Effect of Green and Brown Propolis Extracts on the Expression Levels of microRNAs, mRNAs and Proteins, Related to Oxidative Stress and Inflammation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
- -
- Aqueous extraction, to remove waxes and impurities from raw materials, using a 1:1 solvent/propolis ratio, at 80 °C for 10 h and with 100 Watt ultrasounds. After cooling at 8 °C, the solution was filtered with a 30 μm filter.
- -
- Three hydro-alcoholic extractions, one for each insoluble residue of the preceding extraction step, carried out using different alcoholic degrees and temperatures, from 4 to 36 h, with a fixed 1:1 solvent/propolis residue ratio. Each extraction step was followed by a sample cooling step at circa 15 °C, a filtration step with a 30 to 50 μm filter and a concentration step using a rotating evaporator, to obtain a soft mixture.
- (1)
- The first extraction step used a water/ethanol mixture with an alcoholic concentration ranging from 35 to 40 alcoholic degrees, at 50 °C;
- (2)
- The second extraction step used a mixture with an alcoholic concentration ranging from 55 to 60 alcoholic degrees, at 70 °C;
- (3)
- The third extraction step used a mixture with an alcoholic concentration ranging from 70 to 80 alcoholic degrees, at 80 °C.
- -
- Concentration: the combined extracts were mixed and concentrated to a residual humidity value ranging from 15 to 20% by weight.
- -
- Dissolution in glycerin: the concentrated extracts were evaporated to remove ethanol. The dough was mixed with hot glycerin and water for 2 h within a mixer, and then cooled at 10 °C to give a non-alcoholic liquid. After precipitation the solution was filtered twice, using a 30 μm and a 10 μm filter.
2.2. Analyses of Propolis Extracts (RP-HPLC-PDA-ESI-MSn)
2.3. Cell Culture
2.4. Cell Viability Test
2.5. Cell Treatment with Green and Brown Propolis
2.6. RNA Extraction and miRNA Real-Time PCR
2.7. Protein Analyses
2.8. Statistical Analyses
3. Results
3.1. Propolis Extracts RP-HPLC-PDA-ESI-MSn Analyses
3.2. Cell Viability Test
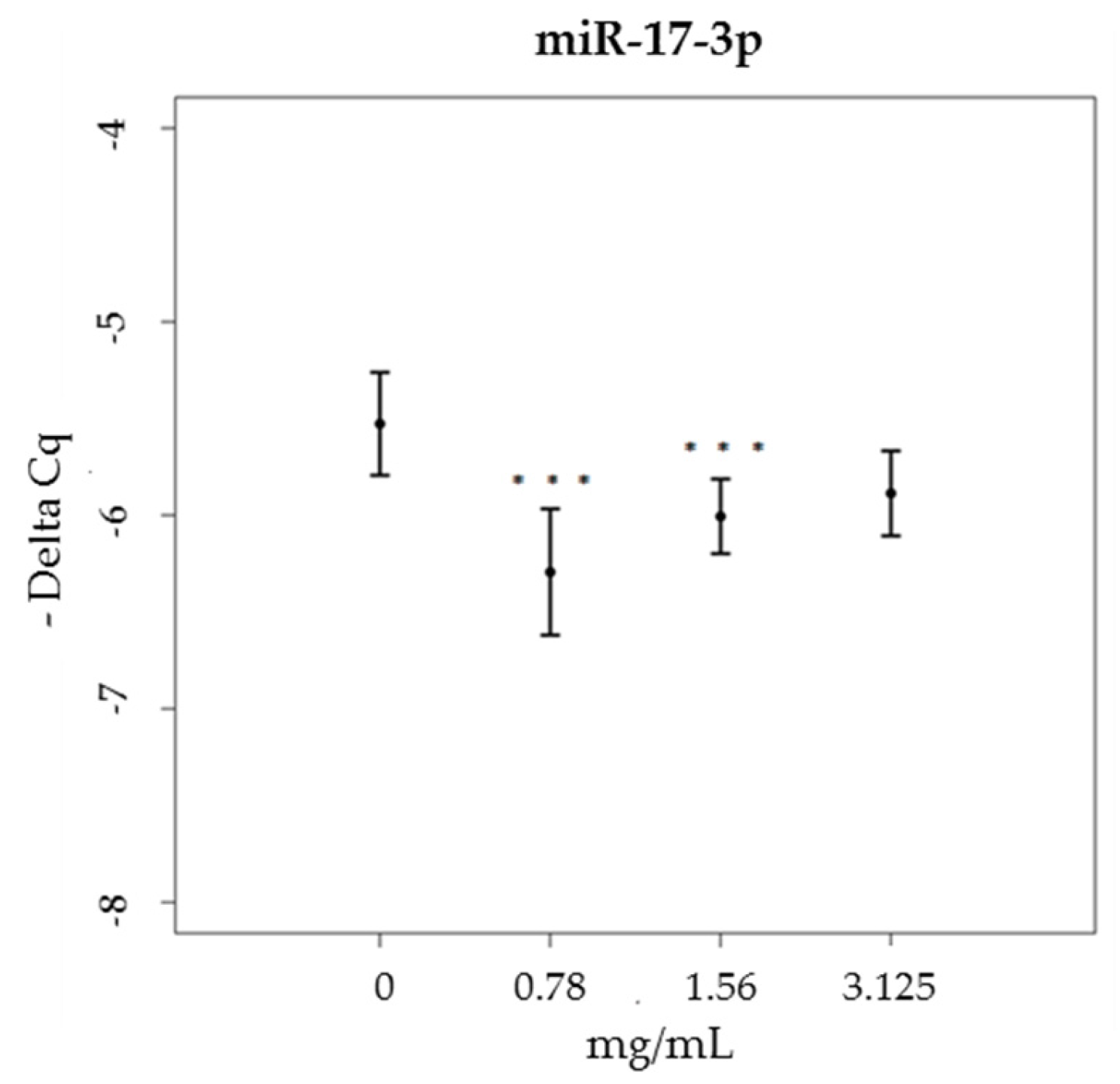
3.3. miRNA
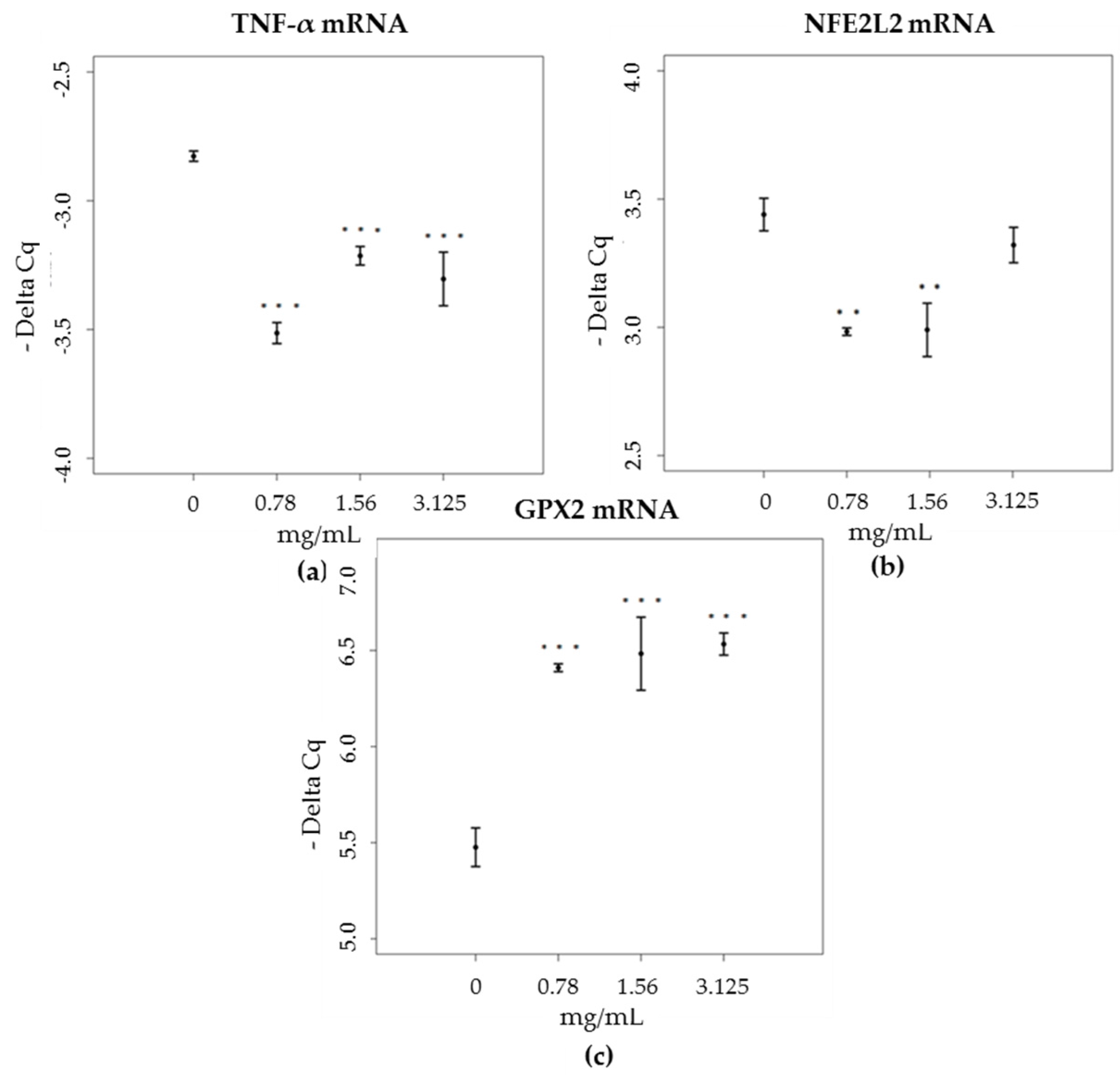
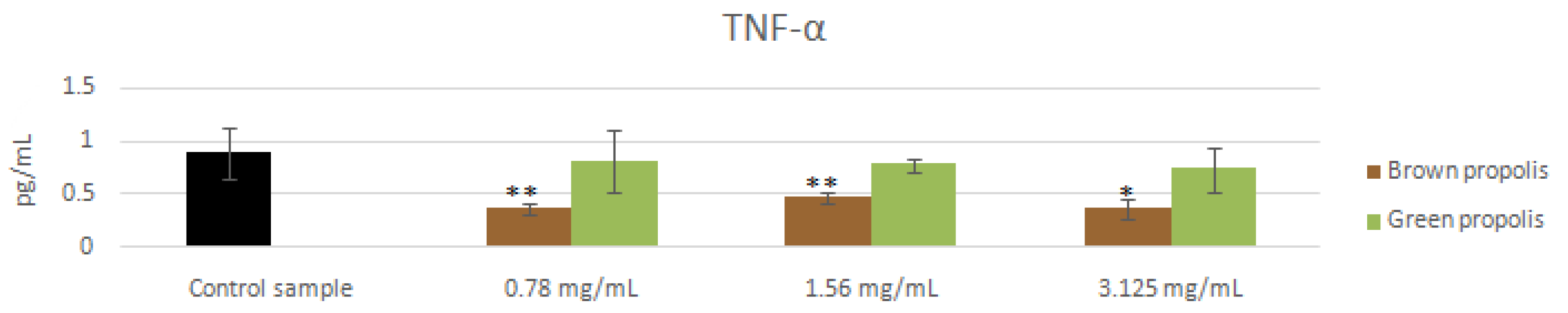
3.4. mRNA and Proteins
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Marcucci, M.C. Propolis: Chemical composition, biological properties and therapeutic activity. Apidologie 1995, 26, 83–99. [Google Scholar] [CrossRef]
- Attia, Y.A.; Abd Al-Hamid, A.E.; Ibrahim, M.S.; Al-Harthi, M.A.; Bovera, F.; Elnaggar, A.S. Productive performance, biochemical and hematological traits of broiler chickens supplemented with propolis, bee pollen, and mannan oligosaccharides continuously or intermittently. Livest. Sci. 2014, 164, 87–95. [Google Scholar] [CrossRef]
- Lotfy, M. Biological activity of bee propolis in health and disease. Asian Pac. J. Cancer Prev. 2006, 7, 22–31. [Google Scholar]
- Cvek, J.; Medid-Saric, M.; Vitali, D.; Vedrina-Dragojevik, I.; Smit, Z.; Tomic, S. The content of essential and toxic elements in Croatian propolis samples and their tinctures. J. Apicult. Res. 2008, 47, 35–45. [Google Scholar] [CrossRef]
- Kuropatnicki, A.K.; Szliszka, E.; Krol, W. Historical aspects of propolis research in modern times. Evid. Based Complement. Altern. Med. 2013, 96, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.F.; Lin, C.C.; Lin, M.H.; Lin, Y.S.; Lin, S.C. Cytoprotection by propolis ethanol extract of acute absolute ethanol-induced gastric mucosal lesions. Am. J. Chin. Med. 2002, 30, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.P.; Lemos, M.; Maistro, E.L.; Leite, M.F.; Sousa, J.P.; Bastos, J.K.; Andrade, S.F. Evaluation of antiulcer activity of the main phenolic acids found in Brazilian green propolis. J. Ethnopharmacol. 2008, 120, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Casaroto, A.R.; Lara, V.S. Phytomedicines for Candida-associated denture stomatitis. Fitoterapia 2010, 81, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Marinotti, S.; Ranzato, E. Propolis: A new frontier for wound healing? Burns Trauma 2015, 2015, 3–9. [Google Scholar]
- Kurek-Gorecka, A.; Rzepecka-Stojko, A.; Gorecki, M.; Stpiko, J.; Sosada, M.; Swierczek-Zieba, G. Structure and antioxidant activity of polyphenols derived from propolis. Molecules 2013, 19, 78–101. [Google Scholar] [CrossRef] [PubMed]
- Abo-Salem, O.M.; El-Edel, R.H.; Harisa, G.E.I.; Halawany, N.E.; Ghonaim, M.M. Experimental diabetic nephropathy can be prevented by propolis: Effect on metabolic disturbance and renal oxidative parameters. Pakistan J. Pharm. Sci. 2009, 22, 205–210. [Google Scholar]
- Hu, F.; Zhu, W.; Chen, M.; Shou, Q.; Li, Y. Biological activities of Chinese propolis and Brazilian propolis on streptozotocin-induced type 1 diabetes mellitus in rat. Evid. Based Complement. Altern. Med. 2011, 2011, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, L.; Wu, W.; Long, Y.; Wang, R. Potential cytoprotection: Antioxidant defence by caffeic acid phenethyl ester against free radical-induced damage of lipids, DNA, and proteins. Can. J. Physiol. Pharmacol. 2008, 86, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Chee, H.Y. In vitro evaluation of the antifungal activity of propolis extract on Cryptococcus neoformans and Candida albicans. Microbiology 2002, 30, 93–95. [Google Scholar]
- Banskota, A.H.; Tezuka, Y.; Adnyana, I.K.; Midorikawa, K.; Matsushige, K.; Message, D.; Huertas, A.A.; Kadota, S. Cytotoxic, hepatoprotective and free radical scavenging effects of propolis from Brazil, Peru, the Netherlands and China. J. Ethnopharmacol. 2000, 72, 239–246. [Google Scholar] [CrossRef]
- Lahouel, M.; Boulkour, S.; Segueni, N.; Fillastre, J.P. The flavonoids effect against vinblastine, cyclophosphamide and paracetamol toxicity by inhibition of lipid-peroxidation and increasing liver glutathione concentration. Pathol. Biol. 2004, 52, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Singh, S.; Burke, T.R.; Grunberger, D.; Aggarwal, B.B. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kappa B. Proc. Natl. Acad. Sci. USA 1996, 93, 9090–9095. [Google Scholar] [CrossRef] [PubMed]
- Rajoo, M.; Parolia, A.; Pau, A.; Amalraj, F.D. The role of propolis in inflammation and orofacial pain: A review. Ann. Res. Rev. Biol. 2004, 4, 651–664. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kumazaki, M. Propolis cinnamic acid derivatives induce apoptosis through both extrinsic and intrinsic apoptosis signaling pathways and modulate of miRNA expression. Phytomedicine 2014, 21, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, A.; Saavedra, N.; Cavalcante, M.F.; Salazar, L.A.; Abdalla, D.S. Identification of microRNAs involved in the modulation of pro-angiogenic factors in atherosclerosis by a polyphenol-rich extract from propolis. Arch. Biochem. Biophys. 2014, 557, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, A.; Saavedra, N.; Rudnicki, M.; Abdalla, D.S.P.; Salazar, L.A. ERK1/2 and HIF1α are involved in antiangiogenic effect of polyphenols-enriched fraction from Chilean propolis. Evid. Based Complement. Altern. Med. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Alencar, S.M.; Aguiar, C.L. Botanical origin and chemical composition of Brazilian propolis. J. Agric. Food Chem. 2002, 50, 2502–2506. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, C.P.; Wang, K.; Li, G.Q.; Hu, F.L. Recent advances in the chemical composition of propolis. Molecules 2014, 19, 19610–19632. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N.; Fachini, A. Procedimento Per L’ottenimento di Estratti Integrali di Propoli Ricchi in Polifenoli e Dotati di Attività Antibatterica e Sua Applicazione Nella Prevenzione e Trattamento di Processi Infettivi di Origine Batterica. Ufficio Italiano Brevetti e Marchi No. 0001425516, 02/02/ 2017.
- Zhang, C.; Huang, S.; Wei, W.; Ping, S.; Shen, X.; Li, Y.; Hu, F. Development of high-performance liquid chromatographic for quality and authenticity control of Chinese propolis. J. Food Sci. 2014, 79, C1315–C1322. [Google Scholar]
- Curti, V.; Di Lorenzo, A.; Rossi, D.; Martino, E.; Capelli, E.; Collina, S.; Daglia, M. Enantioselective modulatory effects of naringenin enantiomers on the expression levels of miR-17-3p involved in endogenous antioxidant defenses. Nutrients 2017, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Curti, V.; Capelli, E.; Boschi, F.; Nabavi, S.F.; Bongiorno, A.I.; Habtemariam, S.; Nabavi, S.M.; Daglia, M. Modulation of human miR-17-3p expression by methyl 3-O-methyl gallate as explanation of its in vivo protective activities. Mol. Nutr. Food Res. 2014, 58, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Machado, B.A.; Silva, R.P.; Barreto Gde, A.; Costa, S.S.; Silva, D.F.; Brandão, H.N.; Rocha, J.L.; Dellagostin, O.A.; Henriques, J.A.; Umsza-Guez, M.A.; et al. Chemical composition and biological activity of extracts obtained by supercritical extraction and ethanolic extraction of brown, green and red propolis derived from different geographic regions in Brazil. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N.; Bergonzini, G. Analysis of flavonoids from propolis by on-line HPLC-electrospray mass spectrometry. J. Pharm. Biomed. Anal. 2006, 42, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Yee, K.L.; Sumbria, R.K. Tumor necrosis factor α inhibition for Alzheimer’s disease. J. Cent. Nerv. Syst. Dis. 2017, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, J.; Ping, S.; Ma, Q.; Chen, X.; Xuan, H.; Shi, J.; Zhang, C.; Hu, F. Anti-inflammatory effects of ethanol extracts of Chinese propolis and buds from poplar (Poplus × canadensis). J. Ethnopharmacol. 2014, 155, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Bufalo, C.M.; Bordon-Graciani, A.P.; Conti, B.J.; Golim, M.A.; Sforcin, J.M. The immunomodulatory effect of propolis on receptors expression, cytokine production and fungicidal activity of human monocytes. J. Pharm. Pharmacol. 2014, 66, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Khayyal, M.T.; El-Hazek, R.M.; El-Ghazaly, M.A. Propolis aqueous extract preserves functional integrity of murine intestinal mucosa after exposure to ionizing radiation. Environ. Toxicol. Pharmacol. 2015, 40, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Ertürküner, S.P.; Saraç, E.Y.; Göçmez, S.S.; Ekmekçi, H.; Öztürk, Z.B.; Seçkin, I.; Sever, Ö.; Keskinbora, K. Anti-inflammatory and ultrastructural effects of Turkish propolis in a rat model of endotoxin-induced uveitis. Folia Histochem. Cytobiol. 2016, 54, 49–57. [Google Scholar]
- Wu, Z.; Zhu, A.; Takayama, F.; Okada, R.; Liu, Y.; Harada, Y.; Wu, S.; Nakanishi, H. Brazilian green propolis suppresses the hypoxia-induced neuroinfammatory responses by inhibiting NF-kB activation in microglia. Oxid. Med. Cell. Longev. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Watanabe, W.; Toyama, S.; Hayashi, Y.; Honda, S.; Sakamoto, S.; Matsuoka, S.; Yoshida, H.; Takeda, S.; Hidaka, M.; et al. Effect of Brazilian propolis on exacerbation of respiratory syncytial virus infection in mice exposed to tetrabromobisphenol A, a brominated flame retardant. Evid. Based Complement. Altern. Med. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Neiva, K.G.; Catalfamo, D.L.; Holliday, S.; Wallet, S.M.; Pileggi, R. Propolis decreases lipopolysaccharide-induced inflammatory mediators in pulp cells and osteoclast. Dent. Traumatol. 2014, 30, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Pu, L.; Wei, J.; Li, J.; Wu, J.; Xin, Z.; Gao, W.; Guo, C. Brazilian green propolis improves antioxidant function in patients with type 2 diabetes mellitus. Int. J. Environ. Res. Public Health 2016, 13. [Google Scholar] [CrossRef] [PubMed]
- Tiveron, A.P.; Rosalen, P.L.; Franchin, M.; Lacerda, R.C.C.; Bueno-Silva, B.; Benso, B.; Denny, C.; Ikegaki, M.; de Alencar, S.M. Chemical characterization and antioxidant, antimicrobial, and anti-inflammatory activities of south Brazilian organic propolis. PLoS ONE 2016. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Lu, Y.; Chen, Y.; Cheng, J. The role of Nrf2 in oxidative stress-induced endothelial injuries. J. Endocrinol. 2015, 225, R83–R99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shen, X.; Wang, K.; Cao, X.; Zhang, C.; Zheng, H.; Hu, F. Antioxidant activities and molecular mechanisms of the ethanol extracts of Baccharis popolis and Eucalyptus propolis in RAW64.7 cells. Pharm. Biol. 2016, 54, 2220–2235. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.F.; Braidy, N.; Habtemariam, S.; Orhan, I.E.; Daglia, M.; Manayi, A.; Gortzi, O.; Nabavi, S.M. Neuroprotective effects of chrysin: From chemistry to medicine. Neurochem. Int. 2015, 90, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Eldutar, E.; Kandemir, F.M.; Kucukler, S.; Caglayan, C. Restorative effects of Chrysin pretreatment on oxidant-antioxidant status, inflammatory cytokine production, and apoptotic and autophagic markers in acute paracetamol-induced hepatotoxicity in rats: An experimental and biochemical study. J. Biochem. Mol. Toxicol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Palacz-Wrobel, M.; Borkowska, P.; Paul-Samojedny, M.; Kowalczyk, M.; Fila-Danilow, A.; Suchanek-Raif, R.; Kowalski, J. Effect of apigenin, kaempferol and resveratrol on the gene expression and protein secretion of tumor necrosis factor alpha (TNF-α) and interleukin-10 (IL-10) in RAW-264.7 macrophages. Biomed. Pharmacother. 2017, 93, 1205–1212. [Google Scholar] [CrossRef] [PubMed]
- Rui, L.; Li, J.-Z.; Song, J.-K.; Sun, J.-L.; Li, Y.-J.; Zhou, S.-B.; Zhang, T.-T.; Du, G.-H. Pinocembrin protects human brain microvascular endothelial cells against fibrillar Amyloid-β1−40 injury by suppressing the MAPK/NF-κB inflammatory pathways. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
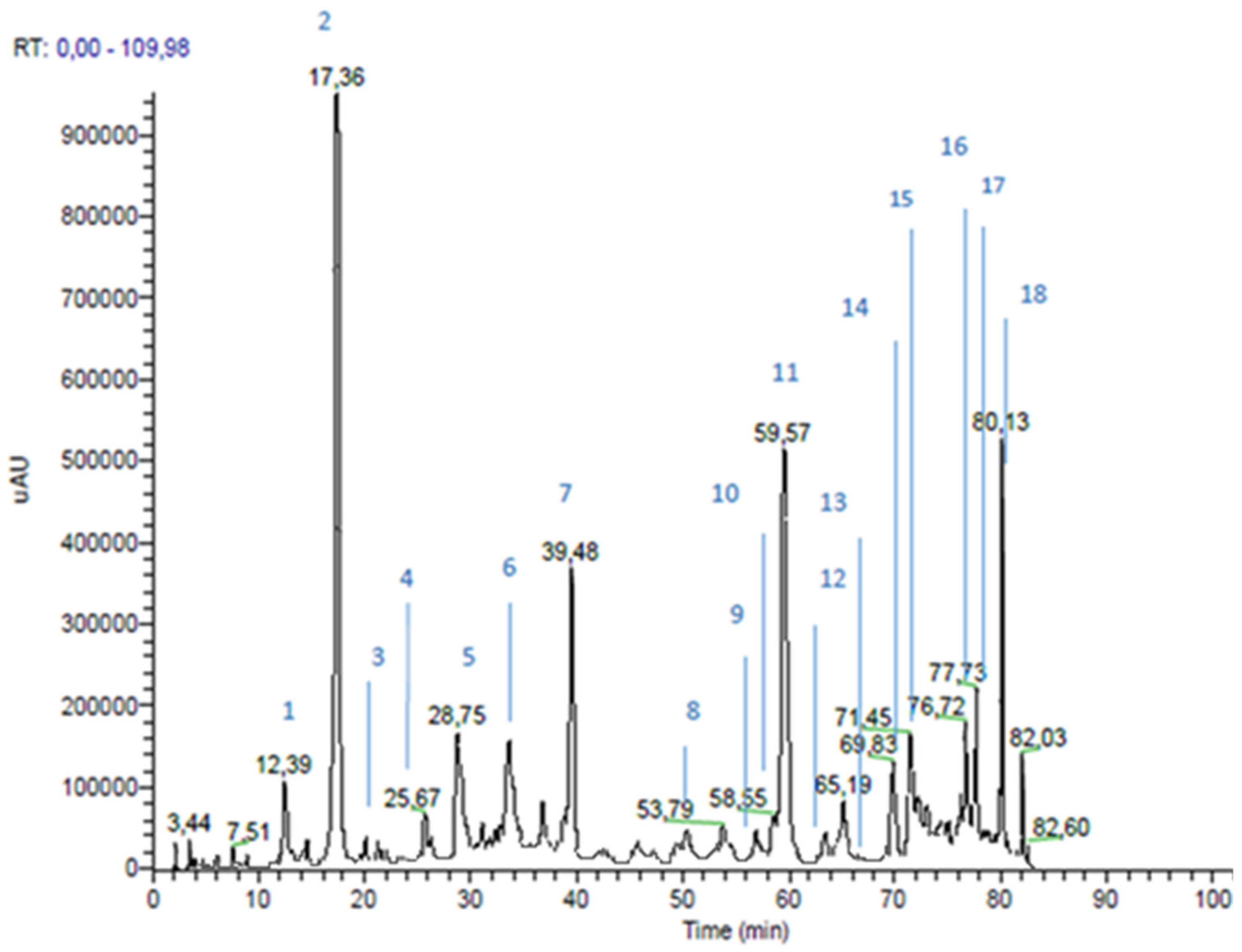
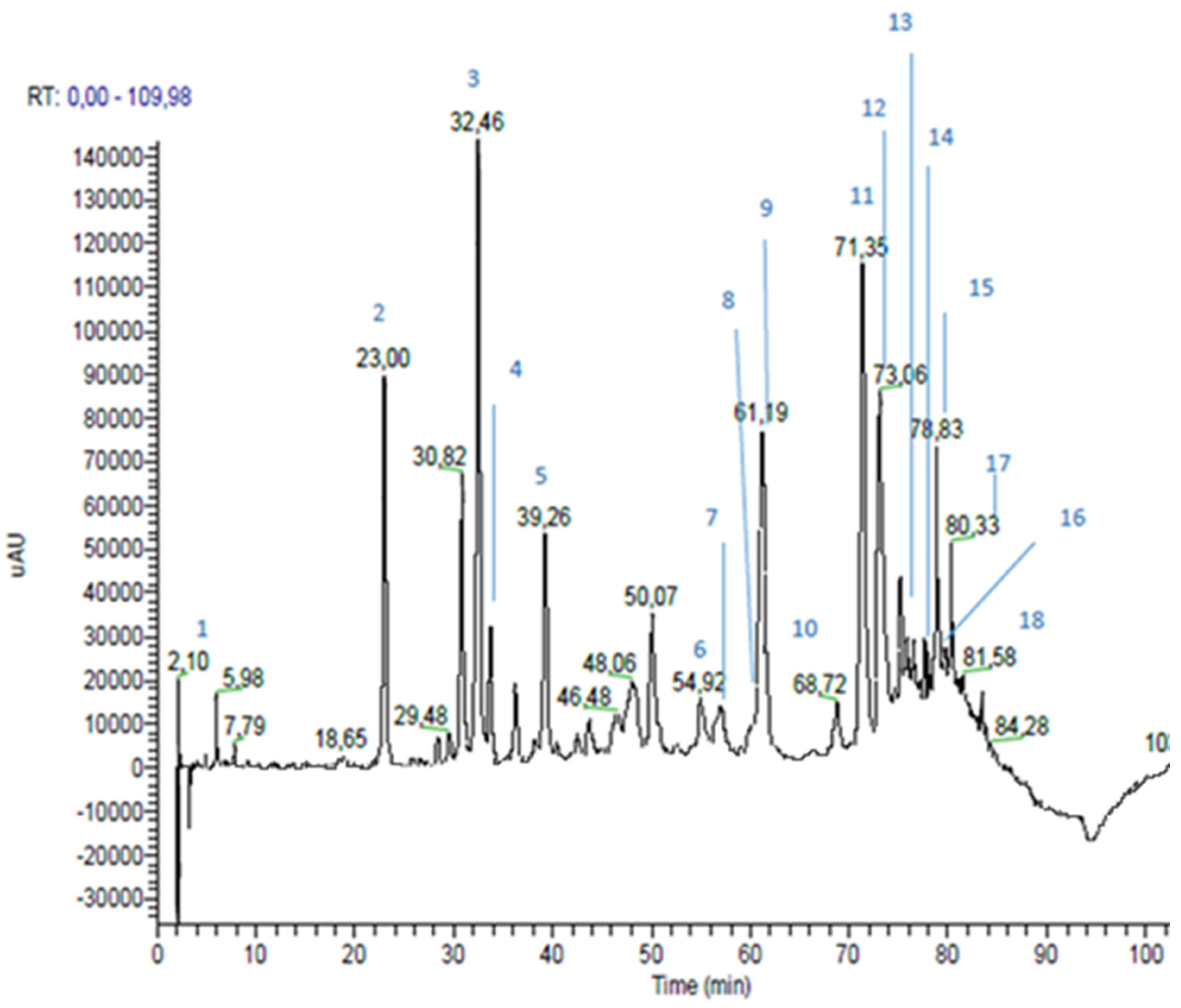
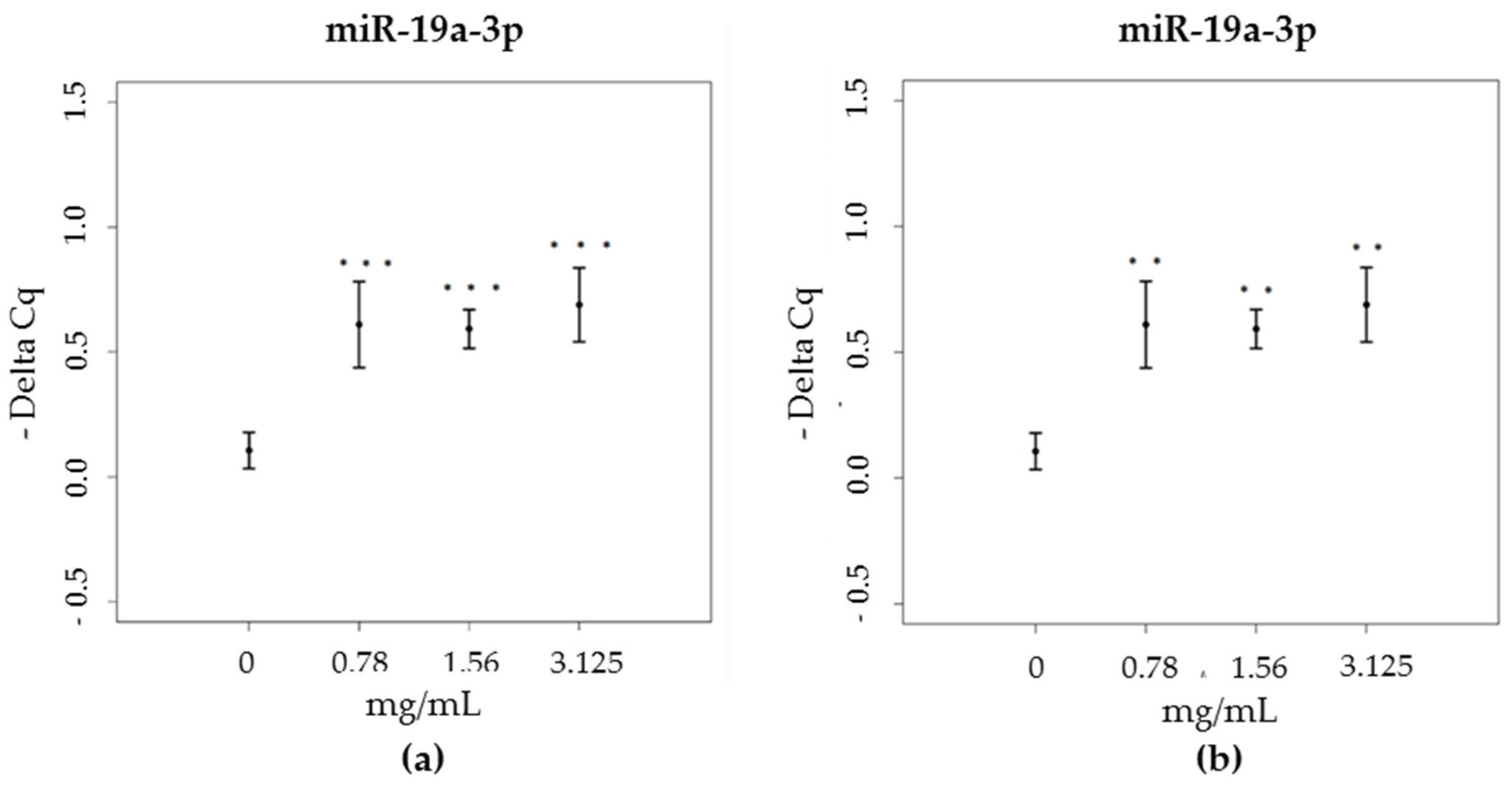
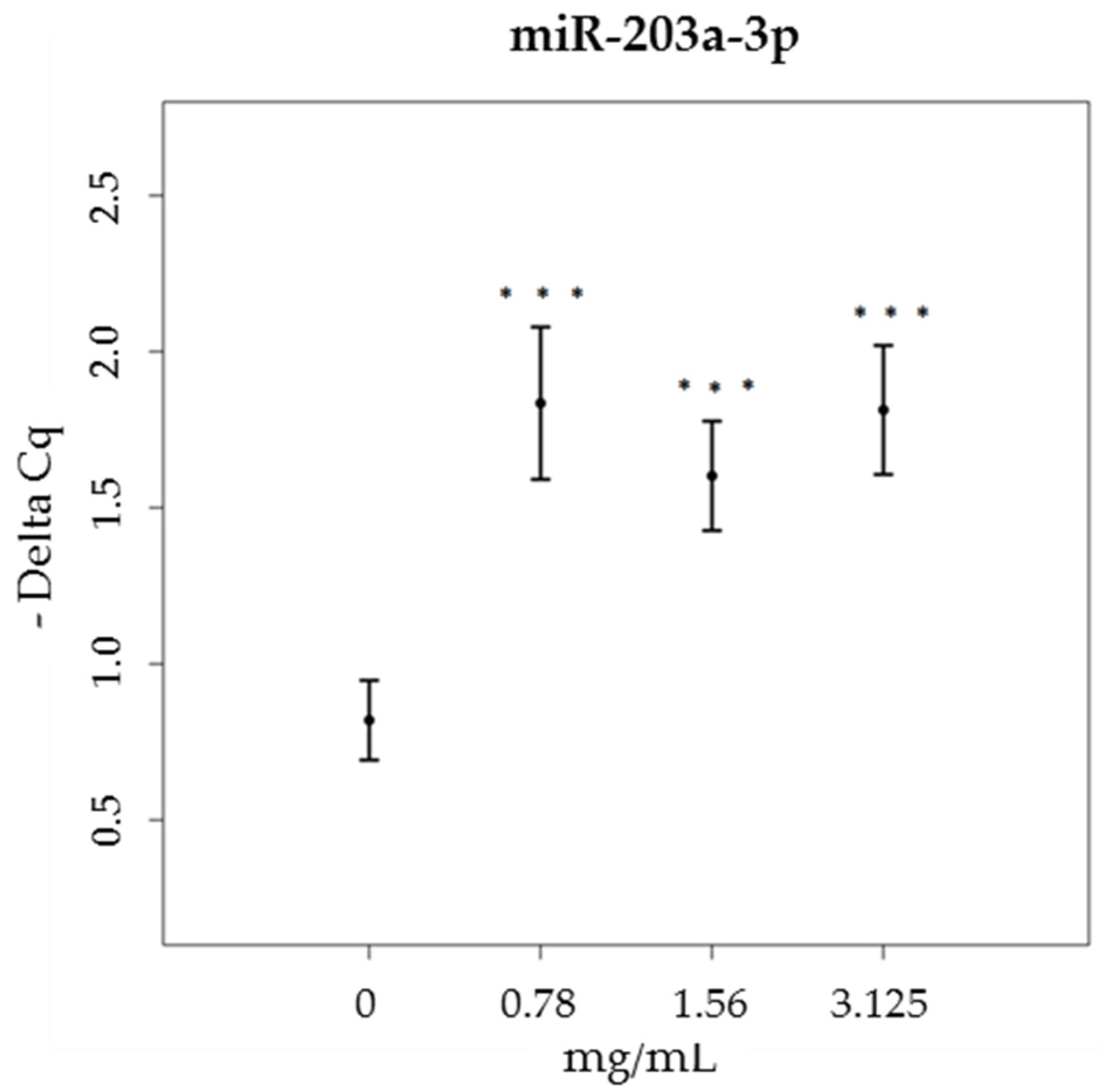


| Time (min) | % Eluent A | % Eluent B |
|---|---|---|
| 0 | 85 | 15 |
| 30 | 60 | 40 |
| 65 | 45 | 55 |
| 70 | 38 | 62 |
| 85 | 0 | 100 |
| 90 | 0 | 100 |
| 100 | 85 | 15 |
| 110 | 85 | 15 |
| Peak n | RT * (min) | m/z | MS2 m/z | Proposed Structure | Area % | |
|---|---|---|---|---|---|---|
| 1 | 12.39 | 179 | 135 | caffeic acid | 2.4 | |
| 2 | 17.36 | 163 | 119 | p-coumaric acid | 10.5 | |
| 3 | 20.12 | 193 | 149; 134 | ferulic acid | 0.2 | |
| 4 | 25.67 | 287 | 269 | dihydrokaempferol | 0.4 | |
| 5 | 28.75 | 515 | 353 | dicaffeoylquinic acid | 2.6 | |
| 6 | 37.21 | 271 | 253; 165; 151; 243 | pinobanksin | 3.4 | |
| 7 | 39.48 | 301 | 283; 255; 215; 187 | not identified | ||
| 8 | 53.79 | 247 | 203; 204 | not identified | ||
| 9 | 57.02 | 255 | 213; 151 | pinocembrin | 0.2 | |
| 10 | 58.55 | 285 | 257 | kaempferol | 2.4 | |
| 11 | 59.57 | 231 | 187 | drupanin | 7.2 | |
| 12 | 63.01 | 313 | 253; 271 | pinobanksin-3-O-acetate | 0.4 | |
| 13 | 65.96 | 253 | 209 | Chrysin | 0.2 | |
| 14 | 69.83 | 315 | 300; 271 | quercetin-3-methyl-ether or quercetin-4-methyl-ether | 2.8 | |
| 15 | 71.45 | 299 | 284 | luteolin-methyl-ether | 3.4 | |
| 16 | 76.72 | 315 | 300; 271 | quercetin-3-methyl-ether or quercetin-4-methyl-ether | 3.6 | |
| 17 | 77.73 | 329 | 314; 299 | quercetin-dimethyl-ether | 4.0 | |
| 18 | 80.13 | 299 | 200; 255 | artepillin C | 7.4 | |
| Cinnamic Acid Derivatives | 30.5 | |||||
| Flavonoids (Area 20.8%) | Flavonols and Dihydroflavonols | 17.0 | ||||
| Flavones and Flavanone | 3.8 | |||||
| Peak n | RT * (min) | m/z | MS2 m/z | Proposed Structure | Area % | |
|---|---|---|---|---|---|---|
| 1 | 2.10 | 225 (179 and 46) | 179 | caffeic acid | 1.1 | |
| 2 | 23.00 | 193 | 193; 149; 134 | ferulic acid | 6.3 | |
| 3 | 32.46 | 137 | 93 | p-hydroxybenzoic acid | 8.5 | |
| 4 | 33.76 | 285 | 241; 257 | kaempferol | 1.9 | |
| 5 | 39.26 | 271 | 253; 243; 151; 165; 107; 225 | pinobanksin | 3.7 | |
| 6 | 54.92 | 269 | 117; 149; 225 | apigenin | 1.8 | |
| 7 | 56.04 | 315 | 300; 228 | quercetin-3-methyl-ether | 0.7 | |
| 8 | 59.98 | 329 | 314; 299; 285 | quercetin-dimethyl-ether | 0.6 | |
| 9 | 61.19 | 255 | 213; 187; 151 | pinocembrin | 5.1 | |
| 10 | 68.72 | 313 | 253; 271; 299 | pinobanksin-3-O-acetate | 1.0 | |
| 11 | 71.35 | 253 | 209 | chrisin | 7.8 | |
| 12 | 73.06 | 269 (538) | 227 | galangin | 7.1 | |
| 13 | 75.20 | 283 | 239; 268 | galangin-5-methyl-ether | 1.1 | |
| 14 | 76.48 | 327 | 253; 271 | pinobanksin-3-O-propionate | 0.9 | |
| 15 | 78.83 | 373 | 279; 161; 277; 256; 305; 258 | not identified | ||
| 16 | 79.71 | 341 | 271; 253 | pinobanksin-3-O-butyrate | 0.5 | |
| 17 | 80.33 | 389 | 295 | not identified | ||
| 18 | 81.58 | 355 | 253; 271; 225 | pinobanksin-3-O-pentanoate | 0.2 | |
| Hydroxycinnamic Acids | 6.4 | |||||
| Flavonoids (Area 32.4%) | Flavonoids and Dihydroflavonoids | 22.8 | ||||
| Flavones | 9.6 | |||||
| Phenolic Acid | 8.5 | |||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaccaria, V.; Curti, V.; Di Lorenzo, A.; Baldi, A.; Maccario, C.; Sommatis, S.; Mocchi, R.; Daglia, M. Effect of Green and Brown Propolis Extracts on the Expression Levels of microRNAs, mRNAs and Proteins, Related to Oxidative Stress and Inflammation. Nutrients 2017, 9, 1090. https://doi.org/10.3390/nu9101090
Zaccaria V, Curti V, Di Lorenzo A, Baldi A, Maccario C, Sommatis S, Mocchi R, Daglia M. Effect of Green and Brown Propolis Extracts on the Expression Levels of microRNAs, mRNAs and Proteins, Related to Oxidative Stress and Inflammation. Nutrients. 2017; 9(10):1090. https://doi.org/10.3390/nu9101090
Chicago/Turabian StyleZaccaria, Vincenzo, Valeria Curti, Arianna Di Lorenzo, Alessandra Baldi, Cristina Maccario, Sabrina Sommatis, Roberto Mocchi, and Maria Daglia. 2017. "Effect of Green and Brown Propolis Extracts on the Expression Levels of microRNAs, mRNAs and Proteins, Related to Oxidative Stress and Inflammation" Nutrients 9, no. 10: 1090. https://doi.org/10.3390/nu9101090






