Toxicodynamics of Mycotoxins in the Framework of Food Risk Assessment—An In Silico Perspective
Abstract
:1. Introduction: Food Toxicology and Risk Assessment
2. Toxicokinetics and Toxicodynamics in Chemicals Risk Assessment
2.1. Toxicokinetics
2.2. Toxicodynamics
3. In Silico Analysis in Risk Assessment: A Toxicodynamics Perspective
3.1. Computational Tools in Assessing the Combined Toxicity
3.2. Computational Tools in Assessing the (Poly)toxicology of Mycotoxins Action
3.3. Computational Tools to Understand Interspecies Variability
3.4. Computational Tools in Accounting Inter-Individual Variability: A First Step in Personalized Risk Assessment?
4. Case Studies
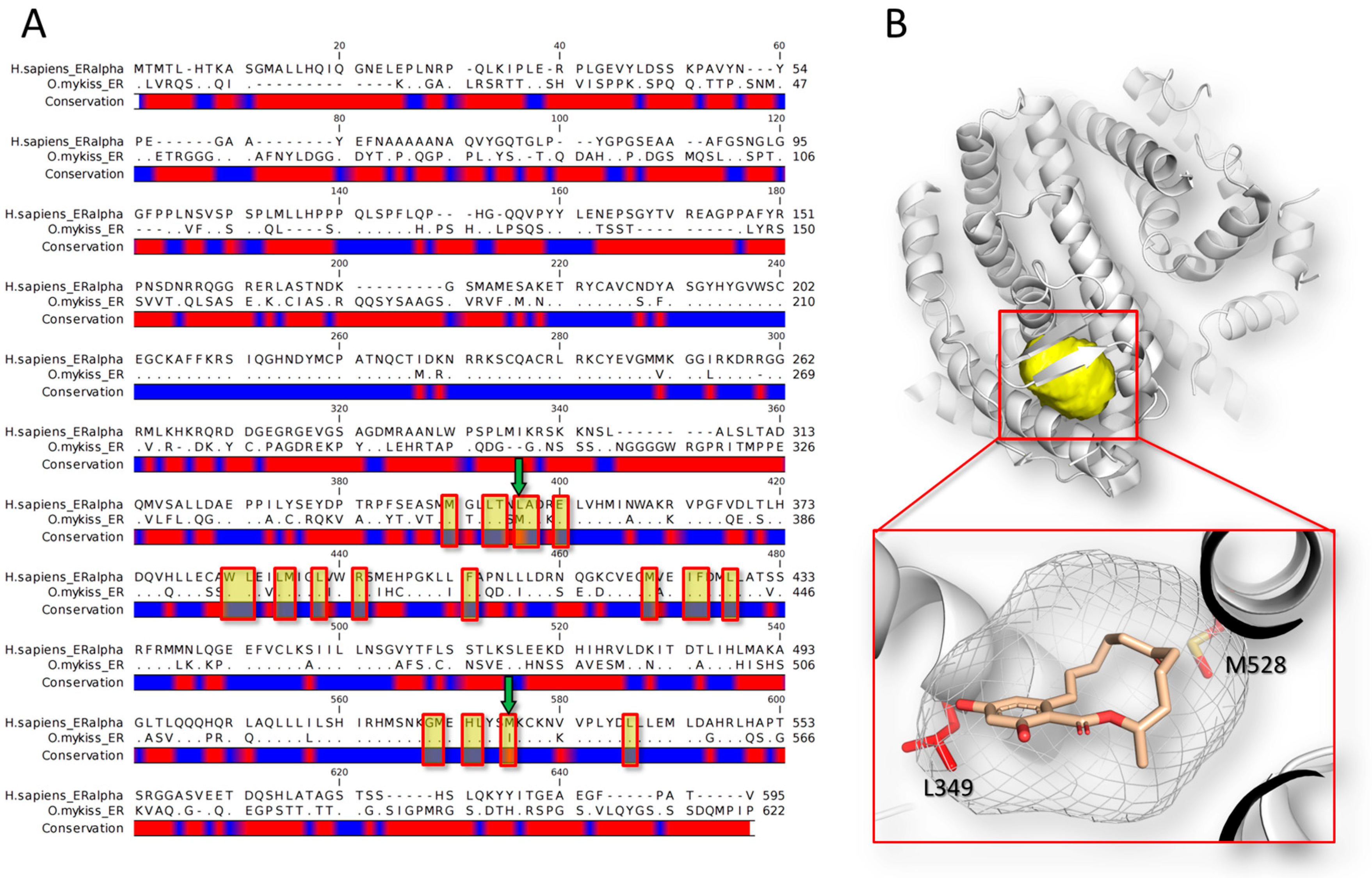
4.1. Estrogenic Activity of Zearalenone Group
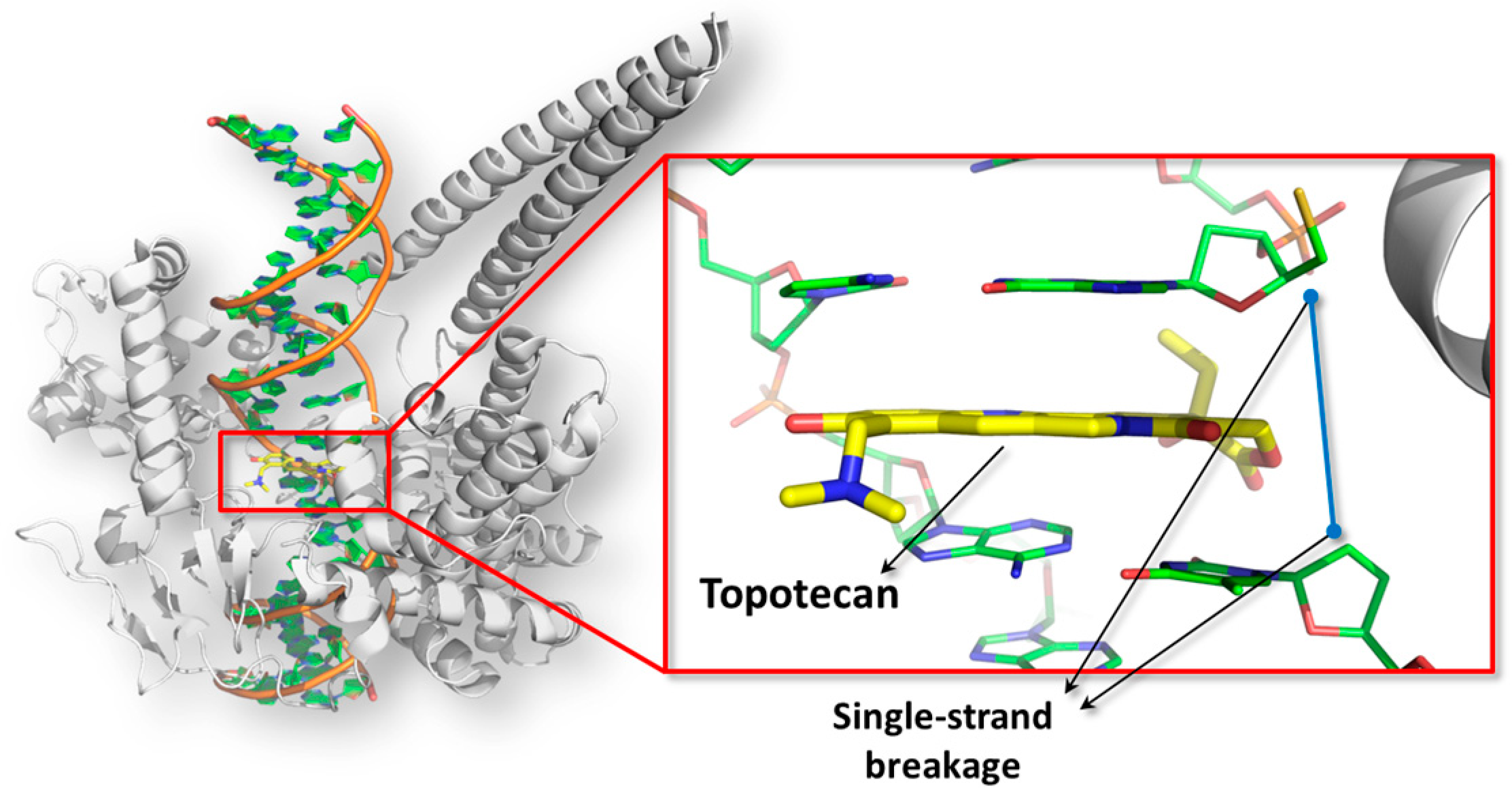
4.2. Topoimoerase I Poisoning by Alternariol
4.3. (Poly)toxicology of Ochratoxin A
4.4. Ribotoxicity of Trichothecenes
5. Conclusions
Author Contributions
Conflicts of Interest
References
- Kotsonis, F.N.; Burdock, G.A. Food toxicology. In Casarett & Doull’s Toxicology: The Basic Science of Poisons, 8th ed.; Klaassen, C.D., Ed.; McGraw-Hill Education: New York, NY, USA, 2013. [Google Scholar]
- Shaw, I.C. Chemical residues, food additives and natural toxicants in food—The cocktail effect. Int. J. Food Sci. Technol. 2014, 49, 2149–2157. [Google Scholar] [CrossRef]
- Svingen, T.; Vinggaard, A.M. The risk of chemical cocktail effects and how to deal with the issue. J. Epidemiol. Commun. Health 2016, 70, 322–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, D.; Tennant, D.R.; Wood, R.; Patel, P.; Hoogenboom, R.; Dixon, S.N.; Harrison, N.; Castle, L.; Shaw, I.; Vannort, R.; et al. Contributors. In Food Chemical Safety; Woodhead Publishing: Cambridge, UK, 2001; pp. xi–xiii. [Google Scholar]
- Silano, M.; Silano, V. Food and feed chemical contaminants in the european union: Regulatory, scientific, and technical issues concerning chemical contaminants occurrence, risk assessment, and risk management in the european union. Crit. Rev. Food Sci. Nutr. 2017, 57, 2161–2217. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Dall’Asta, C. Forthcoming challanges in mycotoxins toxicology research for safer food—A need for multi-omics approach. Toxins (Basel) 2017, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Marroquín-Cardona, A.G.; Johnson, N.M.; Phillips, T.D.; Hayes, A.W. Mycotoxins in a changing global environment—A review. Food Chem. Toxicol. 2014, 69, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Groopman, J.D.; Pestka, J.J. Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 2014, 5, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Guclu, H. Aflatoxin regulations in a network of global maize trade. PLoS ONE 2012, 7, e45151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assunção, R.; Silva, M.J.; Alvito, P. Challenges in risk assessment of multiple mycotoxins in food. World Mycotoxin J. 2016, 9, 791–811. [Google Scholar] [CrossRef]
- Gruber-Dorninger, C.; Novak, B.; Nagl, V.; Berthiller, F. Emerging mycotoxins: Beyond traditionally determined food contaminants. J. Agric. Food Chem. 2017, 65, 7052–7070. [Google Scholar] [CrossRef] [PubMed]
- Dorne, J.L.; Kass, G.E.; Bordajandi, L.R.; Amzal, B.; Bertelsen, U.; Castoldi, A.F.; Heppner, C.; Eskola, M.; Fabiansson, S.; Ferrari, P.; et al. Human risk assessment of heavy metals: Principles and applications. Met. Ions Life Sci. 2011, 8, 27–60. [Google Scholar] [PubMed]
- Tennekes, H.A.; Sánchez-Bayo, F. The molecular basis of simple relationships between exposure concentration and toxic effects with time. Toxicology 2013, 309, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Boonen, J.; Malysheva, S.V.; Taevernier, L.; Diana Di Mavungu, J.; De Saeger, S.; De Spiegeleer, B. Human skin penetration of selected model mycotoxins. Toxicology 2012, 301, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Föllmann, W.; Ali, N.; Blaszkewicz, M.; Degen, G.H. Biomonitoring of mycotoxins in urine: Pilot study in mill workers. J. Toxicol. Environ. Health 2016, 79, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Rubió, L.; Macià, A.; Motilva, M.J. Impact of various factors on pharmacokinetics of bioactive polyphenols: An overview. Curr. Drug Metab. 2014, 15, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Druckrey, H.; Küpfmüller, K. Quantitative analyse der krebsentstehung. Zeitschrift für Naturforschung B 1948, 3, 254–266. [Google Scholar] [CrossRef]
- Taylor, R.D.; Jewsbury, P.J.; Essex, J.W. Fds: Flexible ligand and receptor docking with a continuum solvent model and soft-core energy function. J. Comput. Chem. 2003, 24, 1637–1656. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.Q.; Weaver, L.H.; Ferrari, A.M.; Matthews, B.W.; Shoichet, B.K. Testing a flexible-receptor docking algorithm in a model binding site. J. Mol. Biol. 2004, 337, 1161–1182. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.E.; Goodman, J.M.; Gutsell, S.; Russell, P.J. Defining molecular initiating events in the adverse outcome pathway framework for risk assessment. Chem. Res. Toxicol. 2014, 27, 2100–2112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, A.W.; Kruger, C.L. Hayes’ Principles and Methods of Toxicology; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Grant, R.L.; Combs, A.B.; Acosta, D. Experimental models for the investigation of toxicological mechanisms. In Comprehensive Toxicology, 2nd ed.; McQueen, C.A., Ed.; Elsevier: Oxford, UK, 2010. [Google Scholar]
- Browne, P.; Noyes, P.D.; Casey, W.M.; Dix, D.J. Application of adverse outcome pathways to u.S. Epa’s endocrine disruptor screening program. Environ. Health Perspect. 2017, 25, 096001. [Google Scholar] [CrossRef] [PubMed]
- Kenakin, T.P. Chapter 5—Allosteric drug effects. In Pharmacology in Drug Discovery and Development, 2nd ed.; Academic Press: New York, NY, USA, 2017; pp. 101–129. [Google Scholar]
- Vejdovszky, K.; Hahn, K.; Braun, D.; Warth, B.; Marko, D. Synergistic estrogenic effects of fusarium and alternaria mycotoxins in vitro. Arch. Toxicol. 2017, 91, 1447–1460. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Scientific opinion on the appropriateness to set a group health-based guidance value for zearalenone and its modified forms. EFSA J. 2016, 14, 4425. [Google Scholar]
- Yang, Y.; Yu, S.; Tan, Y.; Liu, N.; Wu, A. Individual and combined cytotoxic effects of co-occurring deoxynivalenol family mycotoxins on human gastric epithelial cells. Toxins (Basel) 2017, 9, E96. [Google Scholar] [CrossRef] [PubMed]
- Frizzell, C.; Ndossi, D.; Verhaegen, S.; Dahl, E.; Eriksen, G.; Sørlie, M.; Ropstad, E.; Muller, M.; Elliott, C.T.; Connolly, L. Endocrine disrupting effects of zearalenone, alpha- and beta-zearalenol at the level of nuclear receptor binding and steroidogenesis. Toxicol. Lett. 2011, 206, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Tatay, E.; Espín, S.; García-Fernández, A.J.; Ruiz, M.J. Estrogenic activity of zearalenone, α-zearalenol and β-zearalenol assessed using the E-screen assay in MCF-7 cells. Toxicol. Mech. Methods 2017, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.; Klich, M. Mycotoxins. Clin. Microb. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef]
- Raies, A.B.; Bajic, V.B. In silico toxicology: Computational methods for the prediction of chemical toxicity. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2016, 6, 147–172. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Liu, G.; Tang, Y. In silico admet prediction: Recent advances, current challenges and future trends. Curr. Top. Med. Chem. 2013, 13, 1273–1278. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Modern methodologies and tools for human hazard assessment of chemicals. EFSA J. 2014, 12, 3638. [Google Scholar]
- Wilson, G.L.; Lill, M.A. Integrating structure-based and ligand-based approaches for computational drug design. Future Med. Chem. 2011, 3, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Fradera, X.; Babaoglu, K. Overview of methods and strategies for conducting virtual small molecule screening. Curr. Protoc. Chem. Biol. 2017, 14, 196–212. [Google Scholar]
- Baig, M.H.; Ahmad, K.; Roy, S.; Ashraf, J.M.; Adil, M.; Siddiqui, M.H.; Khan, S.; Kamal, M.A.; Provazník, I.; Choi, I. Computer aided drug design: Success and limitations. Curr. Pharm. Des. 2016, 22, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Hansch, C.; Fujita, T. Rho-sigmapi analysis: A method for the correlation of biological activity and chemical structure. J. Am. Chem. Soc. 1964, 86, 1616–1626. [Google Scholar] [CrossRef]
- Putz, M.V.; Duda-Seiman, C.; Duda-Seiman, D.; Putz, A.M.; Alexandrescu, I.; Mernea, M.; Avram, S. Chemical structure-biological activity models for pharmacophores’ 3D-interactions. Int. J. Mol. Sci. 2016, 17, E1087. [Google Scholar] [CrossRef] [PubMed]
- Poroikov, V.; Filiminov, D. Pass: Prediction of biological activity spectra for substances. In Predictive Toxicology; Helma, C., Ed.; CRC Press: Boca Raton, FL, USA, 2005; pp. 459–474. [Google Scholar]
- Cozzini, P.; Fornabaio, M.; Marabotti, A.; Abraham, D.J.; Kellogg, G.E.; Mozzarelli, A. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J. Med. Chem. 2002, 45, 2469–2483. [Google Scholar] [CrossRef] [PubMed]
- Michel, J.; Essex, J.W. Prediction of protein-ligand binding affinity by free energy simulations: Assumptions, pitfalls and expectations. J. Comput. Aided Mol. Des. 2010, 24, 639–658. [Google Scholar] [CrossRef] [PubMed]
- Yip, K.Y.; Wan, M.L.Y.; Wong, A.S.T.; Korach, K.S.; El-Nezami, H. Combined low-dose zearalenone and aflatoxin b1 on cell growth and cell-cycle progression in breast cancer MCF-7 cells. Toxicol. Lett. 2017, 281, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.C.; Madec, S.; Pawtowski, A.; Coton, E.; Hymery, N. Individual and combined toxicological effects of deoxynivalenol and zearalenone on human hepatocytes in in vitro chronic exposure conditions. Toxicol. Lett. 2017, 280, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; George, S.; Hay, C.; Lee, J.; Qian, H.; Sun, X. Individual and combined effects of aflatoxin B1, deoxynivalenol and zearalenone on HepG2 and RAW 264.7 cell lines. Food Chem. Toxicol. 2017, 103, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Kovalsky, P.; Kos, G.; Nährer, K.; Schwab, C.; Jenkins, T.; Schatzmayr, G.; Sulyok, M.; Krska, R. Co-occurrence of regulated, masked and emerging mycotoxins and secondary metabolites in finished feed and maize-an extensive survey. Toxins (Basel) 2016, 8, E363. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant. Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berthiller, F.; Lemmens, M.; Werner, U.; Krska, R.; Hauser, M.T.; Adam, G.; Schuhmacher, R. Short review: Metabolism of thefusarium mycotoxins deoxynivalenol and zearalenone in plants. Mycotoxin Res. 2007, 23, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Shoichet, B.K. Virtual screening of chemical libraries. Nature 2004, 432, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Pati, S.P.; Kumar, P.P.; Pradeep, H.N.; Sastry, G.N. Virtual screening in drug discovery—A computational perspective. Curr. Protein Pept. Sci. 2007, 8, 329–351. [Google Scholar] [CrossRef] [PubMed]
- Guardado Yordi, E.; Matos, M.J.; Pérez Martínez, A.; Tornes, A.C.; Santana, L.; Molina, E.; Uriarte, E. In Silico genotoxicity of coumarins: Application of the phenol-explorer food database to functional food science. Food Funct. 2017, 8, 2958–2966. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, V.A.; Dellafiora, L.; Mollergues, J.; Dall’Asta, C.; Serrant, P.; Marin-Kuan, M.; Lo Piparo, E.; Schilter, B.; Cozzini, P. Hazard assessment through hybrid in vitro/in silico approach: The case of zearalenone. ALTEX 2015, 32, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, E.; Hildebrand, A.; Damm, G.; Rapp, A.; Cramer, B.; Humpf, H.-U.; Metzler, M. Aromatic hydroxylation is a major metabolic pathway of the mycotoxin zearalenone in vitro. Mol. Nutr. Food Res. 2009, 53, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, E.; Schebb, N.H.; Podlech, J.; Metzler, M. Novel oxidative in vitro metabolites of the mycotoxins alternariol and alternariol methyl ether. Mol. Nutr. Food Res. 2007, 51, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Fleck, S.C.; Churchwell, M.I.; Doerge, D.R. Metabolism and pharmacokinetics of zearalenone following oral and intravenous administration in juvenile female pigs. Food Chem. Toxicol. 2017, 106, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Molina-Molina, J.M.; Real, M.; Jimenez-Diaz, I.; Belhassen, H.; Hedhili, A.; Torné, P.; Fernández, M.; Olea, N. Assessment of estrogenic and anti-androgenic activities of the mycotoxin zearalenone and its metabolites using in vitro receptor-specific bioassays. Food Chem. Toxicol. 2014, 74, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Matthews, J.; Celius, T.; Halgren, R.; Zacharewski, T. Differential estrogen receptor binding of estrogenic substances: A species comparison. J. Steroid Biochem. Mol. Biol. 2000, 74, 223–234. [Google Scholar] [CrossRef]
- Drzymala, S.S.; Binder, J.; Brodehl, A.; Penkert, M.; Rosowski, M.; Garbe, L.A.; Koch, M. Estrogenicity of novel phase i and phase ii metabolites of zearalenone and cis-zearalenone. Toxicon 2015, 105, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, L.; Wagner, J.; Metzler, M. Estrogenic and clastogenic potential of the mycotoxin alternariol in cultured mammalian cells. Food Chem. Toxicol. 2006, 44, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Stypuła-Trębas, S.; Minta, M.; Radko, L.; Jedziniak, P.; Posyniak, A. Nonsteroidal mycotoxin alternariol is a full androgen agonist in the yeast reporter androgen bioassay. Environ. Toxicol. Pharmacol. 2017, 55, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Fehr, M.; Pahlke, G.; Fritz, J.; Christensen, M.O.; Boege, F.; Altemöller, M.; Podlech, J.; Marko, D. Alternariol acts as a topoisomerase poison, preferentially affecting the iialpha isoform. Mol. Nutr. Food Res. 2009, 53, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Jarolim, K.; Del Favero, G.; Ellmer, D.; Stark, T.D.; Hofmann, T.; Sulyok, M.; Humpf, H.U.; Marko, D. Dual effectiveness of Alternaria but not Fusarium mycotoxins against human topoisomerase ii and bacterial gyrase. Arch. Toxicol. 2017, 91, 2007–2016. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, R.; Tan, Z.; Huang, B.; Zhang, S. Computational polypharmacology: A new paradigm for drug discovery. Expert Opin. Drug Discov. 2017, 12, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Zhou, Q.; Sun, X.; Li, L.; Zhou, B.; Zeng, F.; Zhao, Y.; Shen, W.; Sun, Z. Effect of low-dose zearalenone exposure on reproductive capacity of male mice. Toxicol. Appl. Pharmacol. 2017, 333, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Massart, F.; Saggese, G. Oestrogenic mycotoxin exposures and precocious pubertal development. Int. J. Androl. 2010, 33, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Cozzini, P.; Dellafiora, L. In silico approach to evaluate molecular interaction between mycotoxins and the estrogen receptors ligand binding domain: A case study on zearalenone and its metabolites. Toxicol. Lett. 2012, 214, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.; Pries, V.; Hedberg, C.; Waldmann, H. Target identification for small bioactive molecules: Finding the needle in the haystack. Angew. Chem. Int. Ed. Engl. 2013, 52, 2744–2792. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Kwon, H.J. Target deconvolution of bioactive small molecules: The heart of chemical biology and drug discovery. Arch. Pharm. Res. 2015, 38, 1627–1641. [Google Scholar] [CrossRef] [PubMed]
- Delavan, B.; Roberts, R.; Huang, R.; Bao, W.; Tong, W.; Liu, Z. Computational drug repositioning for rare diseases in the era of precision medicine. Drug Discov. Today 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Y.; Li, S.; Wang, Y.; Peng, J.; Luo, C.; Luo, X.; Zheng, M.; Chen, K.; Jiang, H.; et al. In Silico target fishing: Addressing a “big data” problem by ligand-based similarity rankings with data fusion. J. Cheminform. 2014, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Schiff, P.L. Ergot and its alkaloids. Am. J. Pharm. Educ. 2006, 70, 98. [Google Scholar] [CrossRef] [PubMed]
- Krska, R.; Crews, C. Significance, chemistry and determination of ergot alkaloids: A review. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2008, 25, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Mulac, D.; Humpf, H.-U. Cytotoxicity and accumulation of ergot alkaloids in human primary cells. Toxicology 2011, 282, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Dänicke, S. Ergot alkaloids in fattening chickens (broilers): Toxic effects and carry over depending on dietary fat proportion and supplementation with non-starch-polysaccharide (NSP) hydrolyzing enzymes. Toxins (Basel) 2017, 9, E118. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Dall’Asta, C.; Cozzini, P. Ergot alkaloids: From witchcraft till in silico analysis. Multi-receptor analysis of ergotamine metabolites. Toxicol. Rep. 2015, 2, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, B.; Spiteller, M. Functionalized ergot-alkaloids as potential dopamine D3 receptor agonists for treatment of schizophrenia. J. Mol. Struct. 2012, 1029, 106–118. [Google Scholar] [CrossRef]
- Ivanova, B.; Spiteller, M. Derivatives of ergot-alkaloids: Molecular structure, physical properties, and structure-activity relationships. J. Mol. Struct. 2012, 1024, 18–31. [Google Scholar] [CrossRef]
- Heinrich-Hirsch, B.; Madle, S.; Oberemm, A.; Gundert-Remy, U. The use of toxicodynamics in risk assessment. Toxicol. Lett. 2001, 120, 131–141. [Google Scholar] [CrossRef]
- Grenier, B.; Applegate, T.J. Modulation of intestinal functions following mycotoxin ingestion: Meta-analysis of published experiments in animals. Toxins (Basel) 2013, 5, 396–430. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Risks for animal health related to the presence of zearalenone and its modified forms in feed. EFSA J. 2017, 15, 4851. [Google Scholar]
- Ingles-Prieto, A.; Ibarra-Molero, B.; Delgado-Delgado, A.; Perez-Jimenez, R.; Fernandez, J.M.; Gaucher, E.A.; Sanchez-Ruiz, J.M.; Gavira, J.A. Conservation of protein structure over four billion years. Structure 2013, 21, 1690–1697. [Google Scholar] [CrossRef] [PubMed]
- Forslund, K.; Pekkari, I.; Sonnhammer, E.L. Domain architecture conservation in orthologs. BMC Bioinform. 2011, 12, 326. [Google Scholar] [CrossRef] [PubMed]
- Björklund, Å.K.; Ekman, D.; Light, S.; Frey-Skött, J.; Elofsson, A. Domain rearrangements in protein evolution. J. Mol. Biol. 2005, 353, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Björklund, Å.K.; Ekman, D.; Elofsson, A. Expansion of protein domain repeats. PLoS Comput. Biol. 2006, 2, 114. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Teichmann, S.A.; Pereira-Leal, J. The relationship between domain duplication and recombination. J. Mol. Biol. 2005, 346, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, J.C.; Srinivasan, S.; Bruno, N.E.; Nowak, J.; Wright, N.J.; Minutolo, F.; Rangarajan, E.S.; Izard, T.; Yao, X.Q.; Grant, B.J.; et al. Systems structural biology analysis of ligand effects on erα predicts cellular response to environmental estrogens and anti-hormone therapies. Cell Chem. Biol. 2017, 24, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.S. Molecular modeling: Molecular mechanics. In An Introduction to Computational Biochemistry; John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Manuel, D.G.; Abdulaziz, K.E.; Perez, R.; Beach, S.; Bennett, C. Personalized risk communication for personalized risk assessment: Real world assessment of knowledge and motivation for six mortality risk measures from an online life expectancy calculator. Inform. Health Soc. Care 2017, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dornbos, P.; Crawford, R.B.; Kaminski, N.E.; Hession, S.L.; LaPres, J.J. The influence of human interindividual variability on the low-dose region of dose-response curve induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in primary B cells. Toxicol. Sci. 2016, 153, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.T.; Spector, T.D. A twin approach to unraveling epigenetics. Trends Genet. 2011, 27, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Ankley, G.T.; Bennett, R.S.; Erickson, R.J.; Hoff, D.J.; Hornung, M.W.; Johnson, R.D.; Mount, D.R.; Nichols, J.W.; Russom, C.L.; Schmieder, P.K.; et al. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 2010, 29, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Oldenkamp, R.; Huijbregts, M.A.J.; Ragas, A.M.J. Uncertainty and variability in human exposure limits—A chemical-specific approach for ciprofloxacin and methotrexate. Crit. Rev. Toxicol. 2016, 46, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Alluri, P.G.; Speers, C.; Chinnaiyan, A.M. Estrogen receptor mutations and their role in breast cancer progression. Breast Cancer Res. 2014, 16, 494. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.R.; Wu, Y.M.; Vats, P.; Su, F.Y.; Lonigro, R.J.; Cao, X.H.; Kalyana-Sundaram, S.; Wang, R.; Ning, Y.; Hodges, L.; et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013, 45, 1446–1451. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, J.C.; Tard, A.; Volatier, J.L.; Verger, P. Estimated dietary exposure to principal food mycotoxins from the first french total diet study. Food Addit. Contam. 2005, 22, 652–672. [Google Scholar] [CrossRef] [PubMed]
- European Commission. Commission regulation (ec) no 1881/2006 of 19 december 2006 setting levels for certain contaminants in food stuffs. Off. J. Eur. Union 2006, L364, 5–24. [Google Scholar]
- Pazaiti, A.; Kontos, M.; Fentiman, I. Zen and the art of breast health maintenance. Int. J. Clin. Pract. 2012, 66, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Feyfant, E.; Sali, A.; Fiser, A. Modeling mutations in protein structures. Protein Sci. 2007, 16, 2030–2041. [Google Scholar] [CrossRef] [PubMed]
- Nagasundaram, N.; Zhu, H.L.; Liu, J.M.; Karthick, V.; Doss, C.G.P.; Chakraborty, C.; Chen, L.N. Analysing the effect of mutation on protein function and discovering potential inhibitors of cdk4: Molecular modelling and dynamics studies. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Dellafiora, L.; Galaverna, G.; Reverberi, M.; Dall’Asta, C. Degradation of aflatoxins by means of laccases from trametes versicolor: An in silico insight. Toxins (Basel) 2017, 9, E17. [Google Scholar] [CrossRef] [PubMed]
- Gromadzka, K.; Waskiewicz, A.; Chelkowski, J.; Golinski, P. Zearalenone and its metabolites: Occurrence, detection, toxicity and guidelines. World Mycotoxins J. 2008, 1, 209–220. [Google Scholar] [CrossRef]
- EFSA. Scientific opinion on the risks for public health related to the presence of zearalenone in food. EFSA J. 2011, 9, 2197. [Google Scholar]
- Berthiller, F.; Crews, C.; Dall’Asta, C.; Saeger, S.D.; Haesaert, G.; Karlovsky, P.; Oswald, I.P.; Seefelder, W.; Speijers, G.; Stroka, J. Masked mycotoxins: A review. Mol. Nutr. Food Res. 2013, 57, 165–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shier, W.T.; Shier, A.C.; Xie, W.; Mirocha, C.J. Structure-activity relationships for human estrogenic activity in zearalenone mycotoxins. Toxicon 2001, 39, 1435–1438. [Google Scholar] [CrossRef]
- Delfosse, V.; Grimaldi, M.; Cavaillès, V.; Balaguer, P.; Bourguet, W. Structural and functional profiling of environmental ligands for estrogen receptors. Environ. Health Perspect. 2014, 122, 1306–1313. [Google Scholar] [PubMed]
- Rychlik, M.; Humpf, H.U.; Marko, D.; Dänicke, S.; Mally, A.; Berthiller, F.; Klaffke, H.; Lorenz, N. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res. 2014, 30, 197–205. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Modified mycotoxins in food and feed. EFSA J. 2014, 12, 3916. [Google Scholar]
- Kostelanska, M.; Hajslova, J.; Zachariasova, M.; Malachova, A.; Kalachova, K.; Poustka, J.; Fiala, J.; Scott, P.M.; Berthiller, F.; Krska, R. Occurrence of deoxynivalenol and its major conjugate, deoxynivalenol-3-glucoside, in beer and some brewing intermediates. J. Agric. Food Chem. 2009, 57, 3187–3194. [Google Scholar] [CrossRef] [PubMed]
- Nathanail, A.V.; Syvahuoko, J.; Malachová, A.; Jestoi, M.; Varga, E.; Michlmayr, H.; Adam, G.; Sieviläinen, E.; Berthiller, F.; Peltonen, K. Simultaneous determination of major type a and b trichothecenes, zearalenone and certain modified metabolites in finnish cereal grains with a novel liquid chromatography-tandem mass spectrometric method. Anal. Bioanal. Chem. 2015, 407, 4745–4755. [Google Scholar] [CrossRef] [PubMed]
- Poppenberger, B.; Berthiller, F.; Bachmann, H.; Lucyshyn, D.; Peterbauer, C.; Mitterbauer, R.; Schuhmacher, R.; Krska, R.; Glössl, J.; Adam, G. Heterologous expression of arabidopsis udp-glucosyltransferases in Saccharomyces cerevisiae for production of zearalenone-4-O-glucoside. Appl. Environ. Microbiol. 2006, 72, 4404–4410. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Ruotolo, R.; Perotti, A.; Cirlini, M.; Galaverna, G.; Cozzini, P.; Buschini, A.; Dall’Asta, C. Molecular insights on xenoestrogenic potential of zearalenone-14-glucoside through a mixed in vitro/in silico approach. Food Chem. Toxicol. 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Spyrakis, F.; Cozzini, P. How computational methods try to disclose the estrogen receptor secrecy--modeling the flexibility. Curr. Med. Chem. 2009, 16, 2987–3027. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Galaverna, G.; Righi, F.; Cozzini, P.; Dall’Asta, C. Assessing the hydrolytic fate of the masked mycotoxin zearalenone-14-glucoside—A warning light for the need to look at the “maskedome”. Food Chem. Toxicol. 2016, 99, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Frizzell, C.; Uhlig, S.; Miles, C.O.; Verhaegen, S.; Elliott, C.T.; Eriksen, G.S.; Sørlie, M.; Ropstad, E.; Connolly, L. Biotransformation of zearalenone and zearalenols to their major glucuronide metabolites reduces estrogenic activity. Toxicol. In Vitro 2015, 29, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Fraeyman, S.; Croubels, S.; Devreese, M.; Antonissen, G. Emerging fusarium and alternaria mycotoxins: Occurrence, toxicity and toxicokinetics. Toxins 2017, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Scientific opinion on the risks for animal and public health related to the presence of alternaria toxins in food and feed. EFSA J. 2011, 9, 2407. [Google Scholar]
- Solhaug, A.; Eriksen, G.S.; Holme, J.A. Mechanisms of action and toxicity of the mycotoxin alternariol: A review. Basic Clin. Pharmacol. Toxicol. 2016, 119, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.T.; Qian, Y.Z.; Zhang, P.; Dong, W.H.; Qi, Y.M.; Guo, H.T. Etiological role of alternaria alternata in human esophageal cancer. Chin. Med. J. (Engl.) 1992, 105, 394–400. [Google Scholar] [PubMed]
- Dellafiora, L.; Dall’Asta, C.; Cruciani, G.; Galaverna, G.; Cozzini, P. Molecular modelling approach to evaluate poisoning of topoisomerase i by alternariol derivatives. Food Chem. 2015, 189, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Drugging topoisomerases: Lessons and challenges. ACS Chem. Biol. 2013, 8, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Staker, B.L.; Hjerrild, K.; Feese, M.D.; Behnke, C.A.; Burgin, A.B.J.; Stewart, L. The mechanism of topoisomerase i poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 2002, 99, 15387–15392. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.T.; Qian, Y.Z.; Zhang, P.; Dong, Z.M.; Shi, Z.Y.; Zhen, Y.Z.; Miao, J.; Xu, Y.M. Relationships between alternaria alternata and oesophageal cancer. IARC Sci. Publ. 1991, 105, 258–262. [Google Scholar]
- Chen, T.; Sun, Y.; Ji, P.; Kopetz, S.; Zhang, W. Topoisomerase iiα in chromosome instability and personalized cancer therapy. Oncogene 2015, 34, 4019–4031. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Ashour, M.E.; Atteya, R.; El-Khamisy, S.F. Topoisomerase-mediated chromosomal break repair: An emerging player in many games. Nat. Rev. Cancer 2015, 15, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Tukey, R.H.; Strassburg, C.P. Human udp-glucuronosyltransferases: Metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 581–616. [Google Scholar] [CrossRef] [PubMed]
- Malir, F.; Ostry, V.; Pfohl-Leszkowicz, A.; Malir, J.; Toman, J. Ochratoxin A: 50 years of research. Toxins 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Pfohl-Leszkowicz, A.; Manderville, R.A. Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol. Nutr. Food Res. 2007, 51, 61–99. [Google Scholar] [CrossRef] [PubMed]
- National Toxicology Program. Toxicology and carcinogenesis studies of Ochratoxin A (CAS No. 303-47-9) in f344/n rats (gavage studies). Natl. Toxicol. Program Tech. Rep. Ser. 1989, 358, 1–142. [Google Scholar]
- Zhu, L.Y.; Zhang, B.Y.; Dai, Y.Q.; Li, H.Y.; Xu, W.T. A review: Epigenetic mechanism in ochratoxin A toxicity studies. Toxins 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- IARC. Iarc Monographs; IARC: Lyon, France, 1993; Volume 56. [Google Scholar]
- Limbeck, E.; Vanselow, J.T.; Hofmann, J.; Schlosser, A.; Mally, A. Linking site-specific loss of histone acetylation to repression of gene expression by the mycotoxin ochratoxin A. Arch. Toxicol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Pfohl-Leszkowicz, A.; Manderville, R.A. An update on direct genotoxicity as a molecular mechanism of ochratoxin A carcinogenicity. Chem. Res. Toxicol. 2012, 25, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Manderville, R.A.; Pfohl-Leszkowicz, A. Bioactivation and DNA adduction as a rationale for ochratoxin a carcinogenesis. World Mycotoxin J. 2008, 1, 357–367. [Google Scholar] [CrossRef]
- Tozlovanu, M.; Faucet-Marquis, V.; Pfohl-Leszkowicz, A.; Manderville, R.A. Genotoxicity of the hydroquinone metabolite of ochratoxin A: Structure-activity relationships for covalent DNA adduction. Chem. Res. Toxicol. 2006, 19, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Hadjeba-Medjdoub, K.; Tozlovanu, M.; Pfohl-Leszkowicz, A.; Frenette, C.; Paugh, R.J.; Manderville, R.A. Structure-activity relationships imply different mechanisms of action for ochratoxin A-mediated cytotoxicity and genotoxicity. Chem. Res. Toxicol. 2012, 25, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Mally, A. Ochratoxin A and mitotic disruption: Mode of action analysis of renal tumor formation by ochratoxin A. Toxicol. Sci. 2012, 127, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Sekine, T.; Takeda, M.; Cha, S.H.; Kanai, Y.; Kimura, M.; Endou, H. Transport of ochratoxin A by renal multispecific organic anion transporter 1. J. Pharmacol. Exp. Ther. 1999, 289, 1301–1305. [Google Scholar] [PubMed]
- Ljubojevic, M.; Herak-Kramberger, C.M.; Hagos, Y.; Bahn, A.; Endou, H.; Burckhardt, G.; Sabolic, I. Rat renal cortical oat1 and oat3 exhibit gender differences determined by both androgen stimulation and estrogen inhibition. Am. J. Physiol. Ren. Physiol. 2004, 287, F124–F138. [Google Scholar] [CrossRef] [PubMed]
- Koszegi, T.; Poor, M. Ochratoxin A: Molecular interactions, mechanisms of toxicity and prevention at the molecular level. Toxins 2016, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- McMasters, D.R.; Vedani, A. Ochratoxin binding to phenylalanyl-trna synthetase: Computational approach to the mechanism of ochratoxicosis and its antagonism. J. Med. Chem. 1999, 42, 3075–3086. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikova, L.; Moor, N.; Lavrik, O.; Vassylyev, D.G. Crystal structures of phenylalanyl-trna synthetase complexed with phenylalanine and a phenylalanyl-adenylate analogue. J. Mol. Biol. 1999, 287, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Vedani, A.; Zbinden, P. Target proteins and mechanisms of ochratoxin toxicity. A contribution to the identification of potential ochratoxin antagonists. ALTEX 1997, 14, 155–164. [Google Scholar] [PubMed]
- Creppy, E.E.; Kern, D.; Steyn, P.S.; Vleggaar, R.; Röschenthaler, R.; Dirheimer, G. Comparative study of the effect of ochratoxin A analogues on yeast aminoacyl-trna synthetases and on the growth and protein synthesis of hepatoma cells. Toxicol. Lett. 1983, 19, 217–224. [Google Scholar] [CrossRef]
- Rottkord, U.; Röhl, C.; Ferse, I.; Schulz, M.C.; Rückschloss, U.; Gekle, M.; Schwerdt, G.; Humpf, H.U. Structure-activity relationship of ochratoxin A and synthesized derivatives: Importance of amino acid and halogen moiety for cytotoxicity. Arch. Toxicol. 2017, 91, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Park, G.; Perry, J.L.; Il’ichev, Y.V.; Bow, D.A.; Pritchard, J.B.; Faucet, V.; Pfohl-Leszkowicz, A.; Manderville, R.A.; Simon, J.D. Molecular aspects of the transport and toxicity of ochratoxin A. Acc. Chem. Res. 2004, 37, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Il’ichev, Y.V.; Perry, J.L.; Simon, J.D. Interaction of ochratoxin A with human serum albumin. Preferential binding of the dianion and ph effects. J. Phys. Chem. B 2002, 106, 452–459. [Google Scholar] [CrossRef]
- Simon, J.D.; Perry, J.L.; Il’ichev, Y.V.; Pritchard, J.B.; Bow, D.A.J. Binding of ochratoxin A to human plasma proteins: Implications in toxicity mechanisms. Biophys. J. 2003, 85, 332A. [Google Scholar]
- Wu, Q.H.; Dohnal, V.; Kuca, K.; Yuan, Z.H. Trichothecenes: Structure-toxic activity relationships. Curr. Drug Metab. 2013, 14, 641–660. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.F. The trichothecenes and their biosynthesis. In Progress in the Chemistry of Organic Natural Products; Herz, W., Falk, H., Kirby, G.W., Eds.; Springer: Vienna, Austria, 2007; pp. 63–130. [Google Scholar]
- Pestka, J.J. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 2010, 84, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Payros, D.; Alassane-Kpembi, I.; Pierron, A.; Loiseau, N.; Pinton, P.; Oswald, I.P. Toxicology of deoxynivalenol and its acetylated and modified forms. Arch. Toxicol. 2016, 90, 2931–2957. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Y.I.; Watts, C.; Li, X.Z.; Zhou, T. A novel peptide-binding motifs inference approach to understand deoxynivalenol molecular toxicity. Toxins (Basel) 2015, 7, 1989–2005. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J. Mechanisms of deoxynivalenol-induced gene expression and apoptosis. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2008, 25, 1128–1140. [Google Scholar] [CrossRef] [PubMed]
- De Loubresse, N.G.; Prokhorova, I.; Holtkamp, W.; Rodnina, M.V.; Yusupova, G.; Yusupov, M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014, 513, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Weixlbaumer, A.; Loakes, D.; Kelley, A.C.; Ramakrishnan, V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70s ribosome. Nat. Struct. Mol. Biol. 2009, 16, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Pierron, A.; Mimoun, S.; Murate, L.S.; Loiseau, N.; Lippi, Y.; Bracarense, A.F.; Schatzmayr, G.; He, J.W.; Zhou, T.; Moll, W.D.; et al. Microbial biotransformation of don: Molecular basis for reduced toxicity. Sci. Rep. 2016, 6, 29105. [Google Scholar] [CrossRef] [PubMed]
- Warth, B.; Del Favero, G.; Wiesenberger, G.; Puntscher, H.; Woelflingseder, L.; Fruhmann, P.; Sarkanj, B.; Krska, R.; Schuhmacher, R.; Adam, G.; et al. Identification of a novel human deoxynivalenol metabolite enhancing proliferation of intestinal and urinary bladder cells. Sci. Rep. 2016, 6, 33854. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, W.E.; Rodarte, C.B.; Lin, A. 3D QSAR study of the toxicity of trichothecene mycotoxins. Eur. J. Med. Chem. 2009, 44, 4485–4489. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Galaverna, G.; Dall’Asta, C. In silico analysis sheds light on the structural basis underlying the ribotoxicity of trichothecenes—A tool for supporting the hazard identification process. Toxicol. Lett. 2017, 270, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the fusarium mycotoxin deoxynivalenol by a udp-glucosyltransferase from arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dellafiora, L.; Dall’Asta, C.; Galaverna, G. Toxicodynamics of Mycotoxins in the Framework of Food Risk Assessment—An In Silico Perspective. Toxins 2018, 10, 52. https://doi.org/10.3390/toxins10020052
Dellafiora L, Dall’Asta C, Galaverna G. Toxicodynamics of Mycotoxins in the Framework of Food Risk Assessment—An In Silico Perspective. Toxins. 2018; 10(2):52. https://doi.org/10.3390/toxins10020052
Chicago/Turabian StyleDellafiora, Luca, Chiara Dall’Asta, and Gianni Galaverna. 2018. "Toxicodynamics of Mycotoxins in the Framework of Food Risk Assessment—An In Silico Perspective" Toxins 10, no. 2: 52. https://doi.org/10.3390/toxins10020052




