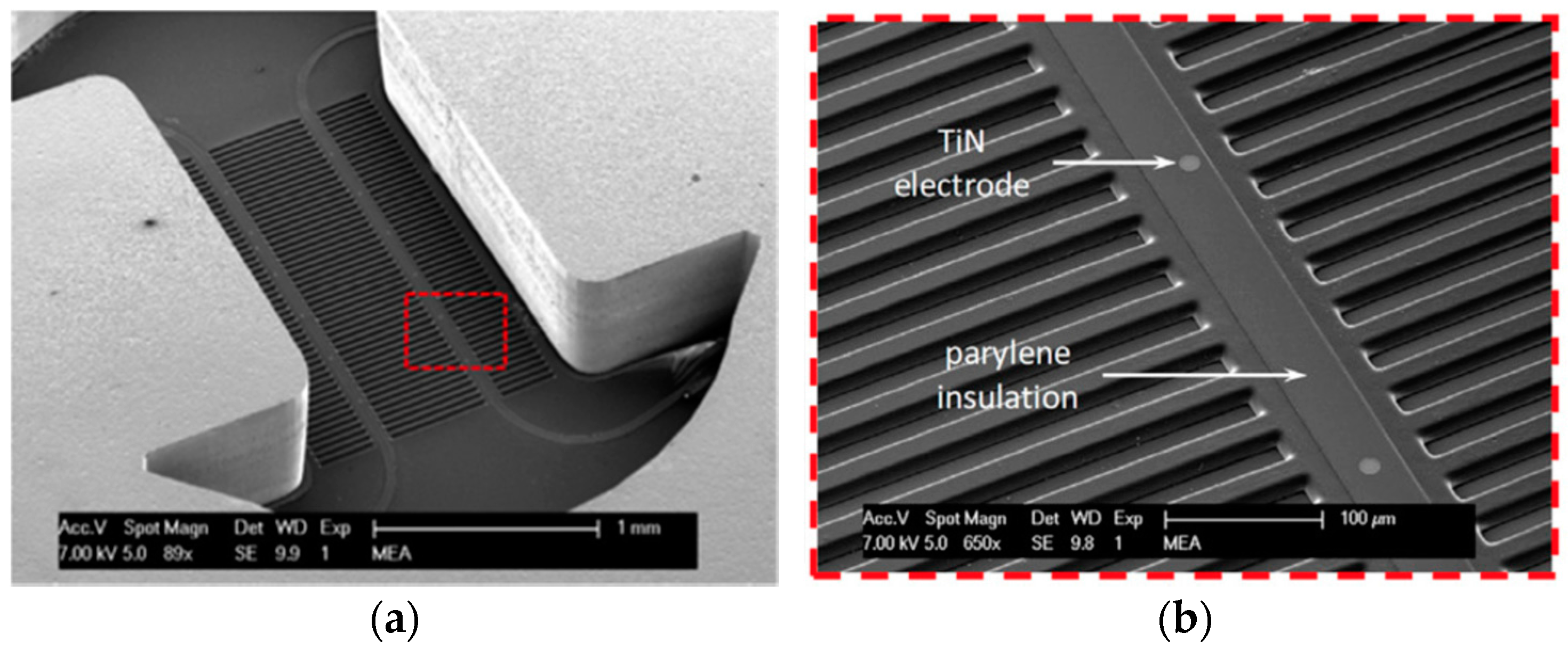
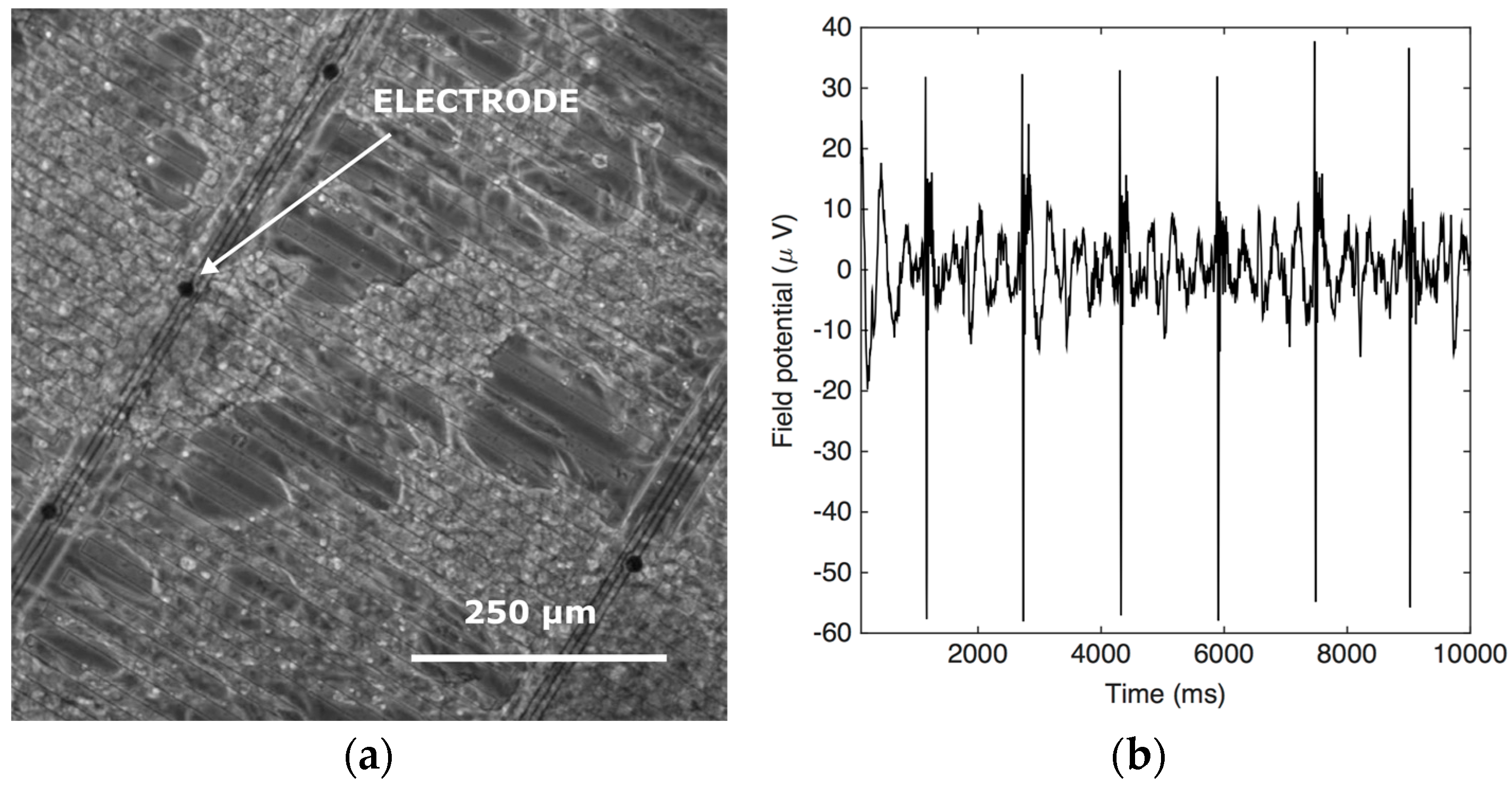
4. Micro-Electrode Array
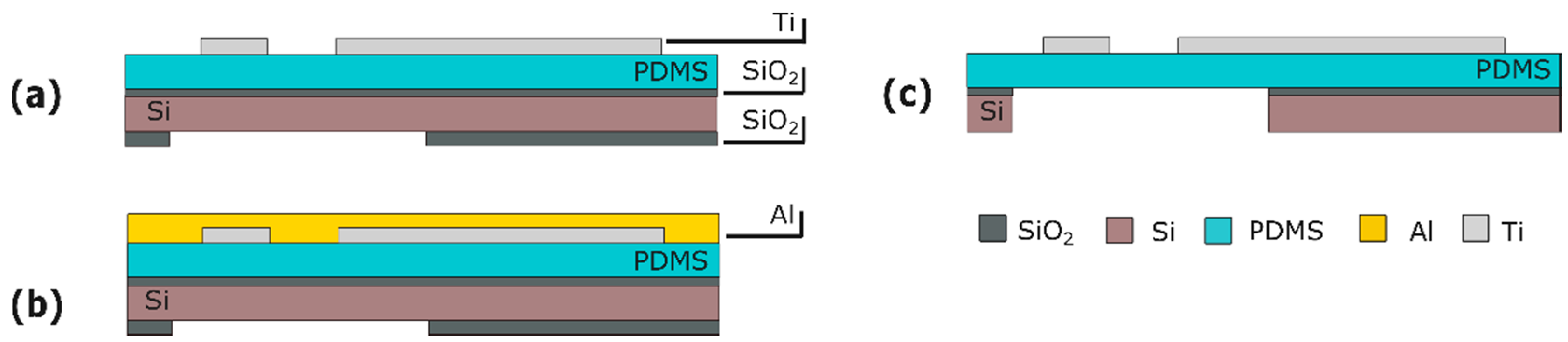
4.1. Fabrication
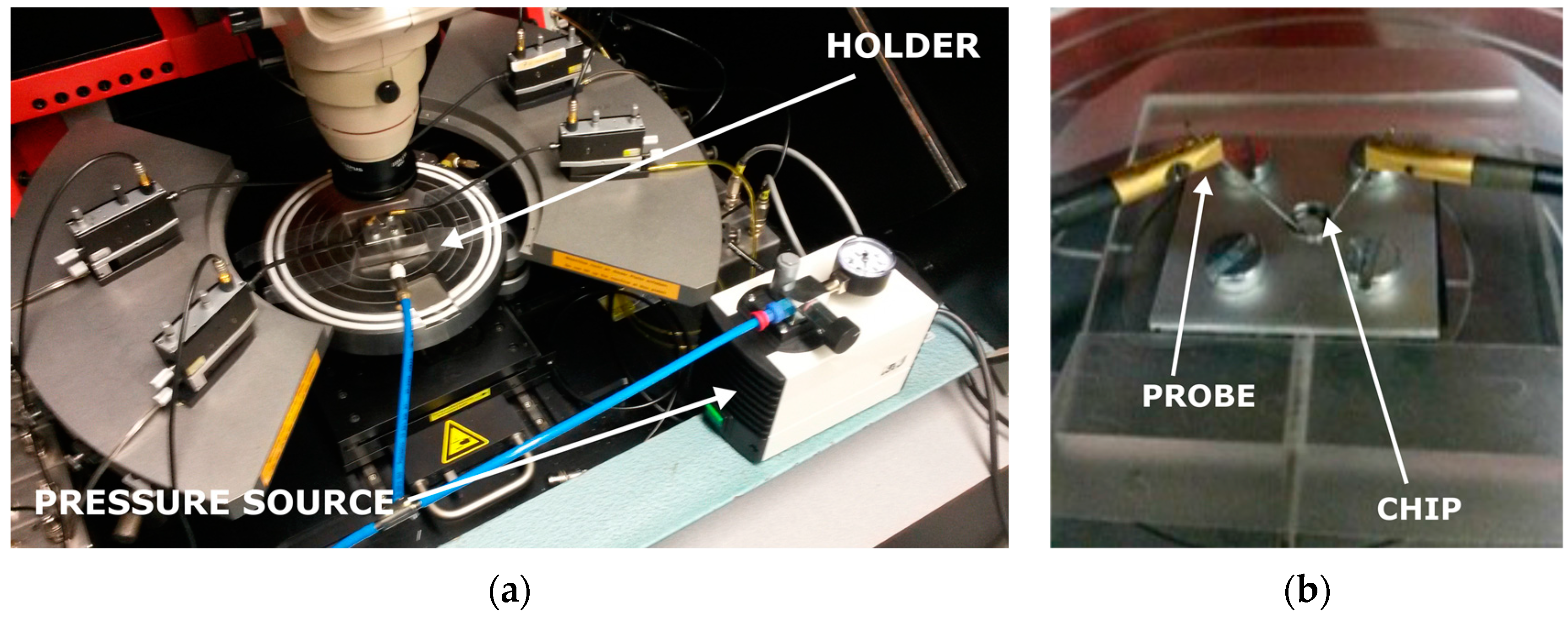
4.2. Characterization
4.3. Results and Discussion
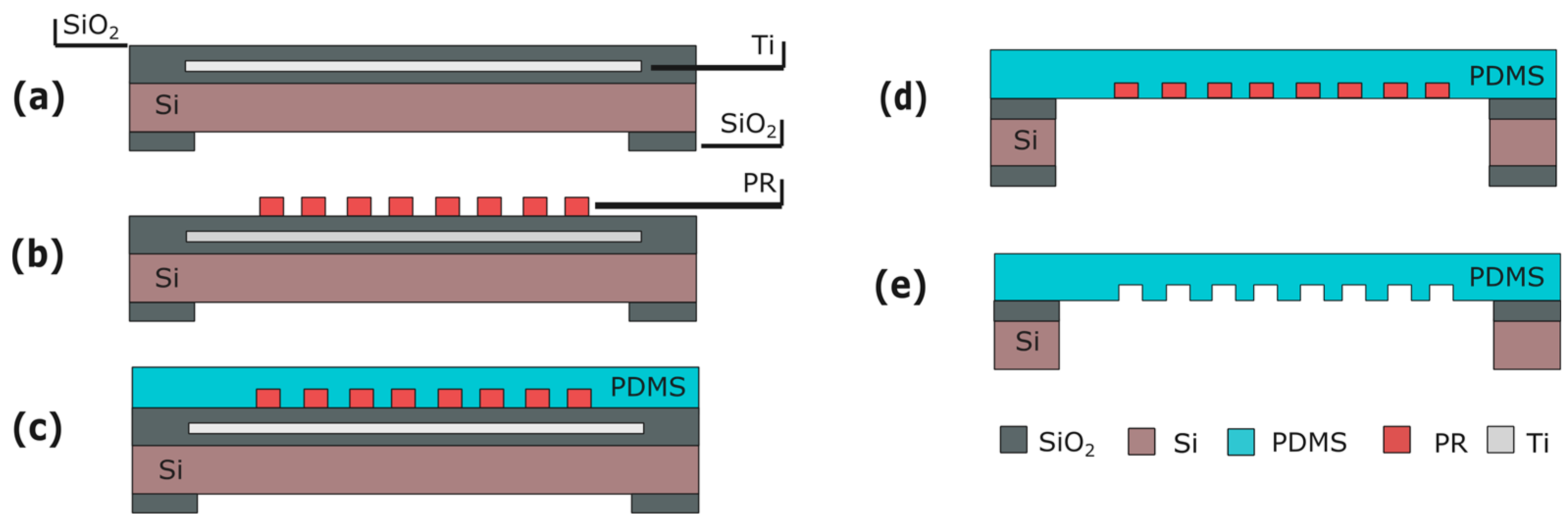
5. Micro-Grooves
5.1. Fabrication
5.2. Characterization
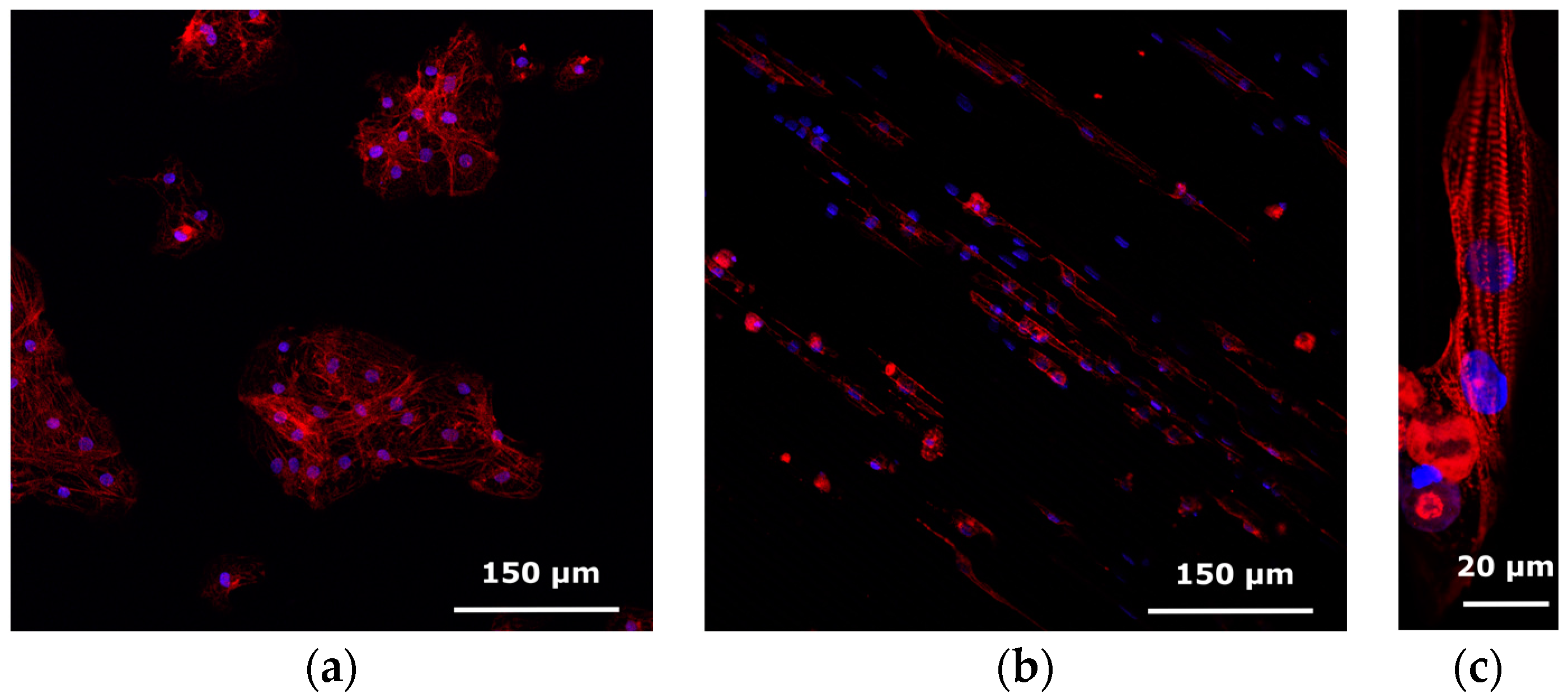
5.3. Results and Discussion
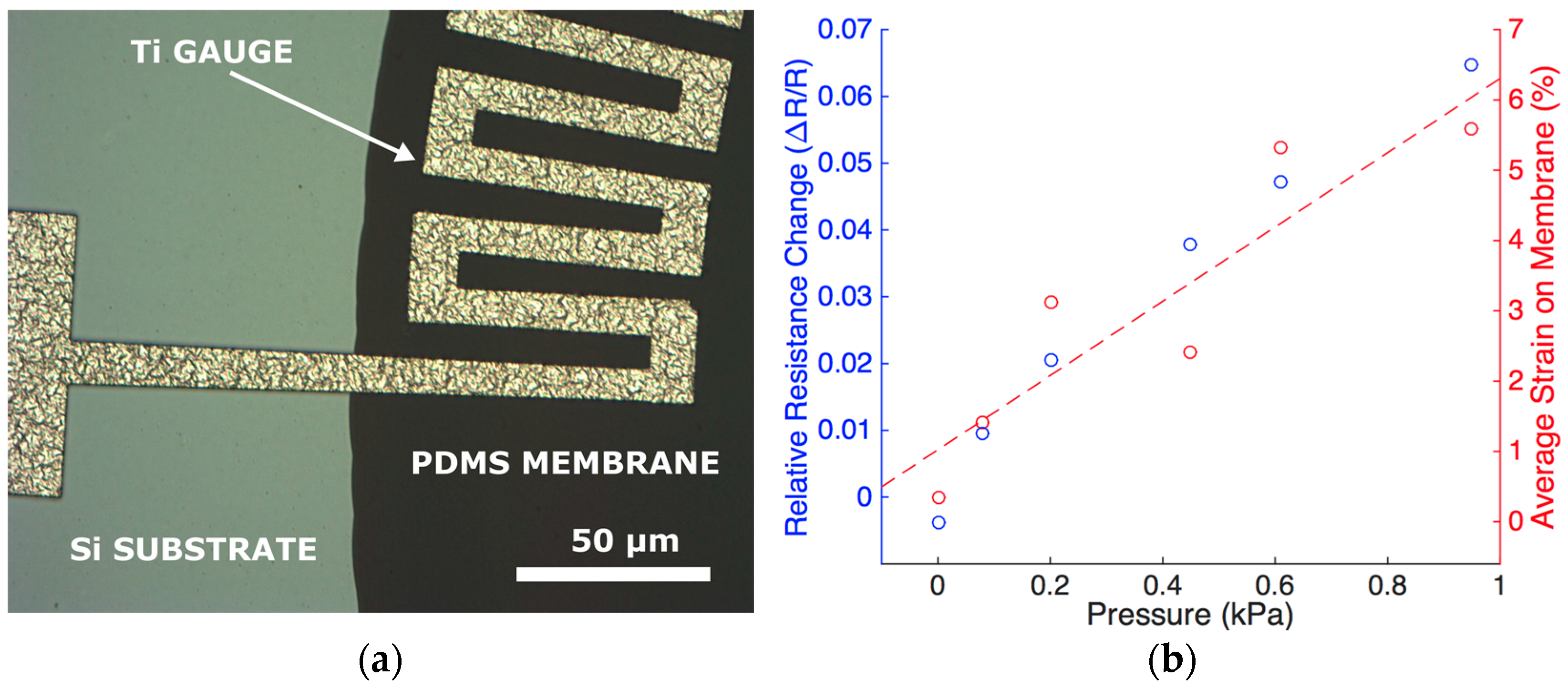
6. Strain Gauges
6.1. Fabrication
6.2. Characterization
6.3. Results and Discussion
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| OOC | Organ-on-Chip |
| PDMS | Polydimethylsiloxane |
| hPSC | Human Pluripotent Stem Cell |
| hPSC-CM | hPSC Derived Cardiomyocytes |
| SEM | Scanning Electron Microscopy |
| MEA | Micro-Electrode Array |
| ECM | Extra Cellular Matrix |
| PR | PhotoResist |
References
- Gassmann, O.; Reepmeyer, G.; von Zedtwitz, M. Trends and drivers for growth in the pharmaceutical industry. In Leading Pharmaceutical Innovation; Springer Science & Business Media: Berlin, Germany, 2008. [Google Scholar]
- Beißner, N.; Lorenz, T.; Reichl, S. Organ on chip. In Microsystems for Pharmatechnology; Springer International Publishing: Gewerbestrasse, Switzerland, 2016; pp. 299–339. [Google Scholar]
- Van der Meer, A.D.; van den Berg, A. Organs-on-chips: Breaking the in vitro impasse. Integr. Biol. 2012, 4, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Van de Stolpe, A.; den Toonder, J. Workshop meeting report Organs-on-Chips: Human disease models. Lab Chip 2013, 13, 3449–3470. [Google Scholar] [CrossRef] [PubMed]
- Jo, B.H.; van Lerberghe, L.M.; Motsegood, K.M.; Beebe, D.J. Three-dimensional micro-channel fabrication in polydimethylsiloxane (PDMS) elastomer. J. Microelectromech. Syst. 2000, 9, 76–81. [Google Scholar] [CrossRef]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Bale, S.S.; Vernetti, L.; Senutovitch, N.; Jindal, R.; Hegde, M.; Gough, A.; McCarty, W.J.; Bakan, A.; Bhushan, A.; Shun, T.Y.; et al. In vitro platforms for evaluating liver toxicity. Exp. Biol. Med. 2014, 239, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Booth, R.; Hanseup, K. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Pakazad, S.K.; Savov, A.; van de Stolpe, A.; Dekker, R. A novel stretchable micro-electrode array (SMEA) design for directional stretching of cells. J. Micromech. Microeng. 2014, 24, 3. [Google Scholar]
- Teixeira, A.I.; Ilkhanizadeh, S.; Wigenius, J.A.; Duckworth, J.K.; Inganäs, O.; Hermanson, O. The promotion of neuronal maturation on soft substrates. Biomaterials 2009, 30, 4567–4572. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, P.; Helen, W.; Engler, A.J.; Levchenko, A.; Kim, D.H. Control of stem cell fate and function by engineering physical microenvironments. Integr. Biol. 2012, 4, 1008–1018. [Google Scholar]
- Discher, D.E.; Mooney, D.J.; Zandstra, P.W. Growth factors, matrices, and forces combine and control stem cells. Science 2009, 324, 1673–1677. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Hameed, F.M.; Yang, B.; Lee, K.; Pan, C.Q.; Park, S.; Sheetz, M. Cyclic stretching of soft substrates induces spreading and growth. Nat. Commun. 2015, 6, 6333. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Nam-Trung, N.; Khoi, S.L.; Peter, P.F.L.; Maohan, S.; Min, W.; Leyla, K.; Benoit, L. Multiarray cell stretching platform for high-magnification real-time imaging. Nanomedicine 2013, 8, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Broek, L.J.; Limandjaja, G.C.; Niessen, F.B.; Gibbs, S. Human hypertrophic and keloid scar models: Principles, limitations and future challenges from a tissue engineering perspective. Exp. Dermatol. 2014, 23, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Bergers, L.; Waaijman, T.; de Gruijl, T.; van de Stolpe, A.; Dekker, R.; Gibbs, S. Skin-on-chip: Integrating skin-tissue and microsystems engineering. In Proceedings of the 2015 4th TERMIS World Congress, Boston, MA, USA, 8–12 September 2015; pp. 8–9.
- Ogawa, R.; Okai, K.; Tokumura, F.; Mori, K.; Ohmori, Y.; Huang, C.; Hyakusoku, H.; Akaishi, S. The relationship between skin stretching/contraction and pathologic scarring: The important role of mechanical forces in keloid generation. Wound Repair Regen. 2012, 20, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Boyden, S. The chemotactin effect of mixtures of antibody and antigen on polymorphonuclear leukocytes. J. Exp. Med. 1962, 115, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Braam, S.R.; Tertoolen, L.; van de Stolpe, A.; Meyer, T.; Passier, R.; Mummery, C.L. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010, 4, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Fishman, H.A.; Dai, H.; Harris, J.S. Neural stimulation with a carbon nanotube microelectrode array. Nano Lett. 2006, 6, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, R.; Okai, K.; Tokumura, F.; Mori, K.; Ohmori, Y.; Huang, C.; Hyakusoku, H.; Akaishi, S.; Kitamura, N.; Kawano, M.; et al. Electrical stimulation modulates fate determination of differentiating embryonic stem cells. Stem Cells 2007, 25, 562–570. [Google Scholar]
- Khoshfetrat, P.S. Stretchable Micro-Electrode Arrays for Electrophysiology. Ph.D. Thesis, Delft University of Technology, Delft, The Netherlands, June 2015. [Google Scholar]
- Gerwig, R.; Fuchsberger, K.; Schroeppel, B.; Link, G.S.; Heusel, G.; Kraushaar, U.; Schuhmann, W.; Stett, A.; Stelzle, M. PEDOT-CNT composite microelectrodes for recording and electrostimulation applications: Fabrication, morphology, and electrical properties. Front. Neuroeng. 2012, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Gabay, T.; Ben-David, M.; Kalifa, I.; Sorkin, R.; Ze’ev, R.A.; Ben-Jacob, E.; Hanein, Y. Electro-chemical and biological properties of carbon nanotube based multi-electrode arrays. Nanotechnology 2007, 18, 035201. [Google Scholar] [CrossRef] [PubMed]
- Gaio, N.; van Meer, B.; Silvestri, C.; Pakazad, S.; Vollebregt, S.; Mummery, C.L.; Dekker, R. Upside-down carbon nanotube (CNT) micro-electrode array (MEA). In Proceedings of the 2015 IEEE on SENSORS, Busan, Korea, 1–4 November 2015.
- Feinberg, A.W.; Feigel, A.; Shevkoplyas, S.S.; Sheehy, S.; Whitesides, G.M.; Parker, K.K. Muscular thin films for building actuators and powering devices. Science 2007, 317, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.A.; Sheehy, S.P.; Parker, K.K. Sarcomere alignment is regulated by myocyte shape. Cell Motil. Cytoskelet. 2008, 65, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Denning, C.; Borgdorff, V.; Crutchley, J.; Firth, K.S.; George, V.; Kalra, S.; Kondrashov, A.; Hoang, M.D.; Mosqueira, D.; Patel, A.; et al. Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochim. Biophys. Acta 2015, 1863, 1728–1748. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, W.K. Introduction to Microsystem Design; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Quiros-Solano, W.F.; Pandraud, G.; Sarro, P.M. Wafer-level fabrication of strain gauges on PDMS membranes for low-pressure sensing. In Proceedings of the 2015 IEEE on SENSORS, Busan, Korea, 1–4 November 2015.


© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaio, N.; Van Meer, B.; Quirós Solano, W.; Bergers, L.; Van de Stolpe, A.; Mummery, C.; Sarro, P.M.; Dekker, R. Cytostretch, an Organ-on-Chip Platform. Micromachines 2016, 7, 120. https://doi.org/10.3390/mi7070120
Gaio N, Van Meer B, Quirós Solano W, Bergers L, Van de Stolpe A, Mummery C, Sarro PM, Dekker R. Cytostretch, an Organ-on-Chip Platform. Micromachines. 2016; 7(7):120. https://doi.org/10.3390/mi7070120
Chicago/Turabian StyleGaio, Nikolas, Berend Van Meer, William Quirós Solano, Lambert Bergers, Anja Van de Stolpe, Christine Mummery, Pasqualina M. Sarro, and Ronald Dekker. 2016. "Cytostretch, an Organ-on-Chip Platform" Micromachines 7, no. 7: 120. https://doi.org/10.3390/mi7070120





