Diversification of Ergot Alkaloids in Natural and Modified Fungi
Abstract
:1. Introduction
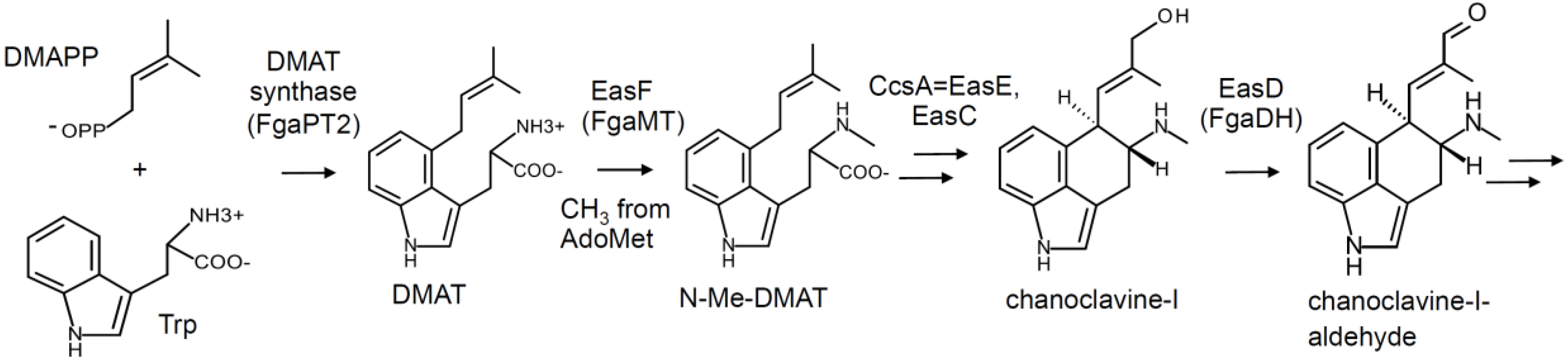
2. Pathway Overview and Branch Points
2.2. Branching Caused by Differences in EasA
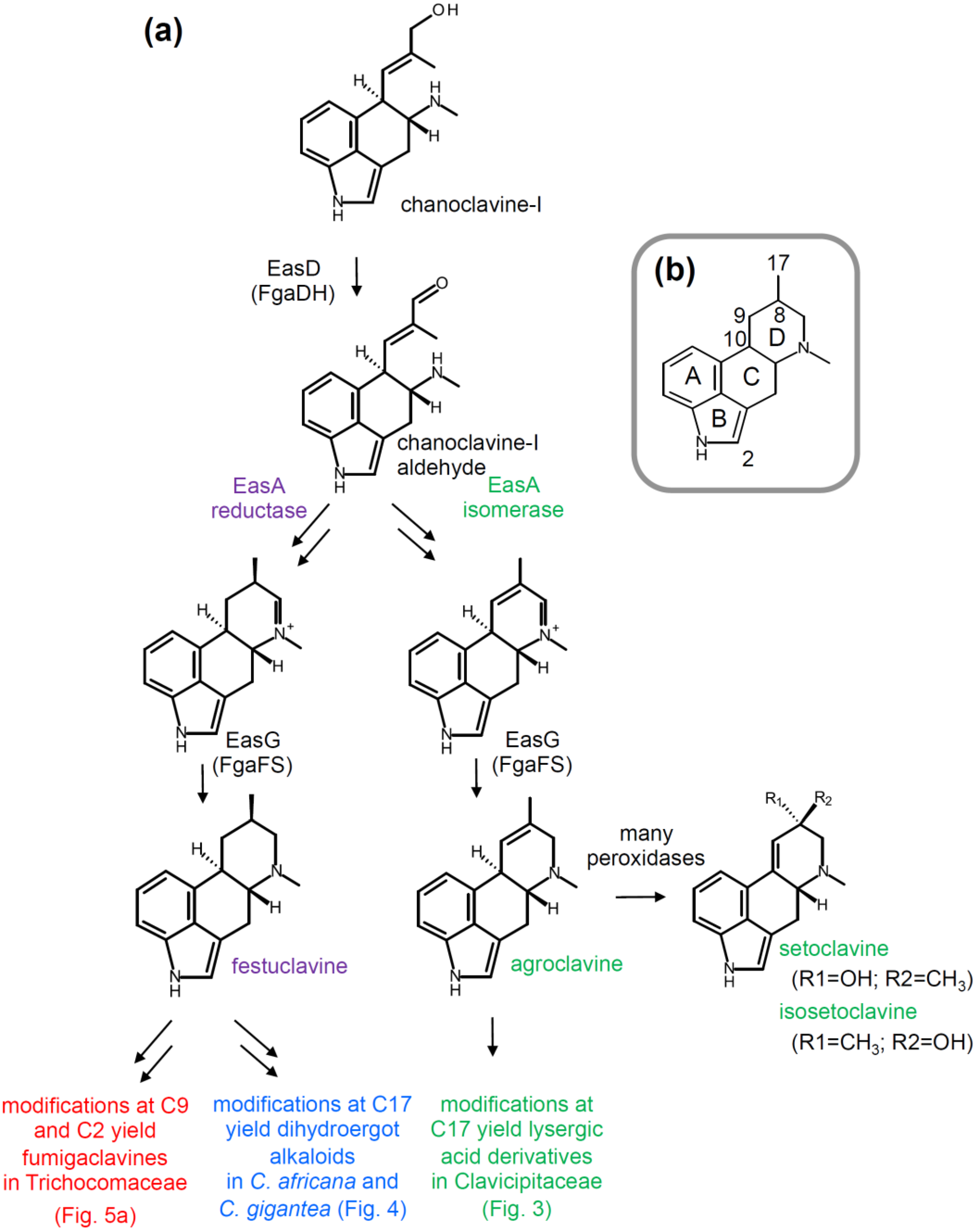
2.3. Branch Point after Festuclavine in Dihydroergot Alkaloid Producers
3. Diversification within the Clavicipitaceae
3.1. Diversity Generated by Peptide Synthetases

| Ergopeptine | Amino acid 1 | Amino acid 2 | Amino acid 3 |
|---|---|---|---|
| Ergocristine | l-Val | l-Phe | l-Pro |
| Ergocryptine | l-Val | l-Leu | l-Pro |
| Ergocornine | l-Val | l-Val | l-Pro |
| Ergotamine | l-Ala | l-Phe | l-Pro |
| Ergovaline | l-Ala | l-Val | l-Pro |
| Ergosine | l-Ala | l-Leu | l-Pro |
| Ergobalansine | l-Ala | l-Leu | l-Ala |
3.2. Clavine vs. Lysergic Acid-Based Pathways
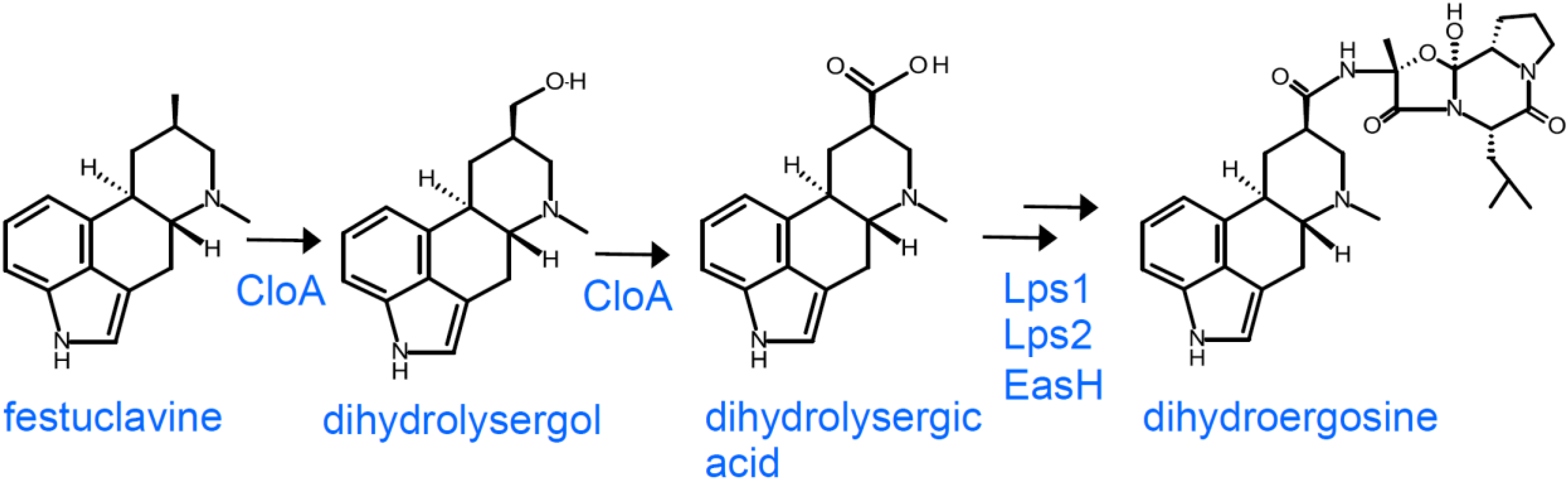
3.3. Dihydroergot Alkaloids in Certain Members of the Clavicipitaceae
4. Diversification within the Trichocomaceae
4.1. Diversification Based on Presence and Relaxed Specificity of Prenyl Transferase EasL
4.2. Stereochemical Diversification Based on Activity of EasG (FgaFS)
4.3. Diversification via Multiple Enzymes Competing for a Common Substrate

5. Diversification of Chemotype via Delayed Flux and Spur Products
5.1. Inefficiency or Delayed Flux Can Yield Multiple de Facto Products from a Single Pathway
5.2. Delayed Flux in the Clavicipitaceae
5.3. Delayed Flux in the Trichocomaceae
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Matossian, M.K. Poisons of the Past; Yale University Press: New Haven, CT, USA, 1989. [Google Scholar]
- Baskys, A.; Hou, A.C. Vascular dementia: Pharmacological treatment approaches and perspectives. Clin. Interv. Aging 2007, 2, 327–335. [Google Scholar] [PubMed]
- Morren, J.A.; Galvez-Jimenez, N. Where is dihydroergotamine mesylate in the changing landscape of migraine therapy? Expert Opin. Pharmacother. 2010, 11, 3085–3093. [Google Scholar] [CrossRef] [PubMed]
- Perez-Lloret, S.; Rascol, O. Dopamine receptor agonists for the treatment of early or advanced Parkinson’s disease. CNS Drugs 2010, 24, 941–968. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.L.; Timpe, E.M.; Petkewicz, K.A. Bromocriptine mesylate for glycemic management in type 2 diabetes mellitus. Ann. Pharmacother. 2010, 44, 1777–1785. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, D.G.; Beaulieu, W.T.; Cook, D. Bioactive alkaloids in vertically transmitted fungal endophytes. Funct. Ecol. 2014, 27, 299–314. [Google Scholar] [CrossRef]
- Hoveland, C.S. Importance and economic significance of the Acremonium endophytes to performance of animals and grass plant. Agric. Ecosyst. Environ. 1993, 44, 3–12. [Google Scholar] [CrossRef]
- Tsai, H.-F.; Wang, H.; Gebler, J.C.; Poulter, C.D.; Schardl, C.L. The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem. Biophys. Res. Commun. 1995, 216, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Machado, C.; Panaccione, D.G.; Tsai, H.-F.; Schardl, C.L. The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Gen. Biol. 2004, 41, 189–198. [Google Scholar] [CrossRef]
- Unsöld, I.A.; Li, S.-M. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 2005, 151, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Coyle, C.M.; Panaccione, D.G. An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl. Environ. Microbiol. 2005, 71, 3112–3118. [Google Scholar] [CrossRef] [PubMed]
- Rigbers, O.; Li, S.-M. Ergot alkaloid biosynthesis in Aspergillus fumigatus: Overproduction and biochemical characterization of a 4-dimethylallyltryptophan N-methyltransferase. J. Biol. Chem. 2008, 283, 26859–26868. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, N.; Olšovská, J.; Šulc, M.; Tudzynski, P. Alkaloid cluster gene ccsA of the ergot fungus Claviceps purpurea encodes chanoclavine I synthase, a flavin adenine dinucleotide-containing oxidoreductase mediating the transformation of N-methyl-dimethylallyltryptophan to chanoclavine I. Appl. Environ. Microbiol. 2010, 76, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Goetz, K.E.; Coyle, C.M.; Cheng, J.Z.; O’Connor, S.E.; Panaccione, D.G. Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Curr. Genet. 2011, 57, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.L.; Moore, C.T.; Panaccione, D.G. Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins 2013, 5, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.A.; Folly, C.; Hatsch, A.; Molt, A.; Schröder, H.; O’Connor, S.E.; Naesby, M. The important ergot alkaloid intermediate chanoclavine-I produced in the yeast Saccharomyces cerevisiae by the combined action of EasC and EasE from Aspergillus japonicus. Microb. Cell Fact. 2014, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- Wallwey, C.; Matuschek, M.; Li, S.-M. Ergot alkaloid biosynthesis in Aspergillus fumigatus: Conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Arch. Microbiol. 2010, 192, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.Z.; Coyle, C.M.; Panaccione, D.G.; O’Connor, S.E. A role for old yellow enzyme in ergot alkaloid biosynthesis. J. Am. Chem. Soc. 2010, 132, 1776–1777. [Google Scholar] [CrossRef] [PubMed]
- Coyle, C.M.; Cheng, J.Z.; O’Connor, S.E.; Panaccione, D.G. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl. Environ. Microbiol. 2010, 76, 3898–3903. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.Z.; Coyle, C.M.; Panaccione, D.G.; O’Connor, S.E. Controlling a structural branch point in ergot alkaloid biosynthesis. J. Am. Chem. Soc. 2010, 132, 12835–12837. [Google Scholar] [CrossRef] [PubMed]
- Wallwey, C.; Matuschek, M.; Xie, X.-L.; Li, S.-M. Ergot alkaloid biosynthesis in Aspergillus fumigatus: Conversion of chanoclavine-I aldehyde to festuclavine by the festuclavine synthase FgaFS in the presence of the old yellow enzyme FgaOx3. Org. Biomol. Chem. 2010, 8, 3500–3508. [Google Scholar] [CrossRef] [PubMed]
- Matuschek, M.; Wallwey, C.; Xie, X.; Li, S.-M. New insights into ergot alkaloid biosynthesis in Claviceps purpurea: An agroclavine synthase EasG catalyses, via a non-enzymatic adduct with reduced glutathione, the conversion of chanoclavine-I aldehyde to agroclavine. Org. Biomol. Chem. 2011, 9, 4328–4335. [Google Scholar] [CrossRef] [PubMed]
- Matuschek, M.; Wallwey, C.; Wollinsky, B.; Xie, X.; Li, S.-M. In vitro conversion of chanoclavine-I aldehyde to the stereoisomers festuclavine and pyroclavine controlled by the second reduction step. RSC Adv. 2012, 2, 3662–3669. [Google Scholar] [CrossRef]
- Kohli, R.M.; Massey, V. The oxidative half-reaction of old yellow enzyme. The role of tyrosine 196. J. Biol. Chem. 1998, 273, 32763–32770. [Google Scholar] [CrossRef] [PubMed]
- Meah, Y.; Massey, V. Old yellow enzyme: Stepwise reduction of nitro-olefins and catalysis of aci-nitro tautomerization. Proc. Natl. Acad. Sci. USA 2000, 97, 10733–10738. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.L.; Panaccione, D.G. Heterologous expression of lysergic acid and novel ergot alkaloids in Aspergillus fumigatus. Appl. Environ. Microbiol. 2014, 80, 6465–6472. [Google Scholar] [CrossRef] [PubMed]
- Wallwey, C.; Li, S.-M. Ergot alkaloids: Structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat. Prod. Rep. 2011, 2, 496–510. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L.; Steffan, N.; Yin, W.B.; Li, S.M. Ergot alkaloid biosynthesis in Aspergillus fumigatus: FgaAT catalyses the acetylation of fumigaclavine B. ChemBioChem 2009, 10, 2325–2328. [Google Scholar] [CrossRef] [PubMed]
- Unsöld, I.A.; Li, S.-M. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: Gene expression, purification, and characterization of fumigaclavine C synthase FgaPT1. ChemBioChem 2006, 7, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Correia, T.; Grammel, N.; Ortel, I.; Keller, U.; Tudzynski, P. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem. Biol. 2003, 10, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Riederer, B.; Han, M.; Keller, U. d-Lysergyl peptide synthetase from the ergot fungus Claviceps purpurea. J. Biol. Chem. 1996, 271, 27524–27530. [Google Scholar] [CrossRef] [PubMed]
- Ortel, I.; Keller, U. Combinatorial assembly of simple and complex d-lysergic acid alkaloid peptide classes in the ergot fungus Claviceps purpurea. J. Biol. Chem. 2009, 284, 6650–6660. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, P.; Hölter, K.; Correia, T.; Arntz, C.; Grammel, N.; Keller, U. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. 1999, 261, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, D.G.; Johnson, R.D.; Wang, J.; Young, C.A.; Damrongkool, P.; Scott, B.; Schardl, C.L. Elimination of ergovaline from a grass-Neotyphodium endophyte symbiosis by genetic modification of the endophyte. Proc. Natl. Acad. Sci. USA 2001, 98, 12820–12825. [Google Scholar] [CrossRef] [PubMed]
- Walton, J.D.; Panaccione, D.G.; Hallen, H. Peptide synthesis without ribosomes. In Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine; Tkacz, J., Lange, L., Eds.; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2004; pp. 127–162. [Google Scholar]
- Haarmann, T.; Machado, C.; Lübbe, Y.; Correia, T.; Schardl, C.L.; Panaccione, D.G.; Tudzynski, P. The ergot alkaloid gene cluster in Claviceps purpurea: Extension of the cluster sequence and intra species evolution. Phytochemistry 2005, 66, 1312–1320. [Google Scholar] [CrossRef] [PubMed]
- Haarmann, T.; Lorenz, N.; Tudzynski, P. Use of a nonhomologous end joining deficient strain (Δku70) of the ergot fungus Claviceps purpurea for identification of a nonribosomal peptide synthetase gene involved in ergotamine biosynthesis. Fungal Genet. Biol. 2008, 45, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-Genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef] [PubMed]
- Flieger, M.; Sedmera, P.; Vokoun, J.; Ricicovā, A.; Rehácek, Z. Separation of four isomers of lysergic acid α-hydroxyethylamide by liquid chromatography and their spectroscopic identification. J. Chromatogr. 1982, 236, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, D.G.; Tapper, B.A.; Lane, G.A.; Davies, E.; Fraser, K. Biochemical outcome of blocking the ergot alkaloid pathway of a grass endophyte. J. Agric. Food Chem. 2003, 51, 6429–6437. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, D.G. Ergot alkaloids. In The Mycota, Industrial Applications, 2nd ed.; Hofrichter, M., Ed.; Springer-Verlag: Berlin-Heidelburg, Germany, 2010; Volume 10, pp. 195–214. [Google Scholar]
- Fleetwood, D.J.; Scott, B.; Lane, G.A.; Tanaka, A.; Johnson, R.D. A complex ergovaline gene cluster in Epichloë endophytes of grasses. Appl. Environ. Microbiol. 2007, 73, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Schardl, C.L.; Young, C.A.; Pan, J.; Florea, S.; Takach, J.E.; Panaccione, D.G.; Farman, M.L.; Webb, J.S.; Jaromczyk, J.; Charlton, N.D.; et al. Currencies of mutualisms: Sources of alkaloid genes in vertically transmitted epichloae. Toxins 2013, 5, 1064–1088. [Google Scholar] [CrossRef] [PubMed]
- Steiner, U.; Leibner, S.; Schardl, C.L.; Leuchtmann, A.; Leistner, E. Periglandula, a new fungal genus within the Clavicipitaceae and its association with Convolvulaceae. Mycologia 2011, 103, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Steiner, U.; Ahimsa-Muller, M.A.; Markert, A.; Kucht, S.; Gross, J.; Kauf, N.; Kuzma, M.; Zych, M.; Lamshoft, M.; Furmanowa, M.; et al. Molecular characterization of a seed transmitted clavicipitaceous fungus occurring on dicotyledoneous plants (Convolvulaceae). Planta 2006, 224, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Markert, A.; Steffan, N.; Ploss, K.; Hellwig, S.; Steiner, U.; Drewke, C.; Li, S.-M.; Boland, W.; Leistner, E. Biosynthesis and accumulation of ergoline alkaloids in a mutualistic association between Ipomoea asarifolia (Convolvulaceae) and a clavicipitalean fungus. Plant Physiol. 2008, 147, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, W.T.; Panaccione, D.G.; Hazekamp, C.S.; McKee, M.C.; Ryan, K.L.; Clay, K. Differential allocation of seed-borne ergot alkaloids during early ontogeny of morning glories (Convolvulaceae). J. Chem. Ecol. 2013, 39, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Gröger, D.; Floss, H.G. Biochemistry of ergot alkaloids-achievements and challenges. Alkaloids: Chem. Biol. 1997, 50, 171–218. [Google Scholar]
- Lorenz, N.; Wilson, E.V.; Machado, C.; Schardl, C.L.; Tudzynski, P. Comparison of ergot alkaloid biosynthesis gene clusters in Claviceps species indicates loss of late pathway steps in evolution of C. fusiformis. Appl. Environ. Microbiol. 2007, 73, 7185–7191. [Google Scholar] [CrossRef] [PubMed]
- Rutschmann, J.; Kobel, H.; Schreier, E. Heterocyclic Carboxylic Acids and their Production. U.S. Patent 3,314,961, 18 April 1967. [Google Scholar]
- Haarmann, T.; Ortel, I.; Tudzynski, P.; Keller, U. Identification of the cytochrome P450 monooxygenase that bridges the clavine and ergoline alkaloid pathways. ChemBioChem 2006, 7, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Agurell, S.; Ramstad, E. A new ergot alkaloid from Mexican maize ergot. Acta Pharm. Suec. 1965, 2, 231–238. [Google Scholar] [PubMed]
- Barrow, K.D.; Mantle, P.G.; Quigley, F.R. Biosynthesis of dihydroergot alkaloids. Tetrahedron Lett. 1974, 16, 1557–1560. [Google Scholar] [CrossRef]
- Blaney, B.J.; Maryam, R.; Murray, S.-A.; Ryley, M.J. Alkaloids of the sorghum ergot pathogen (Claviceps africana): Assay methods for grain and feed and variation between sclerotia/sphacelia. Aust. J. Agric. Res. 2003, 54, 167–175. [Google Scholar] [CrossRef]
- Kozlovsky, A.G. Producers of ergot alkaloids out of Claviceps genus. In Ergot: The Genus Claviceps; Kren, V., Cvak, L., Eds.; Harwood Academic Publishers: Amsterdam, The Netherlands, 1999; pp. 479–499. [Google Scholar]
- Robinson, S.L.; Panaccione, D.G. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia 2012, 104, 804–812. [Google Scholar] [CrossRef] [PubMed]
- O’Hanlon, K.A.; Gallagher, L.; Schrettl, M.; Jöchl, C.; Kavanagh, K.; Larsen, T.O.; Doyle, S. Nonribosomal peptide synthetase genes pesL and pes1 are essential for fumigaclavine C production in Aspergillus fumigatus. Appl. Environ. Microbiol. 2012, 78, 3166–3176. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.M.; Yu, Z.G.; Zhang, J.; Wu, J.H.; Tan, R.X. Bioactive alkaloids from endophytic Aspergillus fumigatus. J. Nat. Prod. 2009, 72, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Song, Y.C.; Guo, Y.; Mei, Y.N.; Tan, R.X. Fumigaclavines D–H, new ergot alkaloids from endophytic Aspergillus fumigatus. Planta Med. 2014, 80, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Béliveau, J.; Ramstad, E. 8-Hydroxylation of agroclavine and elymoclavine by fungi. Llyodia 1967, 29, 234–238. [Google Scholar]
- Panaccione, D.G.; Kotcon, J.B.; Schardl, C.L.; Johnson, R.J.; Morton, J.B. Ergot alkaloids are not essential for endophytic fungus-associated population suppression of the lesion nematode, Pratylenchus scribneri, on perennial ryegrass. Nematology 2006, 8, 583–590. [Google Scholar] [CrossRef]
- Panaccione, D.G.; Cipoletti, J.R.; Sedlock, A.B.; Blemings, K.P.; Schardl, C.L.; Machado, C.; Seidel, G.E. Effects of ergot alkaloids on food preference and satiety in rabbits, as assessed with gene knockout endophytes in perennial ryegrass (Lolium perenne). J. Agric. Food Chem. 2006, 54, 4582–4587. [Google Scholar] [CrossRef] [PubMed]
- Potter, D.A.; Stokes, J.T.; Redmond, C.T.; Schardl, C.L.; Panaccione, D.G. Contribution of ergot alkaloids to suppression of a grass-feeding caterpillar assessed with gene-knockout endophytes in perennial ryegrass. Entomol. Exp. Appl. 2008, 126, 138–147. [Google Scholar] [CrossRef]
- Panaccione, D.G.; Coyle, C.M. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl. Environ. Microbiol. 2009, 71, 3106–3111. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, S.L.; Panaccione, D.G. Diversification of Ergot Alkaloids in Natural and Modified Fungi. Toxins 2015, 7, 201-218. https://doi.org/10.3390/toxins7010201
Robinson SL, Panaccione DG. Diversification of Ergot Alkaloids in Natural and Modified Fungi. Toxins. 2015; 7(1):201-218. https://doi.org/10.3390/toxins7010201
Chicago/Turabian StyleRobinson, Sarah L., and Daniel G. Panaccione. 2015. "Diversification of Ergot Alkaloids in Natural and Modified Fungi" Toxins 7, no. 1: 201-218. https://doi.org/10.3390/toxins7010201






