Systematic Study of Binding of μ-Conotoxins to the Sodium Channel NaV1.4
Abstract
:1. Introduction
2. Computational Methods
2.1. Structures of NaV1.4 and µ-Conotoxins
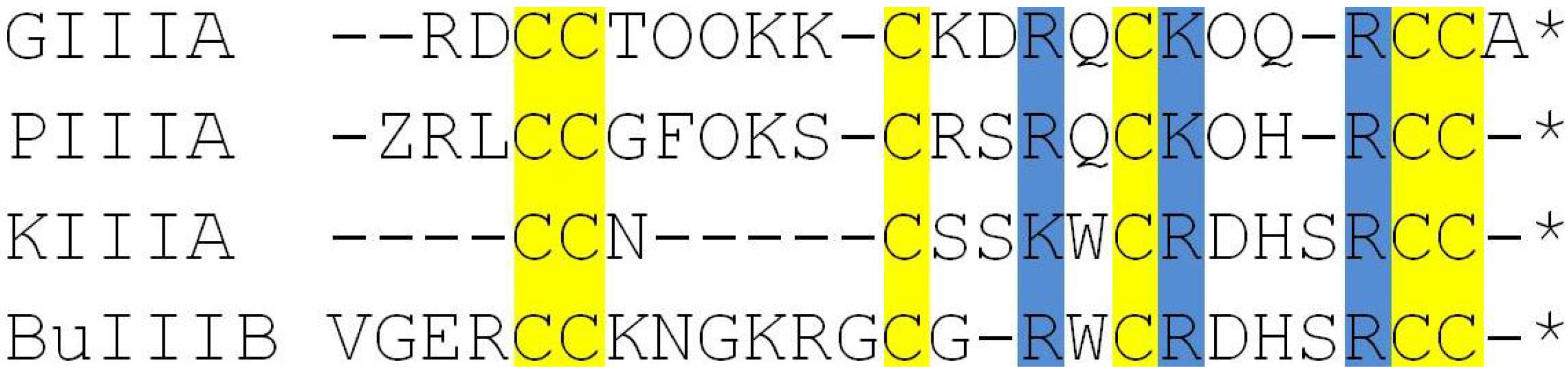
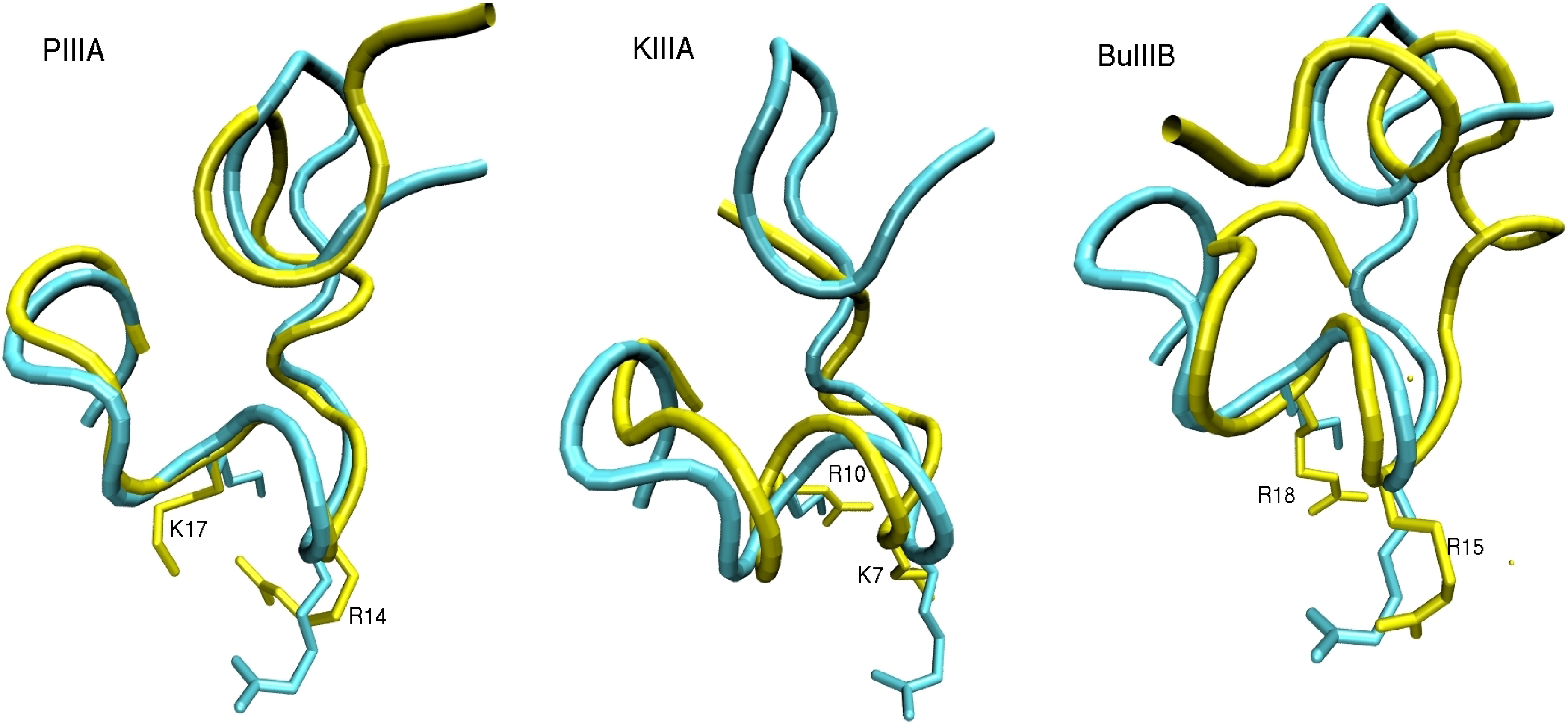
2.2. Complex Structures from Docking and MD Simulations
| Residue | Mutation (Mut.) | Ref. | |
|---|---|---|---|
| PIIIA | |||
| R12 | A | 2.2 | [48] |
| R12 | Q | 1.6 | [48] |
| R12 | K | 1.2 | [48] |
| S13 | D | 6.3 | [48] |
| R14 | A | 13.0 | [48] |
| R14 | Q | 15.5 | [48] |
| R14 | K | 2.2 | [48] |
| K17 | A | 5.0 | [48] |
| K17 | Q | 7.0 | [48] |
| H19 | Q | 0.9 | [48] |
| KIIIA | |||
| K7 | A | 35.7 | [13] |
| K7 | A | 13.4 | [11] |
| K7 | D | 18.3 | [11] |
| W8 | R | 37.3 | [12] |
| W8 | Q | 73.2 | [12] |
| W8 | E | 320 | [12] |
| R10 | A | 27.1 | [13] |
| H12 | A | 2986 | [13] |
| R14 | A | 151 | [13] |
2.3. MD Simulations and Binding Free Energy Calculations
3. Results and Discussion
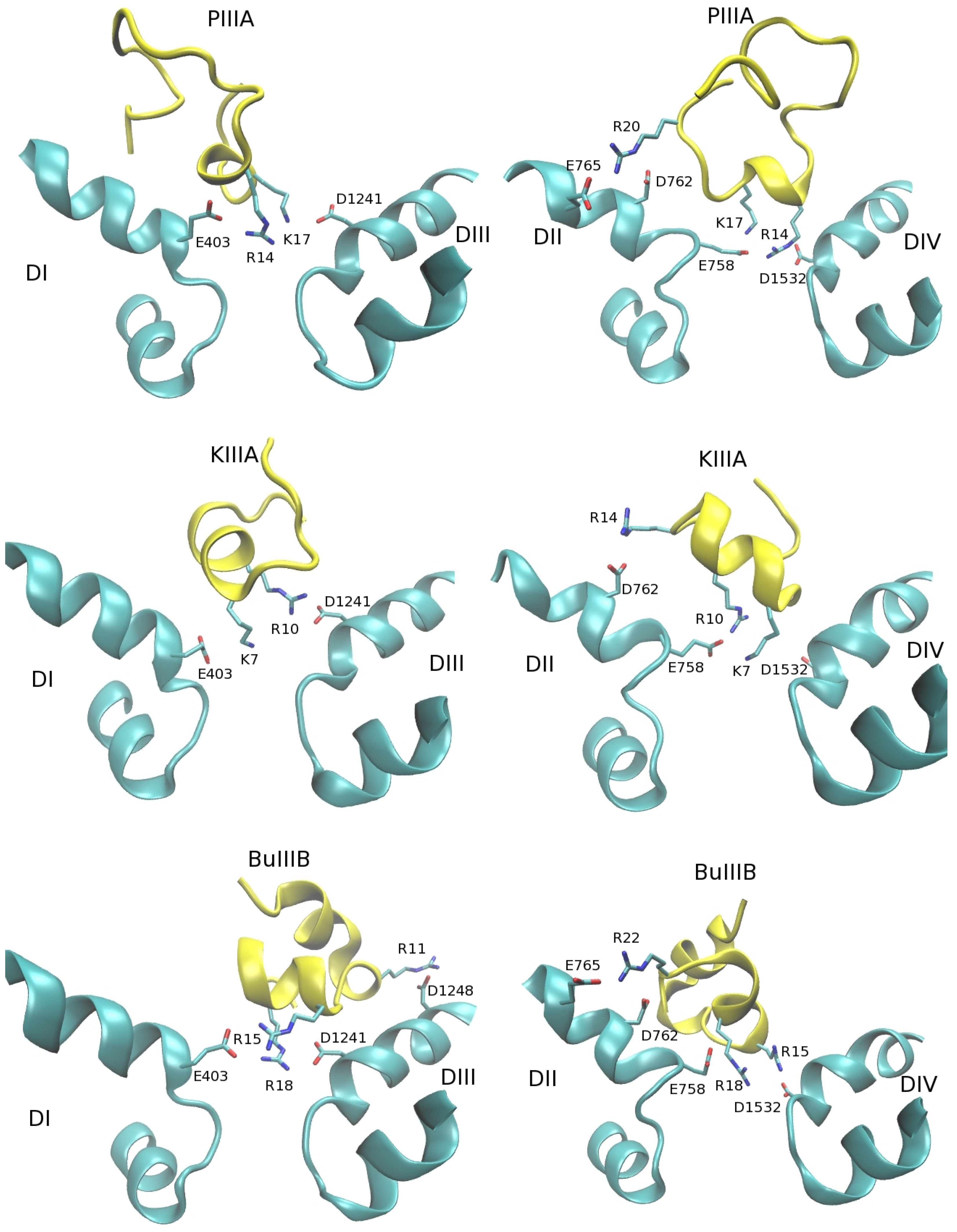
3.1. Binding Modes of the NaV1.4-µ-Conotoxin Complexes
| NaV1.4 | GIIIA | MD | PIIIA | MD | KIIIA | MD | BuIIIB | MD |
|---|---|---|---|---|---|---|---|---|
| E403-O1 | R13-N2 | 2.7 | R14-N1 | 2.7 | − | − | R15-N2 | 2.7 |
| E758-O2 | R13-N1 | 2.8 | R14-N1 | 2.7 | K7-Nz | 2.9 | R18-N1 | 2.7 |
| E758-O1 | K16-Nz | 2.7 | K17-Nz | 2.7 | R10-N1 | 2.7 | R18-N2 | 3.5 |
| D1241-O2 | K16-Nz | 2.7 | K17-Nz | 2.9 | R10-N2 | 2.8 | − | − |
| D1241-O1 | K11-Nz | 2.7 | − | − | − | − | R18-N2 | 2.7 |
| D1532-O1 | K11-Nz | 2.6 | − | − | − | − | − | |
| D1532-O2 | R13-N2 | 2.7 | R14-NE | 2.9 | K7-Nz | 3.0 | R15-N2 | 2.7 |
| D762-O2 | R19-N2 | 2.7 | R20-N2 | 2.7 | R14-N1 | 2.7 | R22-N2 | 2.7 |
| E765-O1 | − | − | − | − | − | R22-N2 | 2.7 | |
| D1248-O1 | K8-Nz | 4.0 | − | − | − | − | R11-N2 | 2.8 |
3.2. Binding Free Energy of PIIIA
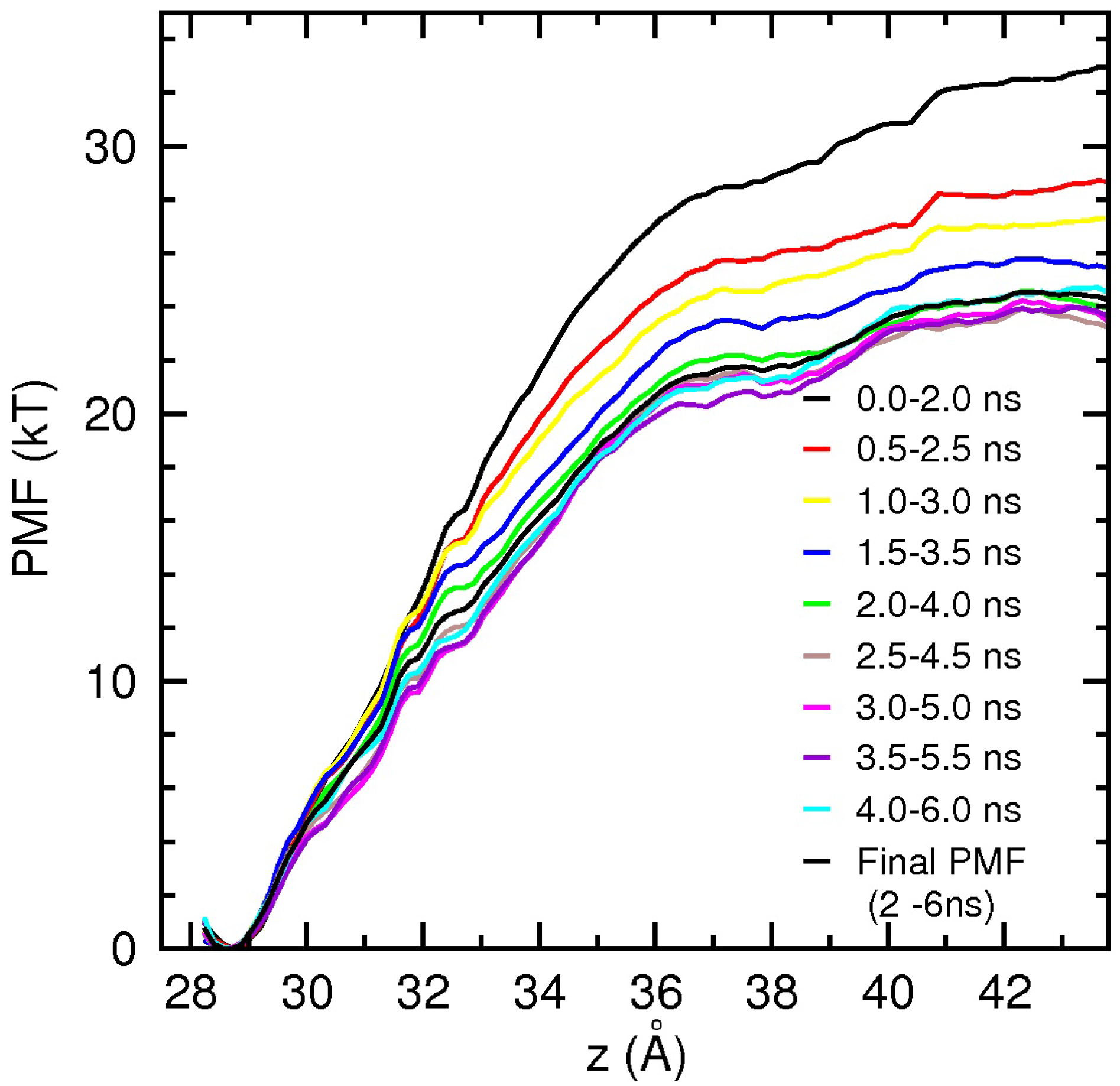
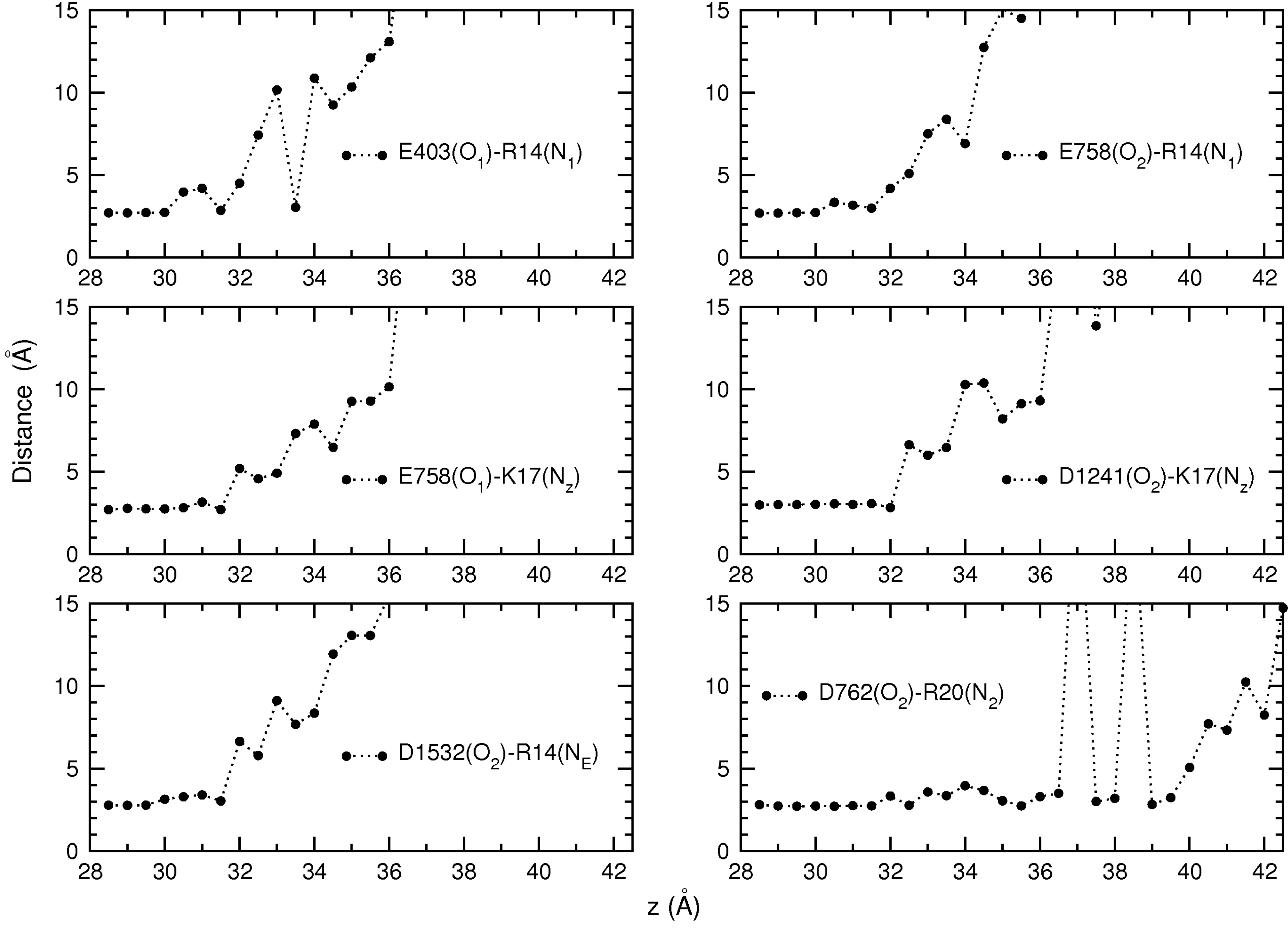
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hille, B. Ionic Channels of Excitable Membranes, 3rd ed.; Sinauer Associates: Sunderland, MA, USA, 2001. [Google Scholar]
- Ashcroft, F.M. Ion Channels and Disease: Channelopathies; Academic Press: San Diego, CA, USA, 2000. [Google Scholar]
- French, R.J.; Terlau, H. Sodium channel toxins—Receptor targeting and therapeutic potential. Curr. Med. Chem. 2004, 11, 3053–3064. [Google Scholar] [CrossRef] [PubMed]
- Twede, V.D.; Miljanich, G.; Olivera, B.M.; Bulaj, G. Neuroprotective and cardioprotective conopeptides: An emerging class of drug leads. Curr. Opin. Drug. Discov. Devel. 2009, 12, 231–239. [Google Scholar] [PubMed]
- Norton, R.S. µ-Conotoxins as leads in the development of new analgesics. Molecules 2010, 15, 2825–2844. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.J.; Yoshikami, D.; Azam, L.; Gajewiak, J.; Olivera, B.M.; Bulaj, G.; Zhang, M.M. µ-Conotoxins that differentially block sodium channels NaV1.1 through 1.8 identify those responsible for action potentials in sciatic nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 10302–10307. [Google Scholar] [CrossRef] [PubMed]
- Mantegazza, M.; Curia, G.; Biagini, G.; Ragsdale, D.S.; Avoli, M. Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet Neurol. 2010, 9, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Peigneur, S.; Dyubankova, N.; Lescrinier, E.; Herdewijn, P.; Tytgat, J. Design of bioactive peptides from naturally occurring µ-conotoxin structures. J. Biol. Chem. 2012, 287, 31382–31392. [Google Scholar] [CrossRef] [PubMed]
- Lampert, A.; O’Reilly, A.O.; Reeh, P.; Leffler, A. Sodium channelopathies and pain. Pflug. Arch. 2010, 460, 249–263. [Google Scholar] [CrossRef]
- Han, T.S.; Zhang, M.M.; Walewska, A.; Gruszczynski, P.; Robertson, C.R.; Cheatham, T.E., III; Yoshikami, D.; Olivera, B.M.; Bulaj, G. Structurally minimized µ-conotoxin analogues as sodium channel blockers: implications for designing conopeptide-based therapeutics. ChemMedChem 2009, 4, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Han, T.S.; Olivera, B.M.; Bulaj, G.; Yoshikami, D. µ-conotoxin KIIIA derivatives with divergent affinities versus efficacies in blocking voltage-gated sodium channels. Biochemistry 2010, 49, 4804–4812. [Google Scholar] [CrossRef] [PubMed]
- Van Der Haegen, A.; Peigneur, S.; Tytgat, J. Importance of position 8 in µ-conotoxin KIIIA for voltage-gated sodium channel selectivity. FEBS J. 2011, 278, 3408–3418. [Google Scholar] [CrossRef] [PubMed]
- McArthur, J.R.; Singh, G.; McMaster, D.; Winkfein, R.; Tieleman, D.P.; French, R.J. Interactions of key charged residues contributing to selective block of neuronal sodium channels by µ-conotoxin KIIIA. Mol. Pharmacol. 2011, 80, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.; Peigneur, S.; Dyubankova, N.; Lescrinier, E.; Herdewijn, P.; Tytgat, J. Design of bioactive peptides from naturally occurring µ-conotoxin structures. J. Biol. Chem. 2012, 287, 31382–31392. [Google Scholar] [CrossRef] [PubMed]
- Khoo, K.K.; Wilson, M.J.; Smith, B.J.; Zhang, M.M.; Gulyas, J.; Yoshikami, D.; Rivier, J.E.; Bulaj, G.; Norton, R.S. Lactam-stabilized helical analogues of the analgesic µ-conotoxin KIIIA. J. Med. Chem. 2012, 54, 7558–7566. [Google Scholar] [CrossRef]
- Cruz, L.J.; Gray, W.R.; Olivera, B.M.; Zeikus, R.D.; Kerr, L.; Yoshikami, D.; Moczydlowski, D. Conus Geographus toxins that discriminate between neuronal and muscle sodium channels. J. Biol. Chem. 1985, 260, 9280–9288. [Google Scholar] [PubMed]
- Terlau, H.; Olivera, B.M. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef] [PubMed]
- Li, R.A.; Tomaselli, G.F. Using the deadly µ-conotoxins as probes of voltage-gated sodium channels. Toxicon 2004, 44, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Lewis, R.J. Use of venom peptides to probe ion channel structure and function. J. Biol. Chem. 2010, 285, 13315–13320. [Google Scholar] [CrossRef] [PubMed]
- Payandeh, J.; Scheuer, T.; Zheng, N.; Catterall, W.A. The crystal structure of a voltage-gated sodium channel. Nature 2011, 475, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Payandeh, J.; Scheuer, T.; Zheng, N.; Catterall, W.A. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 2012, 486, 135–139. [Google Scholar] [PubMed]
- Zhang, X.; Ren, W.; DeCaen, P.; Yan, C.; Tao, X.; Tang, L.; Wang, J.; Hasegawa, K.; Kumasaka, T.; He, J.; et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 2012, 486, 130–134. [Google Scholar] [CrossRef] [PubMed]
- McCusker, E.C.; Bagneris, C.; Naylor, C.E.; Cole, A.R.; D’Avanzo, N.; Nichols, C.G.; Wallace, B.A. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat. Commun. 2012, 3, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Shaya, D.; Findeisen, F.; Abderemane-Ali, F.; Arrigoni, C.; Wong, S.; Nurva1, S.R.; Loussouarn, G.; Minor, D.L., Jr. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J. Mol. Biol. 2014, 426, 467–483. [Google Scholar] [CrossRef] [PubMed]
- Tikhonov, D.B.; Zhorov, B.S. Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NaVAb bacterial sodium channel structure. Mol. Pharmacol. 2012, 82, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Chung, S.H. Mechanism of tetrodotoxin block and resistance in sodium channels. Biochem. Biophys. Res. Commun. 2014, 446, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Robinson, A.; Chung, S.H. Mechanism of µ-conotoxin PIIIA binding to the voltage-gated Na+ channel NaV1.4. PLoS One 2014, 9, e93267. [Google Scholar] [CrossRef] [PubMed]
- Korkosh, V.S.; Zhorov, B.S.; Tikhonov, D.B. Folding similarity of the outer pore region in prokaryotic and eukaryotic sodium channels revealed by docking of conotoxins GIIIA, PIIIA, and KIIIA in a NaVAb-based model of NaV1.4. J. Gen. Physiol. 2014, 144, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, S.; Kuyucak, S. Molecular dynamics study of binding of µ-conotoxin GIIIA to the voltage-gated sodium channel NaV1.4. PLoS One 2014, 9, e105300. [Google Scholar] [CrossRef] [PubMed]
- Shon, K.J.; Olivera, B.M.; Watkins, M.; Jacobsen, R.B.; Gray, W.R.; Floresca, C.Z.; Cruz, L.J.; Hillyard, D.R.; Brink, A.; Terlau, H.; et al. µ-conotoxin PIIIA, a new peptide for discriminating among tetrodotoxin-sensitive Na channel subtypes. J. Neurosci. 1998, 18, 4473–4481. [Google Scholar] [PubMed]
- Bulaj, G.; West, P.J.; Garrett, J.E.; Watkins, M.; Marsh, M.; Zhang, M.M.; Norton, R.S.; Smith, B.J.; Yoshikami, D.; Olivera, B.M. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry 2005, 44, 7259–7265. [Google Scholar] [CrossRef] [PubMed]
- Holford, M.; Zhang, M.M.; Gowd, K.H.; Azam, L.; Green, B.R.; Watkins, M.; Ownby, J.P.; Yoshikami, D.; Bulaj, G.; Olivera, B.M. Pruning nature: Biodiversity-derived discovery of novel sodium channel blocking conotoxins from Conus bullatus. Toxicon 2009, 53, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Kuyucak, S. Accurate determination of the binding free energy for KcsA-Charybdotoxin complex from the potential of mean force calculations with restraints. Biophys. J. 2011, 100, 2466–2474. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Kuyucak, S. Developing a comparative docking protocol for the prediction of peptide selectivity proïnˇA˛les: Investigation of potassium channel toxins. Toxins 2012, 4, 110–138. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Kuyucak, S. Affinity and selectivity of ShK toxin for the Kv1 potassium channels from free energy simulations. J. Phys. Chem. B 2012, 116, 4812–4822. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Heinzelmann, G.; Huq, R.; Tajhya, R.B.; Chang, S.C.; Chhabra, S.; Pennington, M.W.; Beeton, C.; Norton, R.S.; Kuyucak, S. A potent and selective peptide blocker of the Kv1.3 channel: Prediction from free-energy simulations and experimental confirmation. PLoS One 2013, 8, e78712. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, S.; Kuyucak, S. Why Drosophila Shaker K+ channel is not a good model for ligand binding to voltage-gated Kv1 channels. Biochemistry 2013, 52, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Kuyucak, S. Free energy simulations of binding of HsTx1 toxin to Kv1 potassium channels: the basis of Kv1.3/Kv1.1 selectivity. J. Phys. Chem. B 2014, 118, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Huq, R.; Tanner, M.R.; Chhabra, S.; Khoo, K.K.; Estrada, R.; Dhawan, V.; Chauhan, S.; Pennington, M.W.; Beeton, C.; et al. A potent and Kv1.3-selective analogue of the scorpion toxin HsTX1 as a potential therapeutic for autoimmune diseases. Sci. Rep. 2014, 4, 4509. [Google Scholar] [CrossRef] [PubMed]
- Tietze, A.A.; Tietze, D.; Ohlensclager, O.; Leipold, E.; Ulrich, F.; Kuhl, T.; Mischo, A.; Buntkowsky, G.; Gorlach, M.; Heinemnan, S.H.; Imhof, D. Structurally diverse µ-conotoxin PIIIA isomers block sodium channel NaV1.4. Angew. Chem. Int. Ed. 2012, 51, 4058–4061. [Google Scholar] [CrossRef]
- Khoo, K.K.; Gupta, K.; Green, B.R.; Zhang, M.M.; Watkins, M.; Olivera, B.M.; Balaram, P.; Yoshikami, D.; Bulaj, G.; Norton, R.S. Distinct disulfide isomers of µ-conotoxins KIIIA and KIIIB block voltage-gated sodium channels. Biochemistry 2012, 51, 9826–9835. [Google Scholar]
- Kuang, Z.; Zhang, M.M.; Gupta, K.; Gajewiak, J.; Gulyas, J.; Balaram, P.; Rivier, J.E.; Olivera, B.M.; Yoshikami, D.; Bulaj, G.; et al. Mammalian neuronal sodium channel blocker µ-conotoxin BuIIIB has a structured N-terminus that influences potency. ACS Chem. Biol. 2013, 8, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Wakamatsu, K.; Kohda, D.; Hatanaka, H.; Lancelin, J.M.; Ishida, Y.; Oya, M.; Nakamura, H.; Inagaki, F.; Sato, K. Structure-activity relationships of µ-conotoxin GIIIA: Structure determination of active and inactive sodium channel blocker peptides by NMR and simulated annealing calculations. Biochemistry 1992, 31, 12577–12584. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.J.; Watson, M.; Adams, D.J.; Hammarstrom, A.K.; Gage, P.W.; Hill, J.M.; Craik, D.J.; Thomas, L.; Adams, D.; Alewood, P.F.; et al. Solution structure of µ-conotoxin PIIIA, a preferential inhibitor of persistent tetrodotoxin-sensitive sodium channels. J. Biol. Chem. 2002, 277, 27247–27255. [Google Scholar] [CrossRef] [PubMed]
- Khoo, K.K.; Feng, Z.P.; Smith, B.J.; Zhang, M.M.; Yoshikami, D.; Olivera, B.M.; Bulaj, G.; Norton, R.S. Structure of the analgesic µ-conotoxin KIIIA and effects on the structure and function of disulfide deletion. Biochemistry 2009, 48, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.J.; van Dijk, A.D.; Krzeminski, M.; van Dijk, M.; Thureau, A.; Hsu, V.; Wassenaar, T.; Bonvin, A.M.J.J. HADDOCK versus HADDOCK: New features and performance of HADDOCK 2.0 on the CAPRI targets. Proteins 2007, 69, 726–733. [Google Scholar] [CrossRef] [PubMed]
- McArthur, J.R.; Ostroumov, V.; Al-Sabi, A.; McMaster, D.; French, R.J. Multiple, distributed interactions of µ-conotoxin PIIIA associated with broad targeting among voltage-gated sodium channels. Biochemistry 2011, 50, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Chahine, M.; Sirois, J.; Marcotte, P.; Chen, L.Q.; Kallen, R.G. Extrapore residues of the S5-S6 loop of domain 2 of the voltage-gated skeletal muscle sodium channel (rSkM1) contribute to the µ-conotoxin GIIIA binding site. Biophys. J. 1998, 75, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Bastug, T.; Kuyucak, S. Energetics of ion permeation, rejection, binding and block in gramicidin A from free energy simulations. Biophys. J. 2006, 90, 3941–3950. [Google Scholar] [CrossRef] [PubMed]
- Bastug, T.; Kuyucak, S. Importance of the peptide backbone description in modeling the selectivity filter in potassium channels. Biophys. J. 2009, 96, 4006–4012. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force field. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [PubMed]
- Kumar, S.; Bouzida, D.; Swensen, R.H.; Kollman, P.A.; Rosenberg, J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Xue, T.; Ennis, I.L.; Sato, K.; French, R.J.; Li, R.A. Novel interactions identified between µ-conotoxin and the Na+ channel domain I P-loop: Implications for toxin-pore binding geometry. Biophys. J. 2003, 85, 2299–2310. [Google Scholar] [CrossRef] [PubMed]
- Li, R.A.; Ennis, I.L.; French, R.J.; Dudley, S.C., Jr.; Tomaselli, G.F.; Marban, E. Clockwise domain arrangement of the sodium channel revealed by µ-conotoxin (GIIIA) docking orientation. J. Biol. Chem. 2001, 276, 11072–11077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Green, B.R.; Catlin, P.; Fiedler, B.; Azam, L.; Chadwick, A.; Terlau, H.; McArthur, J.R.; French, R.J.; Gulyas, J.; et al. Structure/function characterization of µ-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J. Biol. Chem. 2007, 282, 30699–30706. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Kuyucak, S. Mechanism and energetics of charybdotoxin unbinding from a potassium channel from molecular dynamics simulations. Biophys. J. 2009, 96, 2577–2588. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahdavi, S.; Kuyucak, S. Systematic Study of Binding of μ-Conotoxins to the Sodium Channel NaV1.4. Toxins 2014, 6, 3454-3470. https://doi.org/10.3390/toxins6123454
Mahdavi S, Kuyucak S. Systematic Study of Binding of μ-Conotoxins to the Sodium Channel NaV1.4. Toxins. 2014; 6(12):3454-3470. https://doi.org/10.3390/toxins6123454
Chicago/Turabian StyleMahdavi, Somayeh, and Serdar Kuyucak. 2014. "Systematic Study of Binding of μ-Conotoxins to the Sodium Channel NaV1.4" Toxins 6, no. 12: 3454-3470. https://doi.org/10.3390/toxins6123454




