Discovery of Novel Bacterial Cell-Penetrating Phylloseptins in Defensive Skin Secretions of the South American Hylid Frogs, Phyllomedusa duellmani and Phyllomedusa coelestis
Abstract
:1. Introduction
2. Results
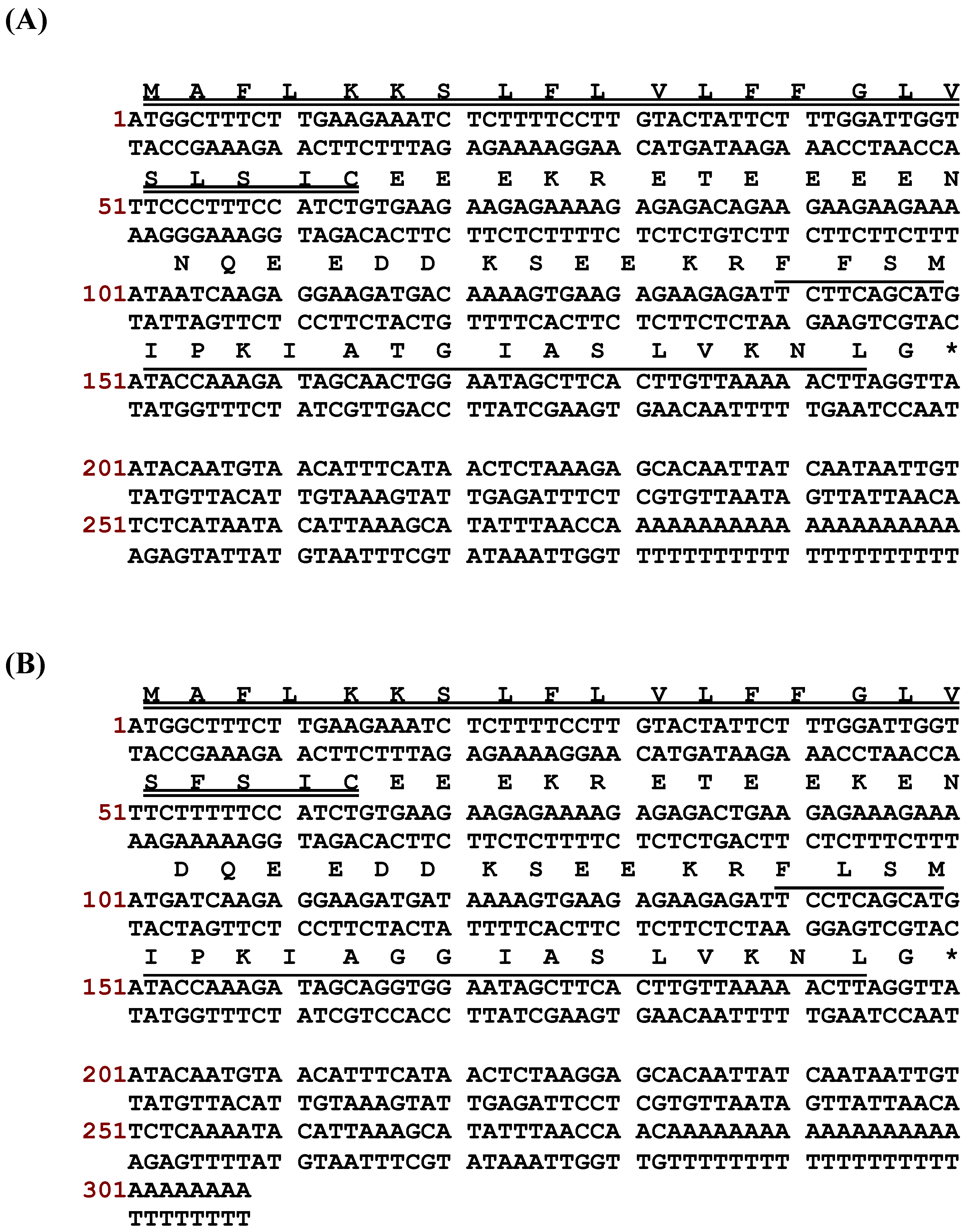
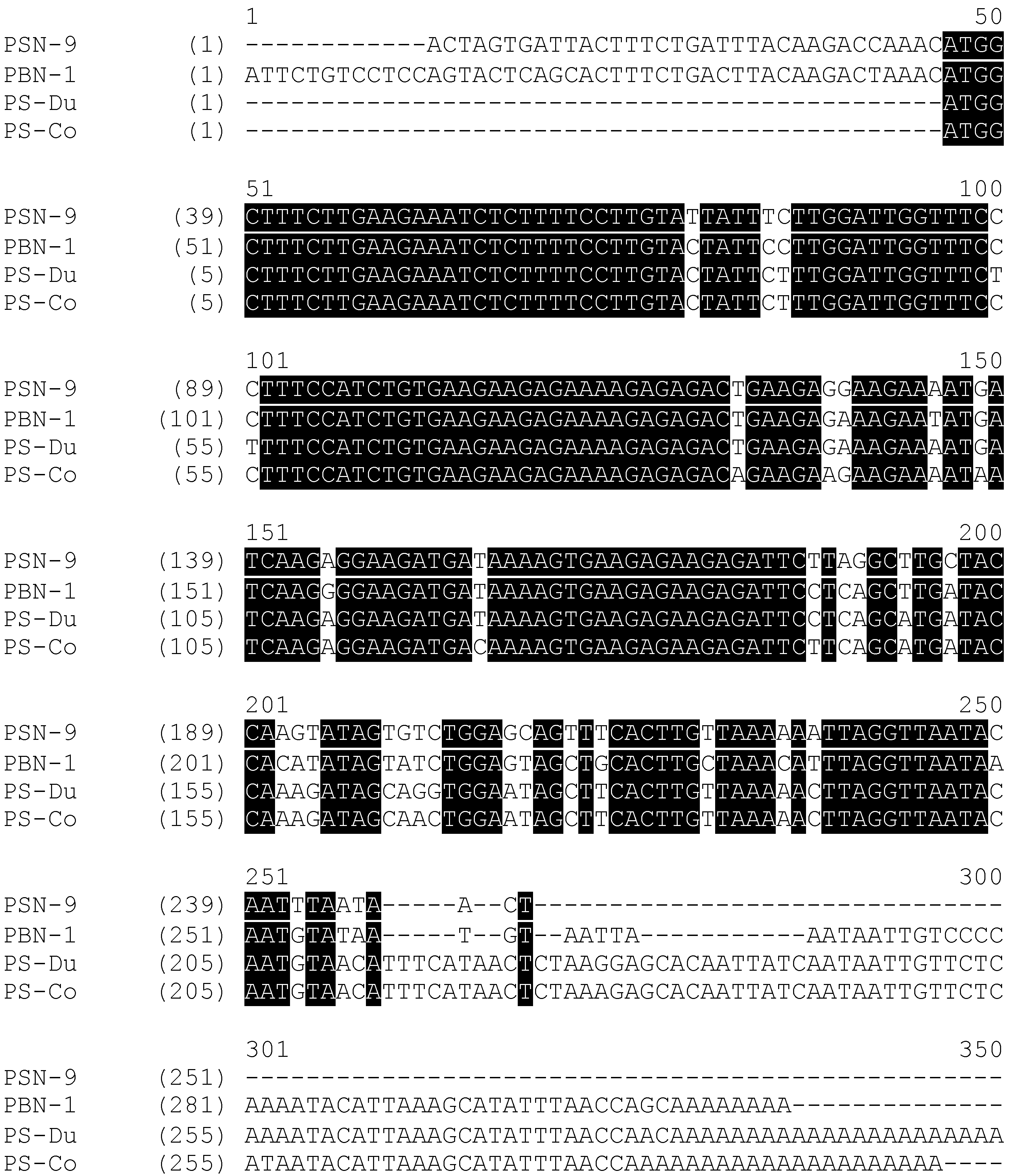
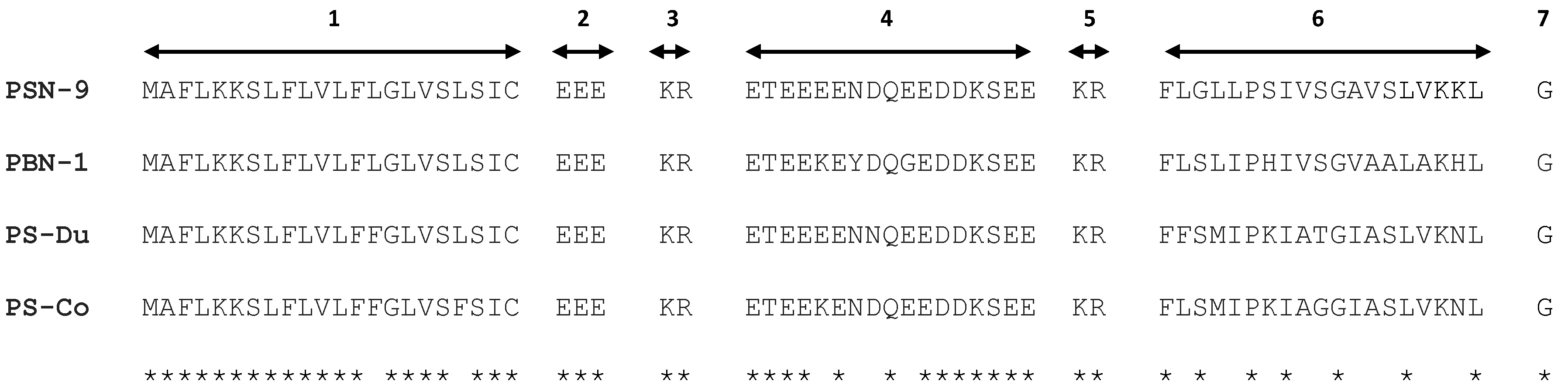
2.1. “Shotgun” Cloning of Novel Peptide Precursor-Encoding cDNAs and Bioinformatic Analyses
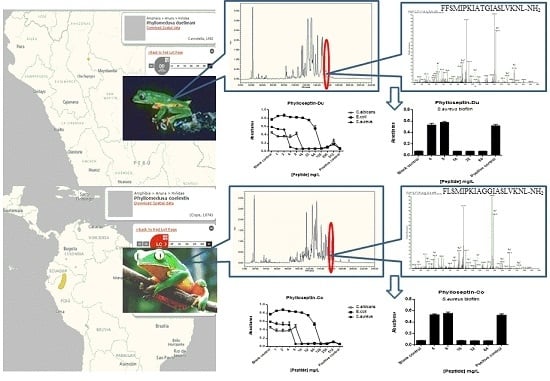
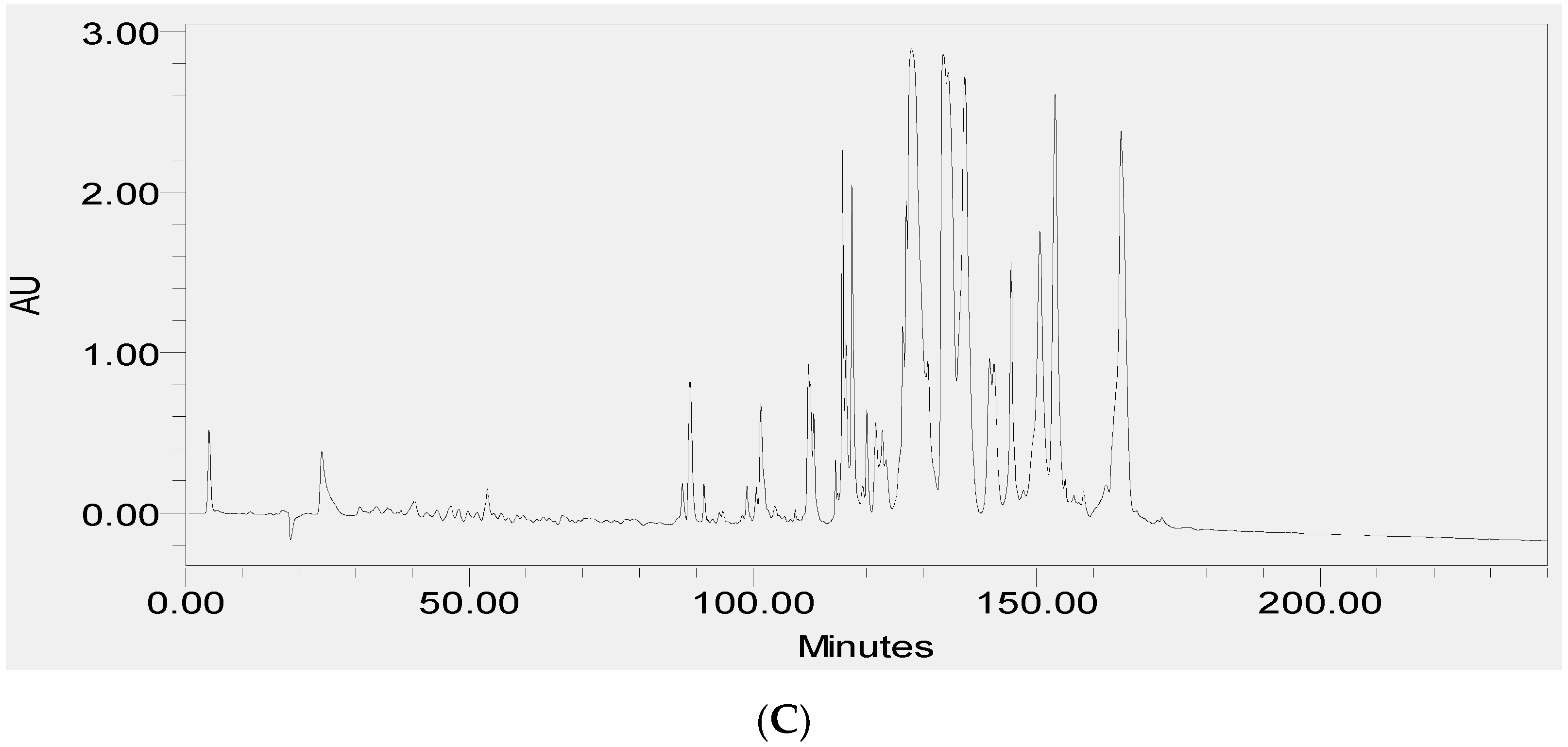
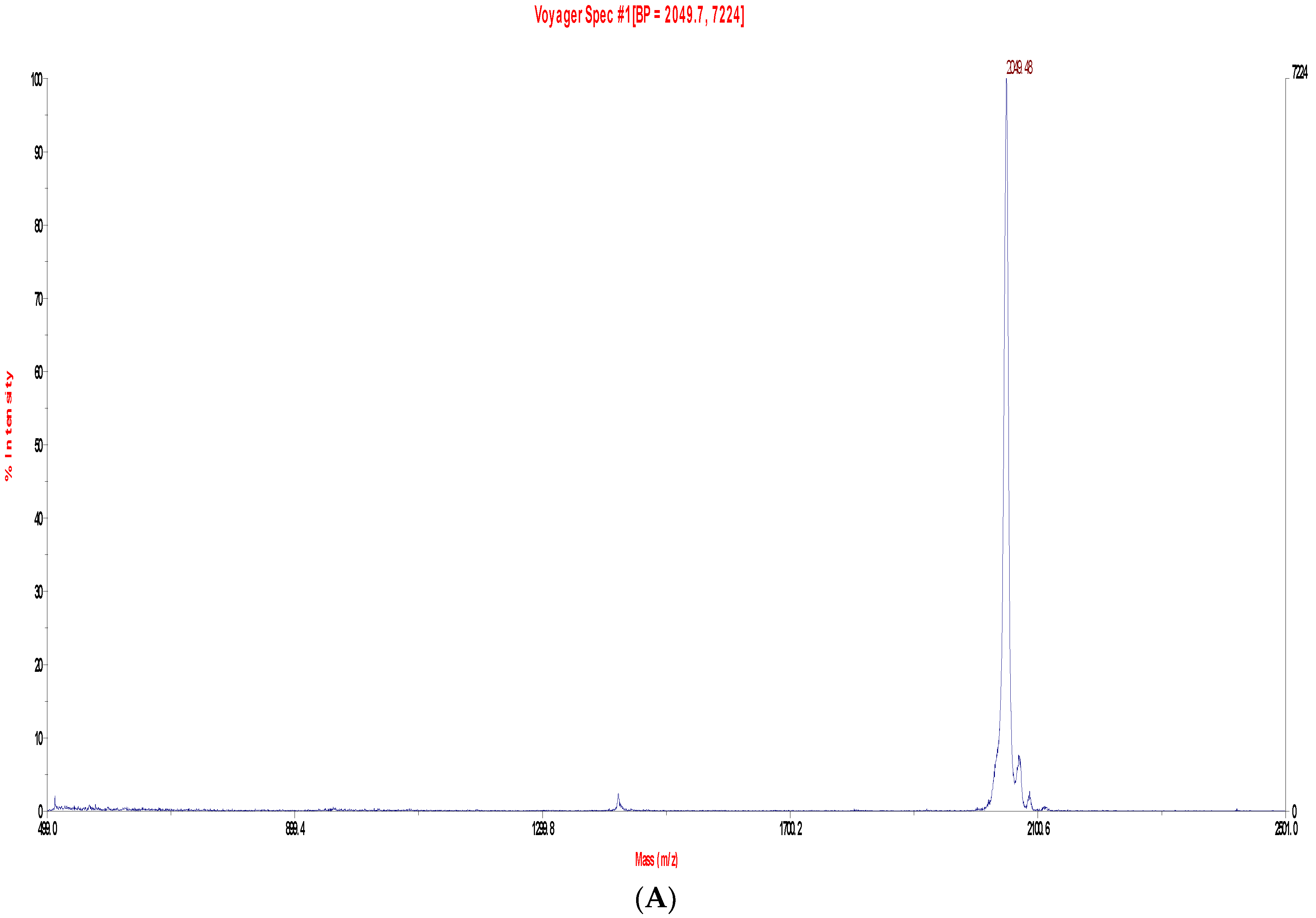
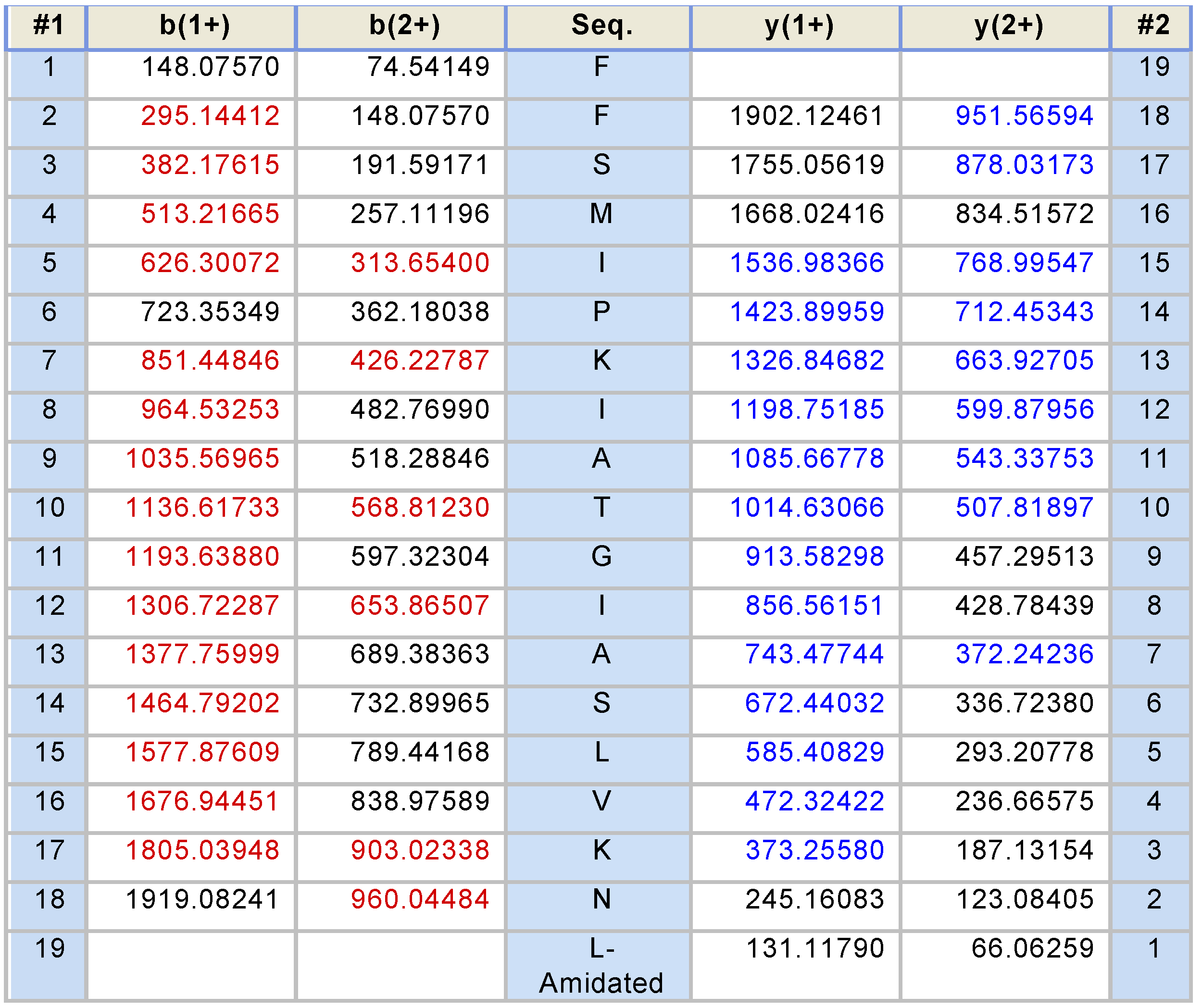
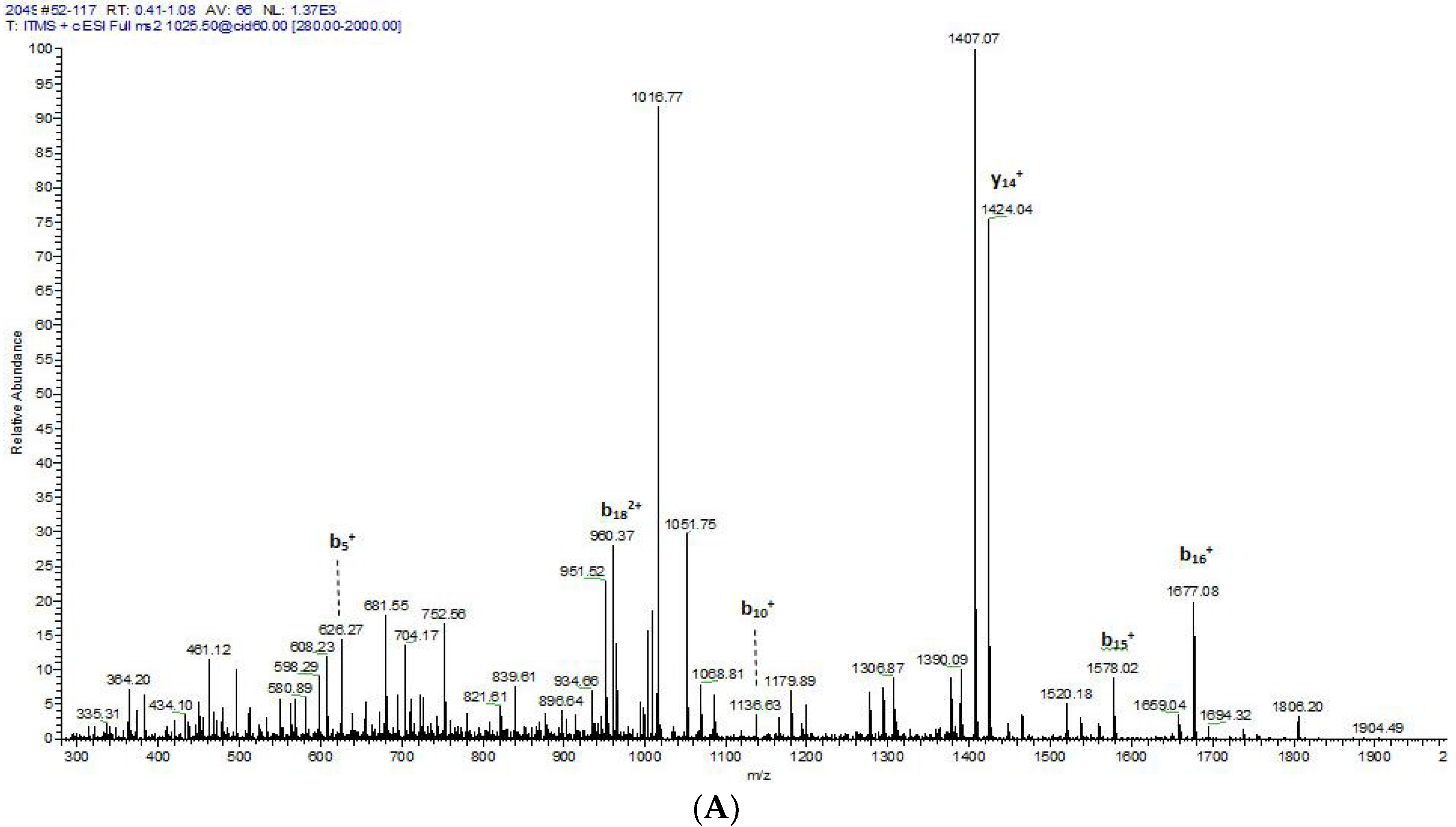
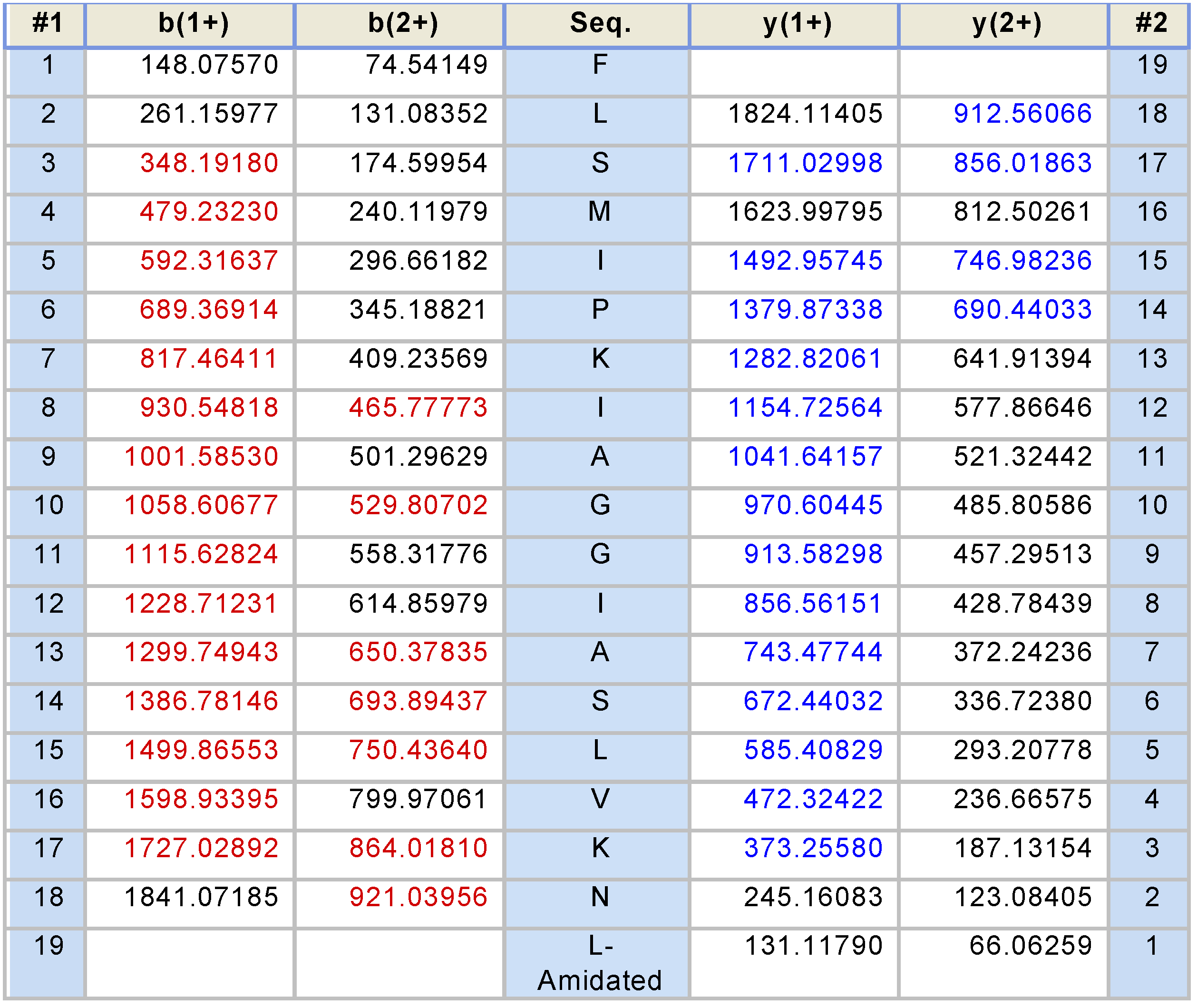
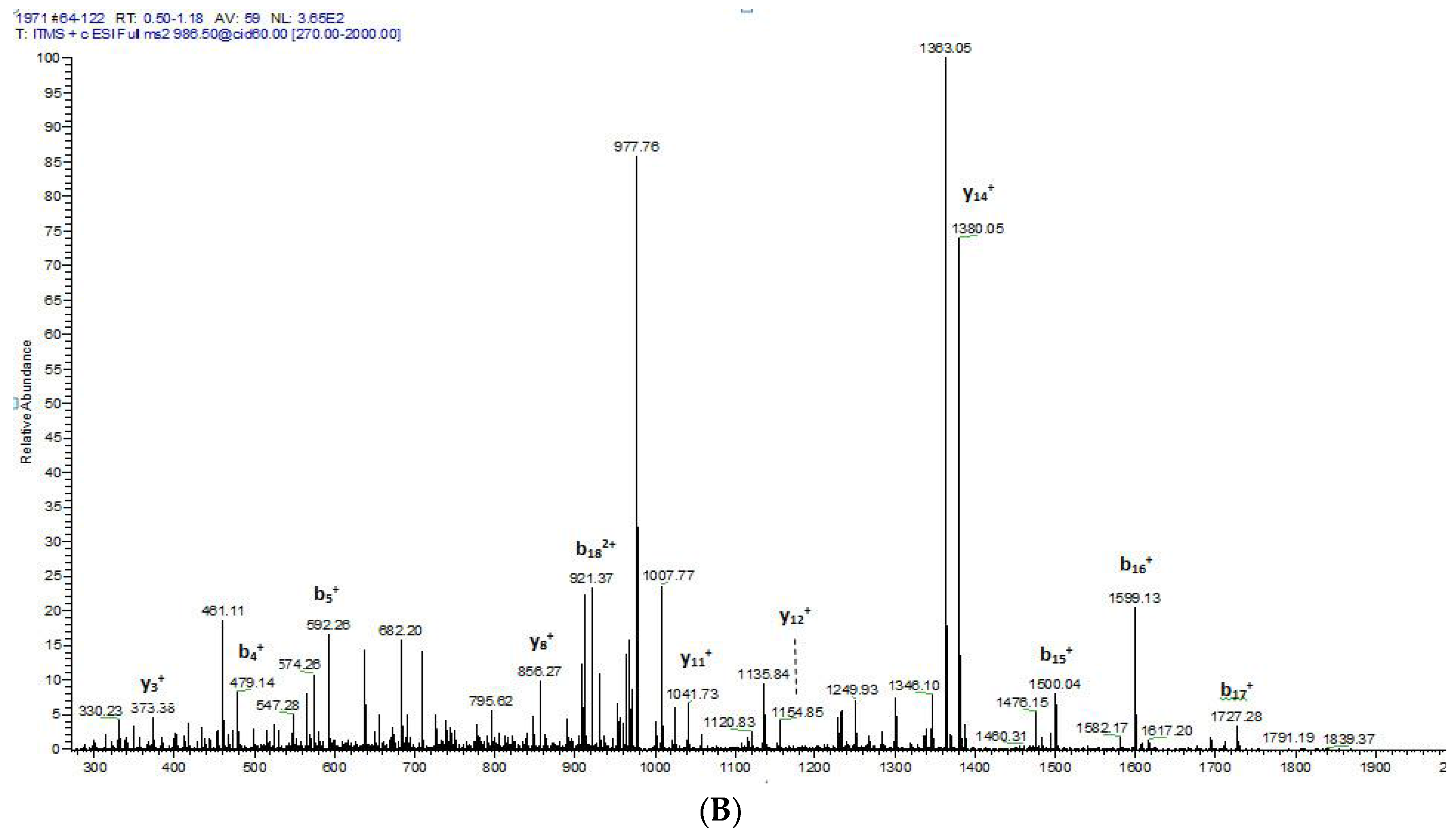
2.2. Fractionation of Skin Secretions, Identification and Structural Characterisation of PS-Du and PS-Co
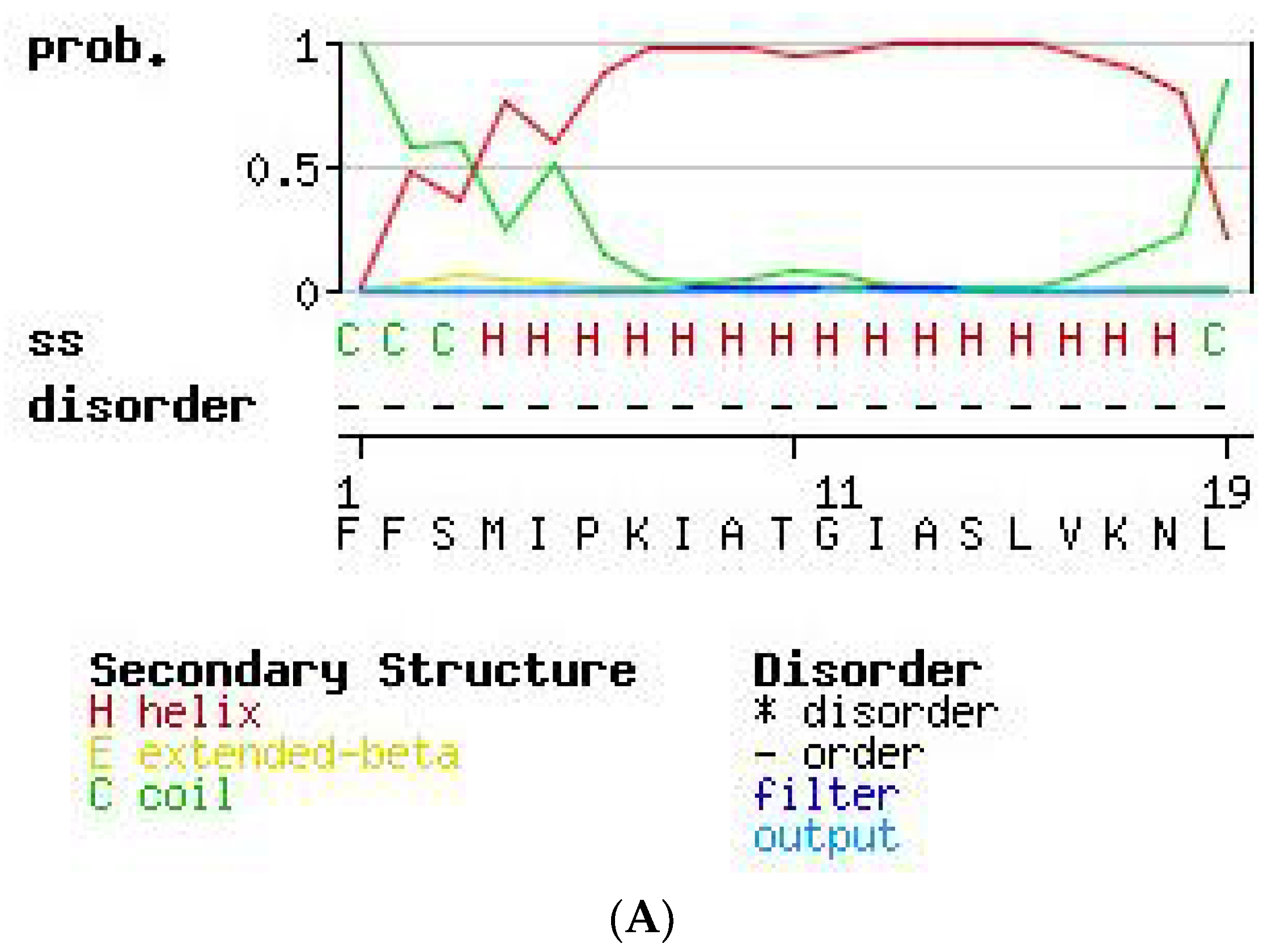
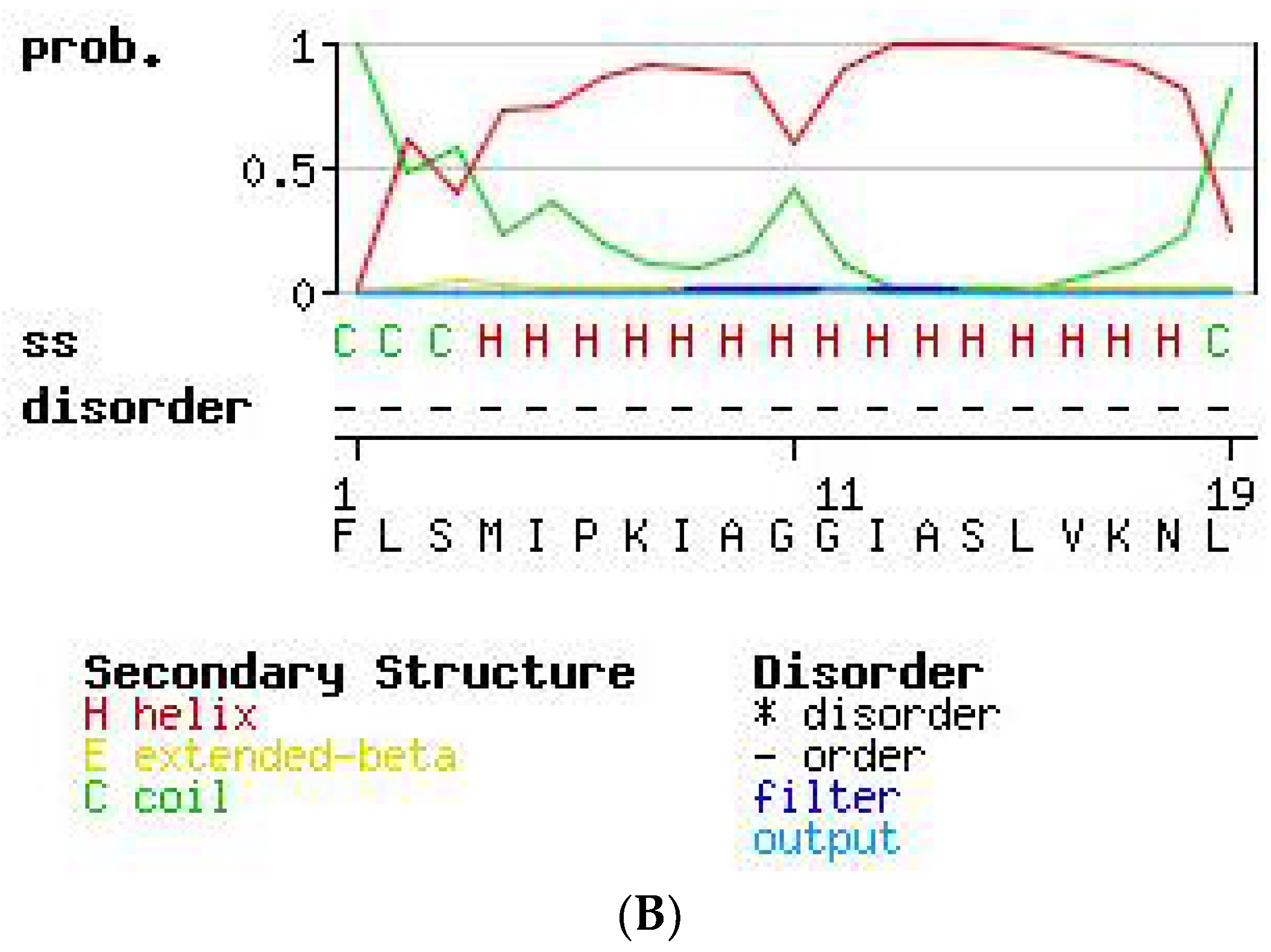
2.3. Secondary Structure Prediction of PS-Du and PS-Co.
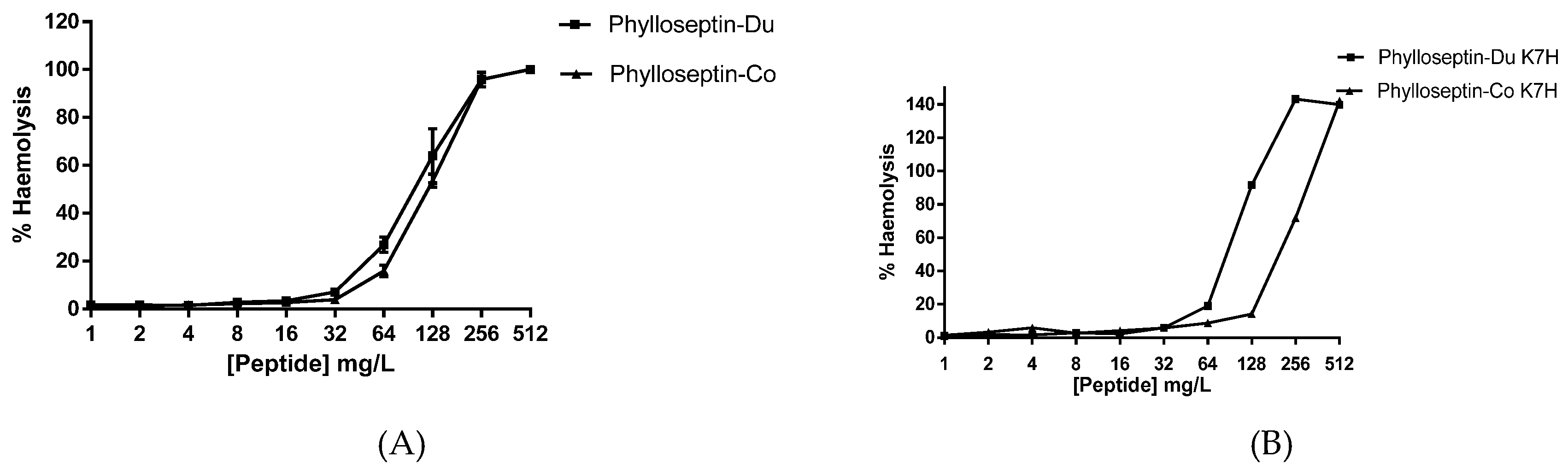
2.4. Antimicrobial and Haemolytic Activities of PS-Du and PS-Co and Those of Their Structurally-Modified Analogues, PS-Du K7H, and PS-Co K7H
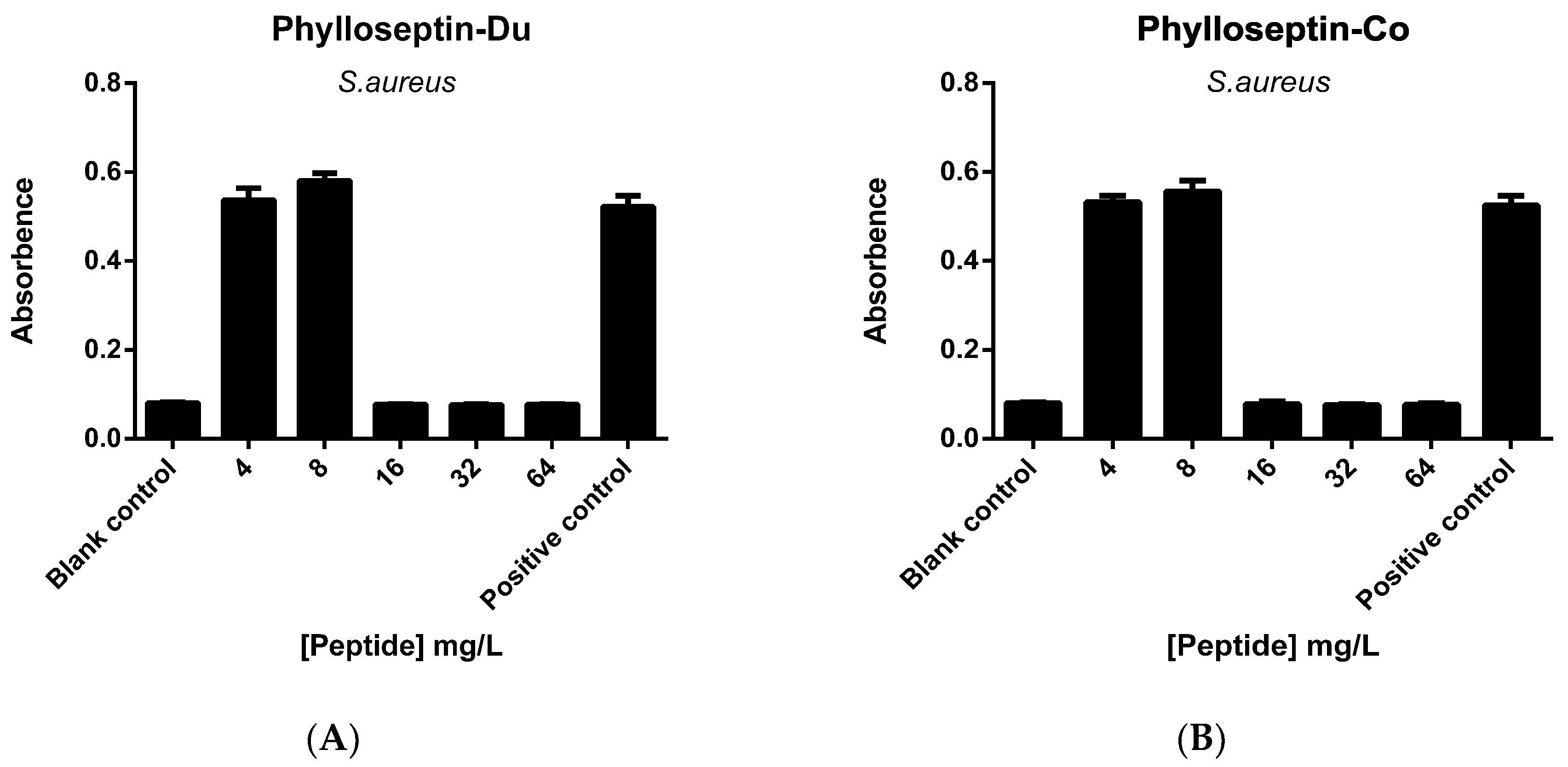
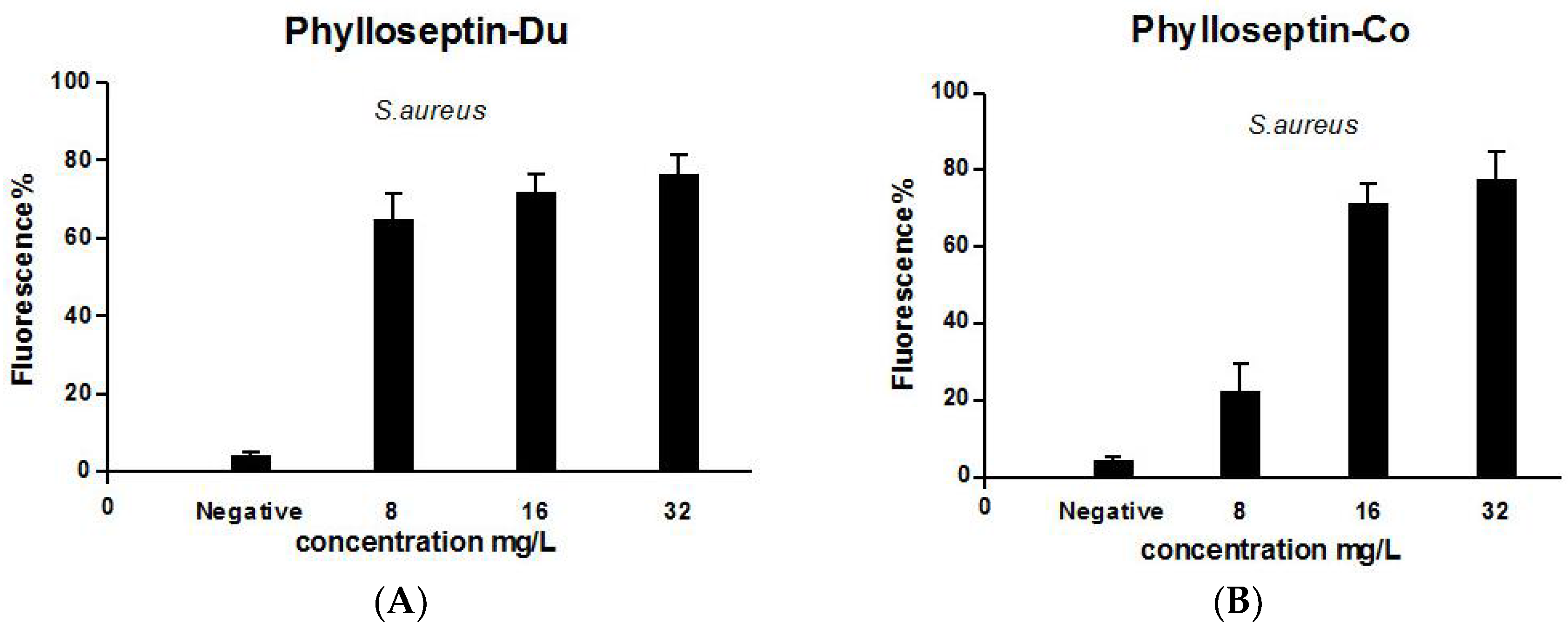
2.5. Anti-Biofilm and Cell-Membrane Permeabilization Activities of Natural Peptides, PS-Du and PS-Co
3. Experimental Section
3.1. “Shotgun” Cloning of Novel Phylloseptin Precursor-Encoding cDNAs from Skin Secretion-Derived cDNA Libraries of Phyllomedusa Duellmani and Phyllomedusa Coelestis
3.2. Chromatographic Isolation and Structural Characterisation of the Two Novel Phylloseptins from the Skin Secretions of Phyllomedusa Duellmani and Phyllomedusa Coelestis
3.3. Solid-Phase Peptide Synthesis of the Two Novel Peptides and Their Structurally- Modified Analogues
3.4. RP-HPLC Analysis of the Two Novel Synthetic Peptides and Co-Elution Profiling of These Two Peptides with Their Respective Skin Secretion Counterparts
3.5. Antimicrobial Activity Assays with the Two Novel Peptides and Their Structurally-Modified Analogues
3.6. Haemolysis Assay of the Two Novel Peptides and Their Modified Analogues
3.7. Anti-Biofilm Activities of the Two Novel Peptides Tested on S. aureus Biofilm
3.8. Bacterial Cell Membrane Permeability Assay of the Two Novel Peptides Using S. aureus
3.9. Statistical Analysis
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Novkovic, M.; Simunic, J.; Bojovic, V.; Tossi, A.; Juretic, D. DADP: The database of anuran defense peptides. Bioinformatics 2012, 28, 1406–1407. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Sonnevend, A. Antimicrobial peptides in frog skin secretions. Methods Mol. Biol. 2010, 618, 3–14. [Google Scholar] [PubMed]
- Barra, D.; Simmaco, M. Amphibian skin: A promising resource for antimicrobial peptides. Trends Biotechnol. 1995, 13, 205–209. [Google Scholar] [CrossRef]
- Nacif-Marçal, L.; Pereira, G.R.; Abranches, M.V.; Costa, N.C.; Cardoso, S.A.; Honda, E.R.; de Paula, S.O.; Feio, R.N.; Oliveira, L.L. Identification and characterization of an antimicrobial peptide of Hypsiboas semilineatus (Spix, 1824) (Amphibia, Hylidae). Toxicon 2015, 99, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Kolodziejek, J.; Nowotny, N. Antimicrobial peptides from ranid frogs: Taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. Biophys. Acta 2004, 1696, 1–14. [Google Scholar] [CrossRef] [PubMed]
- De Azevedo Calderon, L.; Alexandre de Almeida, E.S.; Ciancaglini, P.; Stábeli, R.G. Antimicrobial peptides from Phyllomedusa frogs: From biomolecular diversity to potential nanotechnologic medical applications. Amino Acids 2011, 40, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Simmaco, M.; Kreil, G.; Barra, D. Bombinins, antimicrobial peptides from Bombina species. Biochim. Biophys. Acta 2009, 1788, 1551–1555. [Google Scholar] [CrossRef] [PubMed]
- Zairi, A.; Tangy, F.; Bouassida, K.; Hani, K. Dermaseptins and Magainins: Antimicrobial peptides from frogs’ skin—New sources for a promising spermicides microbicides—A mini review. Biomed. Biotechnol. 2009, 2009, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Radosavljevic, G.; Attoub, S.; King, J.D.; Lukic, M.L.; McClean, S. A family of antimicrobial and immunomodulatory peptides related to the frenatins from skin secretions of the Orinoco lime frog Sphaenorhynchus lacteus (Hylidae). Peptides 2014, 56, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M. Host-defense peptides with therapeutic potentialfrom skin secretions of frogs from the family Pipidae. Pharmaceuticals 2014, 7, 58–77. [Google Scholar] [CrossRef] [PubMed]
- Mechkarska, M.; Attoub, S.; Sulaiman, S.; Pantic, J.; Lukic, M.L.; Conlon, J.M. Anti-cancer, immunoregulatory, and antimicrobial activities of the frog skin host-defense peptides pseudhymenochirin-1Pb and pseudhymenochirin-2Pa. Regul. Pept. 2014, 194, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Frost, D.R.; Grant, T.; Faivovich, J.; Bain, R.H.; Haas, A.; Haddad, C.F.; Sa, R.O.; Channing, A.; Wilkinson, M.; Donnellan, S.C.; et al. The Amphibian Tree of Life; American Museum of Natural History: New York, NY, USA, 2006. [Google Scholar]
- Amiche, M.; Ladram, A.; Nicolas, P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides 2008, 29, 2074–2082. [Google Scholar] [CrossRef] [PubMed]
- Leite, J.R.S.A.; Silca, L.P.; Rodrigues, M.I.S.; Prates, M.V.; Brand, G.D.; Lacava, B.M.; Azevedo, R.B.; Bocca, A.L.; Albuquerque, S.; Bloch, C., Jr. Phyllosptins: A novel class of anti-bacterial and anti-protozoan peptides from the Phyllomedusa genus. Peptides 2005, 26, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.B.; Zhou, M.; Gagliardo, R.; Walker, B.; Shaw, C. Elements of the granular gland peptidome and transcriptome persist in air-dried skin of the South American orange-legged leaf frog, Phyllomedusa hypocondrialis. Peptides 2006, 27, 2129–2136. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.H.; Bjourson, A.J.; Orr, D.F.; Shaw, C.; McClean, S. A combined mass spectrometric and cDNA sequencing approach to the isolation and characterization of novel antimicrobial peptides from the skin secretions of Phyllomedusa hypochondrialis azurea. Peptides 2007, 28, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.W.; Zhou, M.; Wang, L.; McGrath, S.; Chen, T.B.; Chen, X.H.; Shaw, C. PS-1 (PSN-1) from Phyllomedusa sauvagei skin secretion: A novel broad-spectrum antimicrobial peptide with antibiofilm activity. Mol. Immunol. 2010, 47, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Konig, E.; Clark, V.C.; Shaw, C.; Bininda-Emonds, O.R. Molecular cloning of skin peptide precursor-encoding cDNAs from tibial gland secretion of the Giant Monkey Frog, Phyllomedusa bicolor (Hylidae, Anura). Peptides 2012, 38, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Conceicao, K.; Konno, K.; Richardson, M.; Antoniazzi, M.M.; Jared, C.; Daffre, S.; Camargo, A.C.; Pimenta, D.C. Isolation and biochemical characterization of peptides presenting antimicrobial activity from the skin of Phyllomedusa hypochondrialis. Peptides 2006, 27, 3092–3099. [Google Scholar] [CrossRef] [PubMed]
- Angulo, A.; Arizabal, W.; Lehr, E.; Martinez, J.L. Phyllomedusa duellmani, The IUCN Red List of Threatened Species, Version 2014.2; 2004. Available online: www.iucnredlist.org (accessed on 7 December 2014).
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. 1), S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Roth, B.L.; Poot, M.; Yue, S.T.; Millard, P.J. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 1997, 63, 2421–2431. [Google Scholar] [PubMed]
- Clarke, B.T. The natural history of amphibian skin secretions, their normal functioning and potential medical applications. Biol. Rev. 1997, 72, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.B.; Walker, B.; Zhou, M.; Shaw, C. Dermatoxin and phylloxin from the waxy frog, Phyllomedusa sauvagei: Cloning of precursor cDNAs and structural characterization from lyophilized skin secretion. Peptides 2005, 129, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Van Zoggel, H.; Hamma-Kourbali, Y.; Galanth, C.; Ladram, A.; Nicolas, P.; Courty, J.; Amiche, M.; Delbé, J. Antitumor and angiostatic peptides from frog skin secretions. Amino Acids 2012, 42, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, H.; Li, S.; Tian, L.; Shang, D. Antitumor effects and cell selectivity of temporin-1CEa, an antimicrobial peptide from the skin secretions of the Chinese brown frog (Rana chensinensis). Biochimie 2012, 94, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Erspamer, V.; Melchiorri, P.; Falconieri Erspamer, G.; Montecucchi, P.C.; De Castiglione, R. Phyllomedusa skin: A huge factory and store-house of a variety of active peptides. Peptides 1985, 6, 7–12. [Google Scholar] [CrossRef]
- Xi, X.; Li, R.; Jiang, Y.; Lin, Y.; Wu, Y.; Zhou, M.; Xu, J.; Wang, L.; Chen, T.; Shaw, C. Medusins: A new class of antimicrobial peptides from the skin secretions of phyllomedusine frogs. Biochimie 2013, 95, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Zhou, M.; Wang, L.; Chen, T.B.; Walker, B.; Shaw, C. Antimicrobial/cytolytic peptides from the venom of the North African scorpion, Androctonus amoreuxi: Biochemical and functional characterization of natural peptides and a single site-substituted analog. Peptides 2012, 35, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Resende, J.; Verly, R.; Aisenbrey, C.; Cesar, A.; Bertani, P.; Piló-Veloso, D.; Bechinger, B. Membrane Interactions of Phylloseptin-1, -2, and -3 Peptides by Oriented Solid-State NMR Spectroscopy. Biophys. J. 2014, 107, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Resende, J.M.; Moraes, C.M.; Prates, M.V.; Cesar, A.; Almeida, F.C.; Mundim, N.C.; Valente, A.P.; Bemquerer, M.P.; Piló-Veloso, D.; Bechinger, B. Solution NMR structures of the antimicrobial peptides phylloseptin-1,-2, and-3 and biological activity: the role of charges and hydrogen bonding interactions in stabilizing helix conformations. Peptides 2008, 29, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Lambert, P.A.; Rycroft, A.N. The Bacterial Cell Surface. Croom Helm 1984, 234, 389–392. [Google Scholar]
- Mah, T.F.; Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Licking, E. Getting a grip on bacterial slime. Business Week, 13 September 1999; 78–79. [Google Scholar]
- Balaban, N.; Cirioni, O.; Giacometti, A.; Ghiselli, R.; Braunstein, J.B.; Silvestri, C.; Mocchegiani, F.; Saba, V.; Scalise, G. Treatment of Staphylococcus aureus biofilm infection by the Quorum-Sensing inhibitor RIP. Antimicrob. Agents Chemother. 2007, 51, 2226–2229. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.G.; Kluytmans, J.A.; Wertheim, H.F.; Bogaers, D.; Vandenbroucke-Grauls, C.M.; Roosendaal, R.; Troelstra, A.; Box, A.T.; Voss, A.; Van der Tweel, I.; et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N. Engl. J. Med. 2010, 362, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.H.; Arenas, G. Antimicrobial peptides: A natural alternative to chemical antibiotics and a potential for applied biotechnology. Electron. J. Biotechnol. 2003, 6, 271–284. [Google Scholar] [CrossRef]
- Yount, N.Y.; Bayer, A.S.; Xiong, Y.Q.; Yeaman, M.R. Advances in antimicrobial peptide immunobiology. Pept. Sci. 2006, 84, 435–458. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Blecha, F. Antimicrobial peptides and bacteriocins: Alternatives to traditional antibiotics. Anim. Health Res. Rev. 2008, 9, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin (1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell. Mol. Life Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef] [PubMed]
- Altman, H.; Steinberg, D.; Porat, Y.; Mor, A.; Fridman, D.; Friedman, M.; Bachrach, G. In vitro assessment of antimicrobial peptides as potential agents against several oral bacteria. J. Antimicrob. Chemother. 2006, 58, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]






| Peptide Name | Molecular Mass(Da) | MIC | ||
|---|---|---|---|---|
| S. aureus | E. coli | C. albicans | ||
| PS-Du | 2049.5 | 8 mg/L | 128 mg/L | 16 mg/L |
| (3.90 µM) | (62.45 µM) | (7.81 µM) | ||
| PS-Co | 1971.5 | 8 mg/L | 128 mg/L | 16 mg/L |
| (4.06 µM) | (64.93 µM) | (8.12 µM) | ||
| PS-Du K7H | 2057.1 | 32 mg/L | 512 mg/L | 64 mg/L |
| (15.56 µM) | (248.89 µM) | (31.12 µM) | ||
| PS-Co K7H | 1979.1 | 32 mg/L | 512 mg/L | 64 mg/L |
| (16.17 µM) | (258.70 µM) | (32.34 µM) | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, N.; Li, L.; Wu, D.; Gao, Y.; Xi, X.; Zhou, M.; Wang, L.; Chen, T.; Shaw, C. Discovery of Novel Bacterial Cell-Penetrating Phylloseptins in Defensive Skin Secretions of the South American Hylid Frogs, Phyllomedusa duellmani and Phyllomedusa coelestis. Toxins 2016, 8, 255. https://doi.org/10.3390/toxins8090255
Yang N, Li L, Wu D, Gao Y, Xi X, Zhou M, Wang L, Chen T, Shaw C. Discovery of Novel Bacterial Cell-Penetrating Phylloseptins in Defensive Skin Secretions of the South American Hylid Frogs, Phyllomedusa duellmani and Phyllomedusa coelestis. Toxins. 2016; 8(9):255. https://doi.org/10.3390/toxins8090255
Chicago/Turabian StyleYang, Nan, Lei Li, Di Wu, Yitian Gao, Xinping Xi, Mei Zhou, Lei Wang, Tianbao Chen, and Chris Shaw. 2016. "Discovery of Novel Bacterial Cell-Penetrating Phylloseptins in Defensive Skin Secretions of the South American Hylid Frogs, Phyllomedusa duellmani and Phyllomedusa coelestis" Toxins 8, no. 9: 255. https://doi.org/10.3390/toxins8090255