Inhibitory Effects of Culinary Herbs and Spices on the Growth of HCA-7 Colorectal Cancer Cells and Their COX-2 Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Culinary Herb and Spice (CHS) Extracts
2.2. Growth Inhibition Studies: Effect of Herb and Spice Extracts (Individual and in Combination) on Growth of HCA-7 Cells
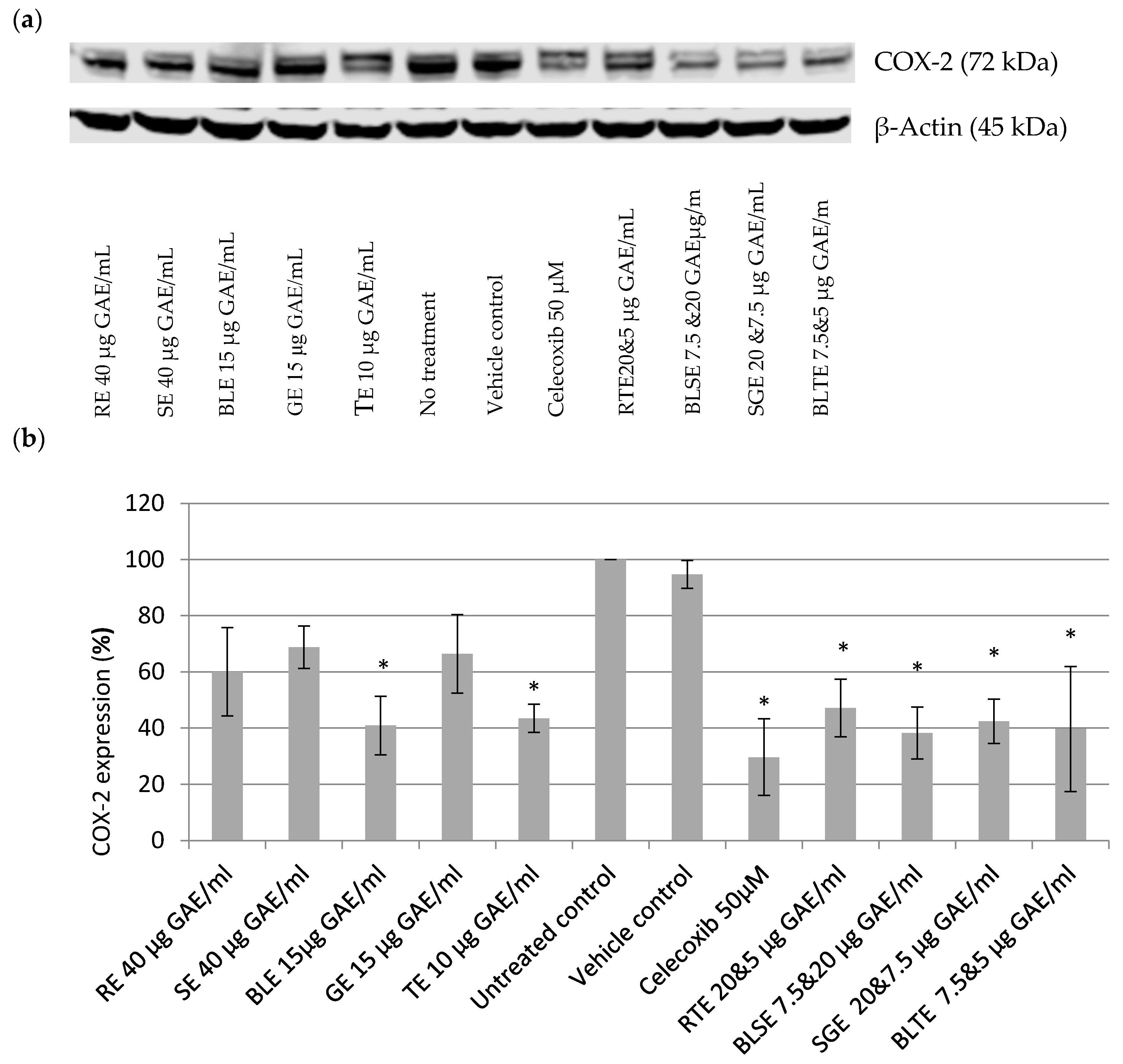
2.3. The Effect of Culinary Herb and Spice Extracts on COX-2 Expression in HCA-7 CRC Cells
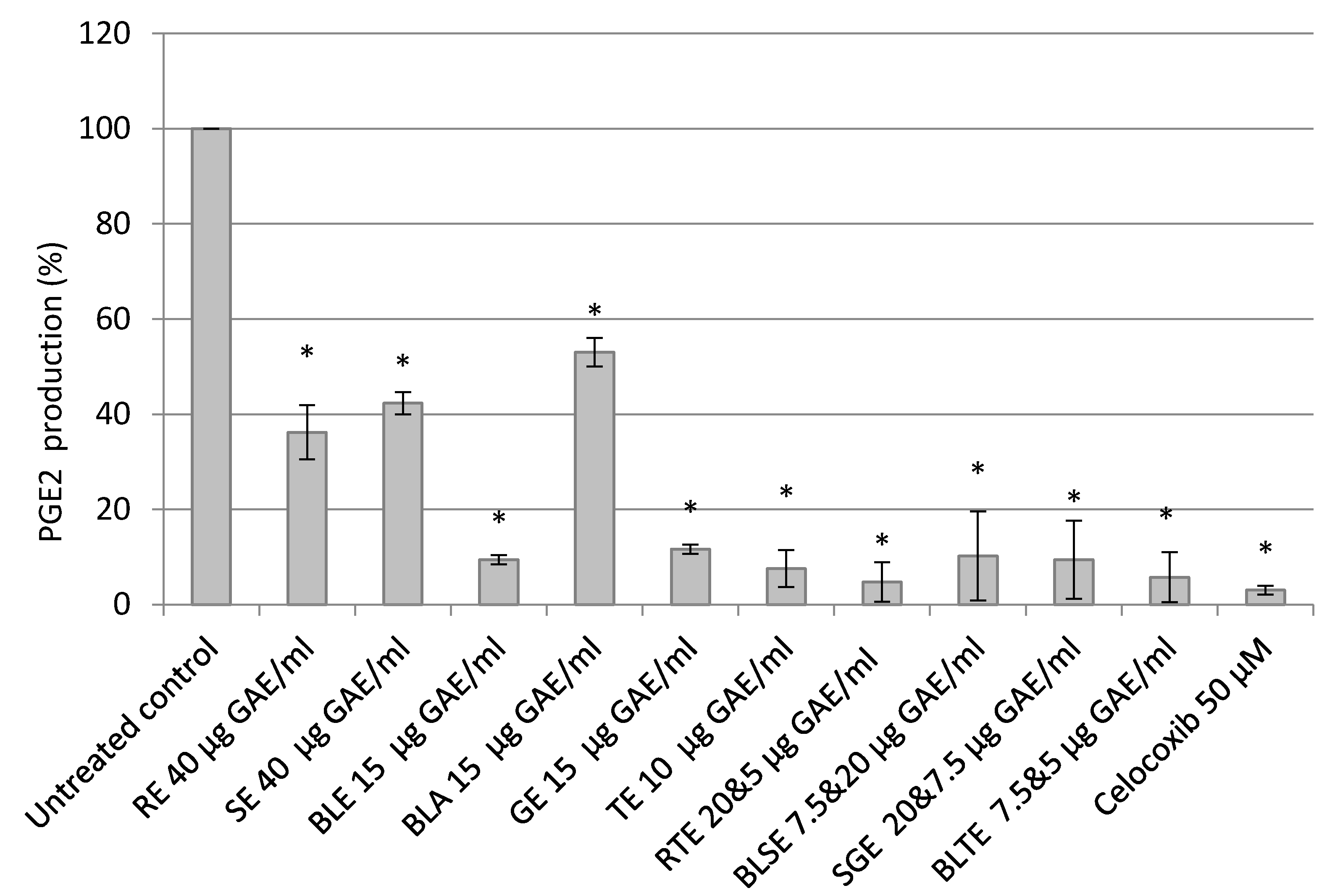
2.4. The Effect of Culinary Herb and Spice Extracts on COX-2 Activity, Based on PGE-2 Release, in HCA-7 CRC Cells
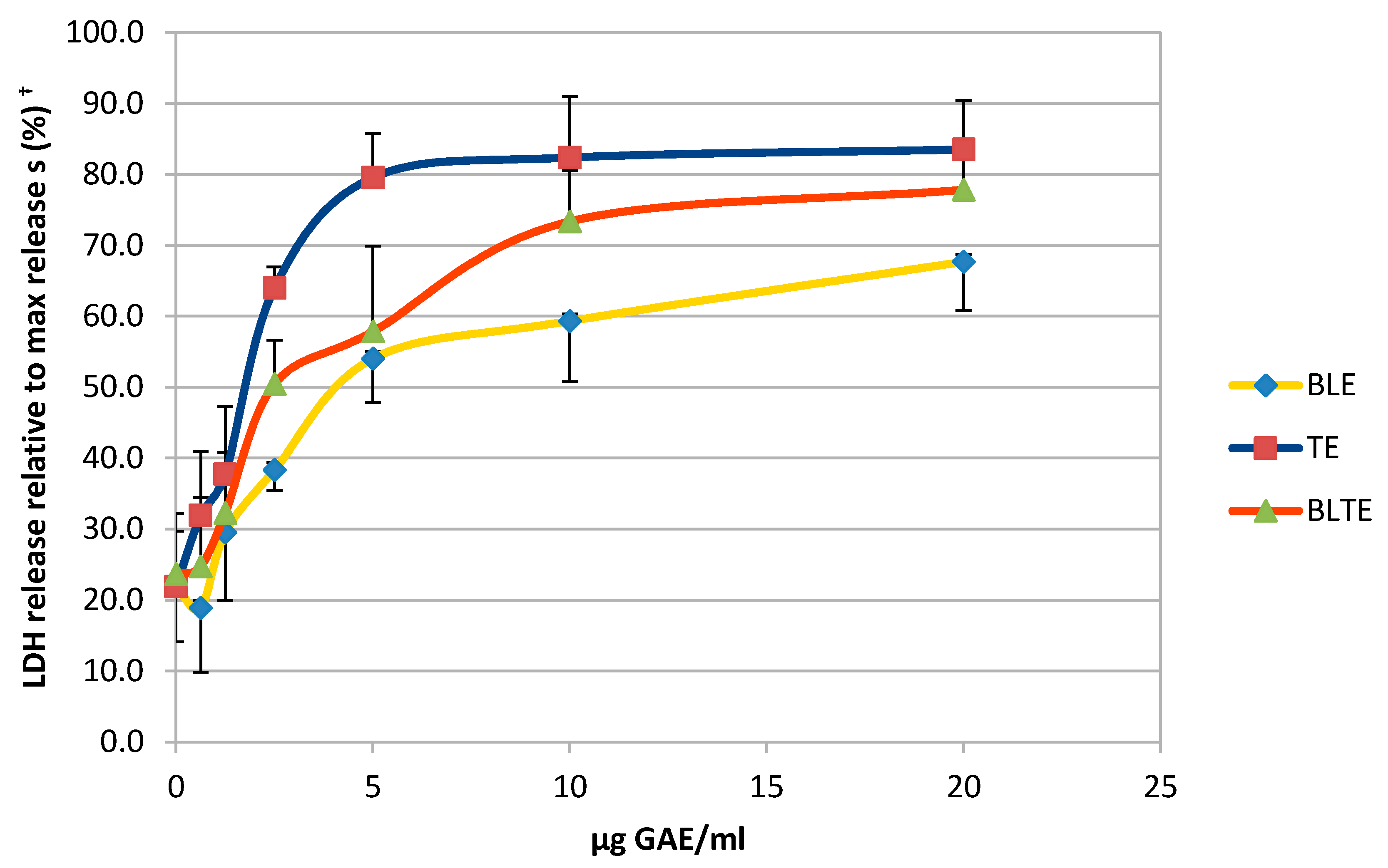
2.5. The Effect of CHS Extracts on HCA-7 Cell Viability at 24, 48 and 72 h
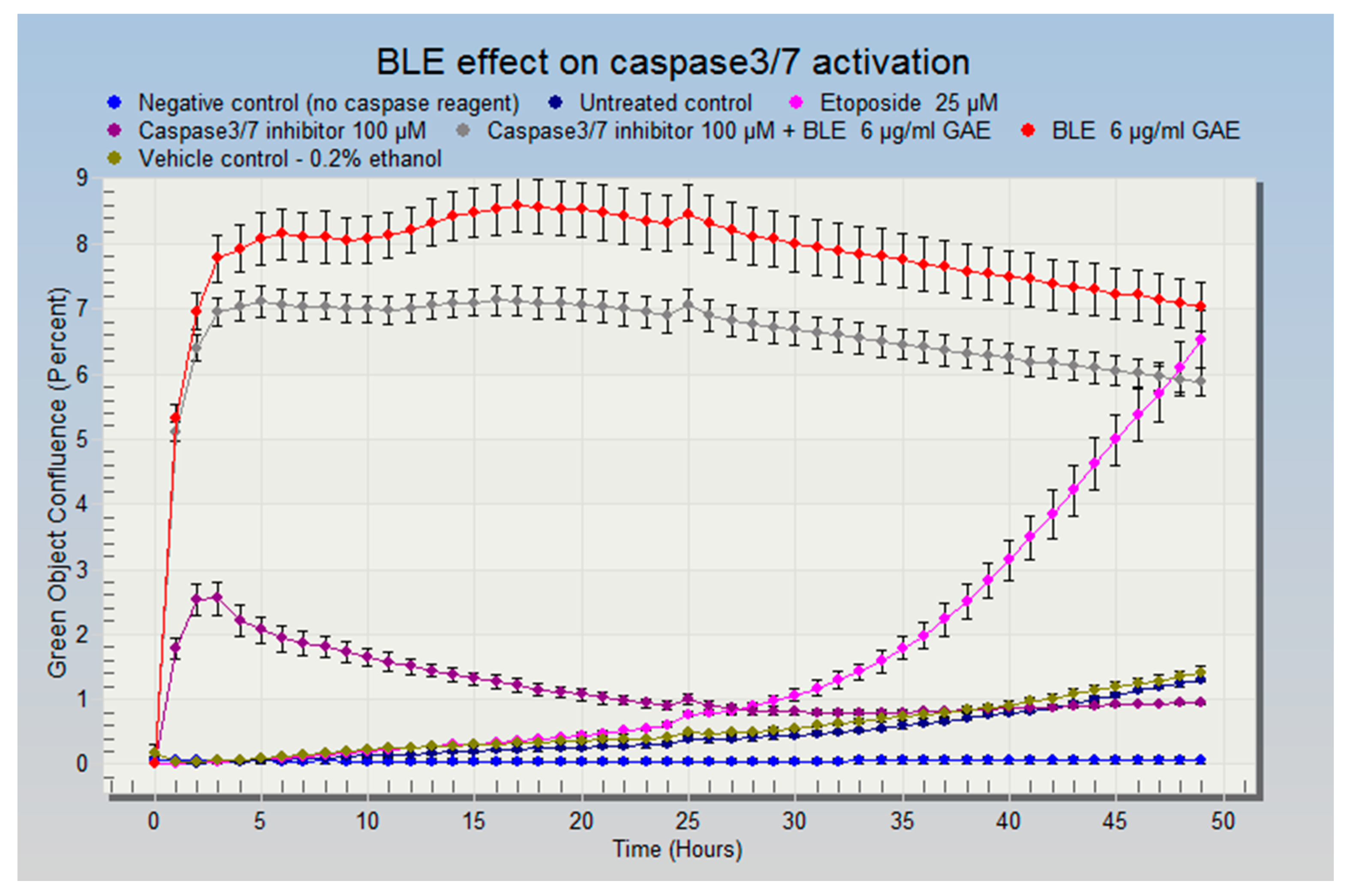
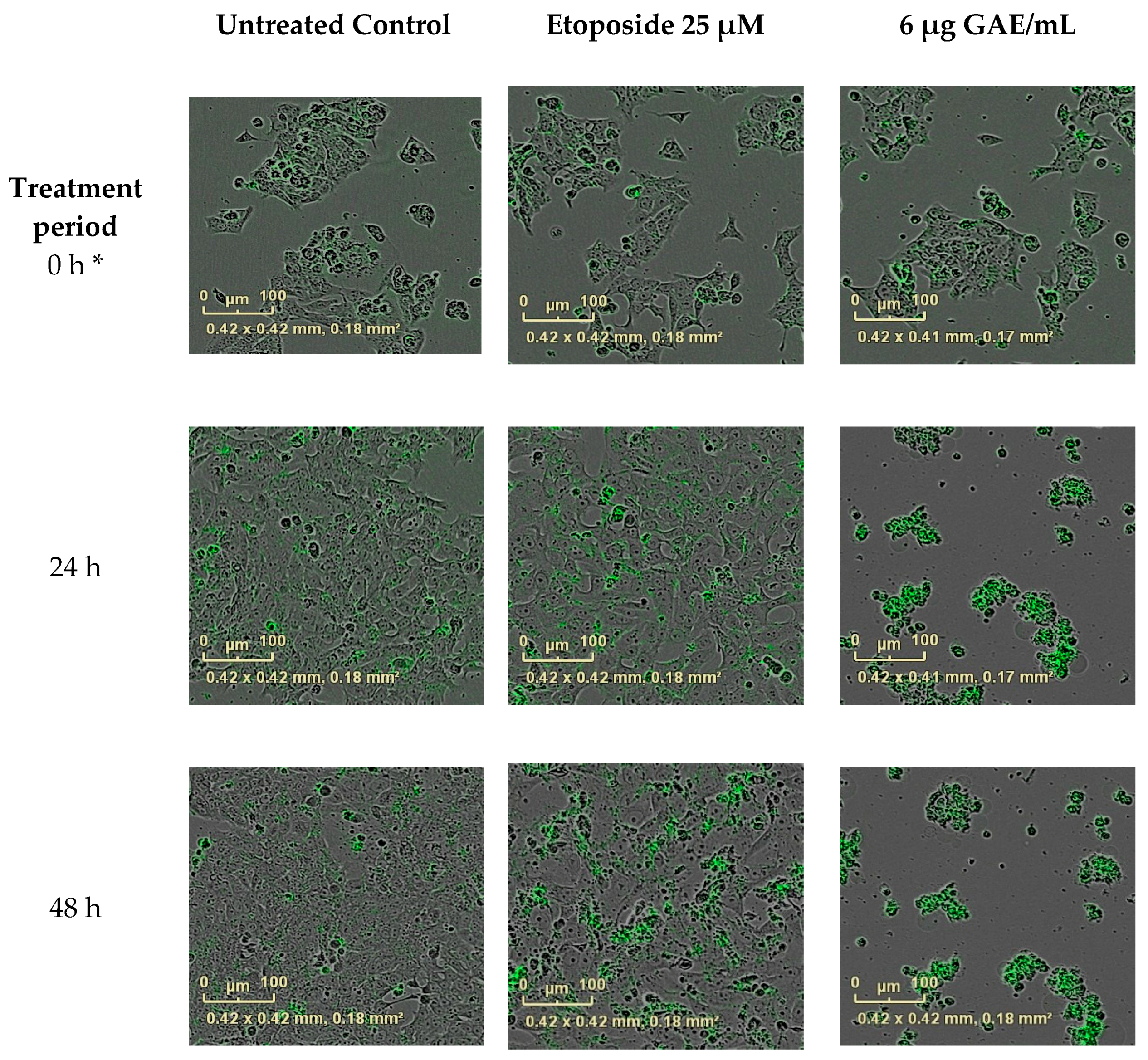
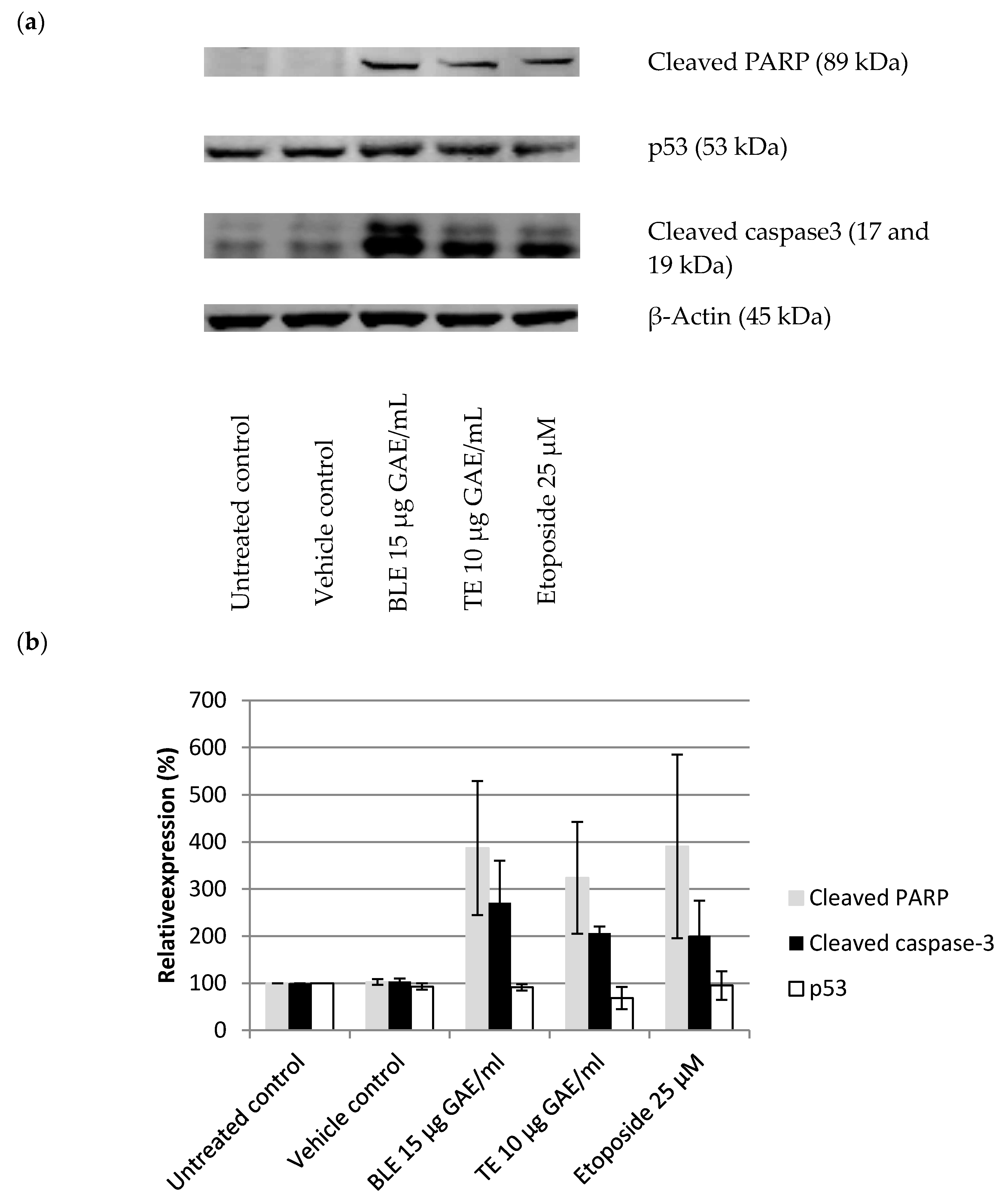
2.6. The Effect of CHS on the Cell Cycle and Apoptosis in HCA-7 CRC Cells
2.7. Data Expression and Statistical Analysis
3. Results
3.1. Effect of the CHS and Their Combinations on HCA-7 Cell Growth Using the SRB Assay
3.2. The Effect of CHS Extracts on COX-2 Expression in HCA-7 CRC Cells
3.3. The Effect of Culinary Herb and Spice Extracts on COX-2 Activity, Based on PGE-2 Release, in HCA-7 CRC Cells
3.4. The Effect of CHS Extracts on HCA-7 Cell Viability at 24, 48, and 72 h
3.5. The Effect of CHS on the Cell Cycle and Apoptosis in HCA-7 CRC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| A | aqueous |
| BLE | bay leaf ethanol |
| BLTE | bay leaf ethanol and turmeric ethanol |
| COX-2 | Cyclo-ooxygenase 2 |
| DMEM | Dulbecco’s modified Eagle’s medium |
| E | ethanol |
| GAE | gallic acid equivalent |
| GE | ginger ethanol |
| PGE-2 | prostaglandin E2 |
| RA | rosemary aqueous |
| RE | rosemary ethanol |
| RTE | rosemary ethanol and turmeric ethanol |
| SA | sag aqueous |
| SBLE | sage ethanol and bay leaf ethanol |
| SE | sage ethanol |
| SGE | sage ethanol and ginger ethanol |
References
- Pourhoseingholi, M.A. Epidemiology and burden of colorectal cancer in Asia-Pacific region: What shall we do now? Transl. Gastrointest. Cancer 2014, 2005, 169–173. [Google Scholar]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal Cancer Statistics, 2014. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar]
- Aggarwal, B.B.; Shishodia, S.; Sandur, S.K.; Pandey, M.K.; Sethi, G. Inflammation and cancer: How hot is the link? Biochem. Pharmacol. 2006, 72, 1605–1621. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xia, D.; DuBois, R.N. The Crosstalk of PTGS2 and EGF Signaling Pathways in Colorectal Cancer. Cancers 2011, 3, 3894–3908. [Google Scholar]
- Greenhough, A.; Smartt, H.J.M.; Moore, A.E.; Roberts, H.R.; Williams, A.C.; Paraskeva, C.; Kaidi, A. The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 2009, 30, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.; Gill, S. Role of cyclooxygenase-2 in colorectal cancer. Cancer Metastasis Rev. 2004, 23, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, X.-F. Overexpression of cyclooxygenase-2 correlates with advanced stages of colorectal cancer. Am. J. Gastroenterol. 2002, 97, 1037–1041. [Google Scholar] [CrossRef] [PubMed]
- Hahn, E.; Kraus, S.; Arber, N. Role of cyclooxygenase-2 in pathogenesis and prevention of colorectal cancer. Dig. Dis. 2010, 28, 585–589. [Google Scholar] [CrossRef]
- Sano, H.; Kawahito, Y.; Wilder, R.L.; Hashiramoto, A.; Mukai, S.; Asai, K.; Kimura, S.; Kato, H.; Kondo, M.; Hla, T. Expression of Cyclooxygene-1 and -2 in Human Colorectal Cancer. Cancer Res. 1995, 55, 3785–3790. [Google Scholar] [PubMed]
- Thun, M.J.; Jacobs, E.J.; Patrono, C. The role of aspirin in cancer prevention. Nat. Rev. Clin. Oncol. 2012, 9, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Friis, S.; Riis, A.H.; Erichsen, R.; Baron, J.A.; Sørensen, H.T. Low-Dose Aspirin or Nonsteroidal Anti-inflammatory Drug Use and Colorectal Cancer Risk. Ann. Int. Med. 2015, 163, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, E.J.; Newton, C.C.; Gapstur, S.M.; Thun, M.J. Daily Aspirin Use and Cancer Mortality in a Large US Cohort. J. Natl. Cancer Inst. 2012, 104, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Saloheimo, P.; Ahonen, M.; Juvela, S.; Pyhtinen, J.; Savolainen, E.; Hillbom, M. Regular aspirin-use preceding the onset of primary intracerebral hemorrhage is an independent predictor for death. Stroke. 2006, 37, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Shishodia, S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem. Pharmacol. 2006, 71, 1397–1421. [Google Scholar] [CrossRef] [PubMed]
- Aravindaram, K.; Yang, N.-S. Anti-inflammatory plant natural products for cancer therapy. Planta Med. 2010, 76, 1103–1117. [Google Scholar] [CrossRef] [PubMed]
- Jungbauer, A.; Medjakovic, S. Anti-inflammatory properties of culinary herbs and spices that ameliorate the effects of metabolic syndrome. Maturitas 2012, 71, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.-H.; Su, J.-D.; Chyau, C.-C.; Sung, T.-Y.; Ho, S.-S.; Peng, C.-C.; Peng, R.Y. Supercritical Fluid Extracts of Rosemary Leaves Exhibit Potent Anti-Inflammation and Anti-Tumor Effects. Biosci. Biotechnol. Biochem. 2007, 71, 2223–2232. [Google Scholar] [CrossRef] [PubMed]
- Baker, I.; Chohan, M.; Opara, E.I. Impact of cooking and digestion, in vitro, on the antioxidant capacity and anti-inflammatory activity of cinnamon, clove and nutmeg. Plant Foods Hum. Nutr. 2013, 68, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Rémésy, C.; Jiménez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [PubMed]
- Opara, E.I.; Chohan, M. Culinary Herbs and Spices: Their Bioactive Properties, the Contribution of Polyphenols and the Challenges in Deducing Their True Health Benefits. Int. J. Mol. Sci. 2014, 15, 19183–19202. [Google Scholar] [CrossRef] [PubMed]
- Yesil-Celiktas, O.; Sevimli, C.; Bedir, E.; Vardar-Sukan, F. Inhibitory effects of rosemary extracts, carnosic acid and rosmarinic acid on the growth of various human cancer cell lines. Plant Foods Hum. Nutr. 2010, 65, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Dilas, S.; Knez, Ž.; Četojević-Simin, D.; Tumbas, V.; Škerget, M.; Čanadanović-Brunet, J.; Ćetković, G. In Vitro antioxidant and antiproliferative activity of three rosemary (Rosmarinus officinalis L.) extract formulations. Int. J. Food Sci. Technol. 2012, 47, 2052–2062. [Google Scholar] [CrossRef]
- Yi, W.; Wetzstein, H.Y. Anti-tumorigenic activity of five culinary and medicinal herbs grown under greenhouse conditions and their combination effects. J. Sci. Food Agric. 2011, 91, 1849–1854. [Google Scholar] [CrossRef] [PubMed]
- Plummer, S.M.; Holloway, K.A.; Manson, M.M.; Munks, R.J.L.; Kaptein, A.; Farrow, S.; Howells, L. Inhibition of cyclo-oxygenase 2 expression in colon cells by the chemopreventive agent curcumin involves inhibition of NF-kB activation via the NIK/IKK signalling complex. Current 1999, 18, 6013–6020. [Google Scholar]
- Zhang, F.; Altorki, N.K.; Mestre, J.R.; Subbaramaiah, K.; Dannenberg, A.J. Curcumin inhibits cyclooxygenase-2 transcription in bile acid- and phorbol ester-treated human gastrointestinal epithelial cells. Carcinogenesis 1999, 20, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Boland, C.R.; Chauhan, D.P. Specific inhibition of cyclooxygenase-2 (COX-2) expression by dietary curcumin in HT-29 human colon cancer cells. Cancer Lett. 2001, 172, 111–118. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Takada, Y.; Oommen, O.V. From chemoprevention to chemotherapy: Common targets and common goals. Expert Opin. Investig. Drugs 2004, 13, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xue, A.; Niu, H.; Jia, Z.; Wang, J. Optimised ultrasonic-assisted extraction of flavonoids from Folium eucommiae and evaluation of antioxidant activity in multi-test systems in vitro. Food Chem. 2009, 114, 1147–1154. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Tang, S.Y.; Whiteman, M.; Peng, Z.F.; Jenner, A.; Yong, E.L.; Halliwell, B. Characterization of antioxidant and antiglycation properties and isolation of active ingredients from traditional Chinese medicines. Free Radic. Biol. Med. 2004, 36, 1575–1587. [Google Scholar] [CrossRef] [PubMed]
- Khelwatty, S.A.; Essapen, S.; Seddon, A.M.; Modjtahedi, H. Growth response of human colorectal tumour cell lines to treatment with afatinib (BIBW2992), an irreversible erbB family blocker, and its association with expression of HER family members. Int. J. Oncol. 2011, 39, 483–491. [Google Scholar] [PubMed]
- Lev-Ari, S. Celecoxib and Curcumin Synergistically Inhibit the Growth of Colorectal Cancer Cells. Clin. Cancer Res. 2005, 11, 6738–6744. [Google Scholar] [CrossRef] [PubMed]
- Arber, N.; Eagle, C.J.; Spicak, J.; Rácz, I.; Dite, P.; Hajer, J.; Zavoral, M.; Lechuga, M.J.; Gerletti, P.; Tang, J.; et al. Celecoxib for the prevention of colorectal adenomatous polyps. N. Engl. J. Med. 2006, 355, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Wlodkowic, D.; Skommer, J.; Darzynkiewicz, Z. Flow cytometry-based apoptosis detection Donald. Methods Mol. Biol. 2009, 559, 1–14. [Google Scholar]
- Nouri, K.; Yazdanparast, R. Proliferation inhibition, cell cycle arrest and apoptosis induced in HL-60 cells by a natural diterpene ester from Daphne mucronata. Daru 2011, 19, 145–153. [Google Scholar] [PubMed]
- Dimas, K.; Tsimplouli, C.; Houchen, C.; Pantazis, P.; Sakellaridis, N.; George, T.; Tsangaris, E.; Anastasiadou, R. An Ethanol Extract of Hawaiian Turmeric: Extensive in Vitro Anticancer Activity against Human Colon Cancer Cells. Altern. Ther. 2015, 21, 46–54. [Google Scholar]
- Lim, D.Y.; Cho, H.J.; Kim, J.; Nho, C.W.; Lee, K.W.; Park, J.H.Y. Luteolin decreases IGF-II production and downregulates insulin-like growth factor-I receptor signaling in HT-29 human colon cancer cells. BMC Gastroenterol. 2012, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Yue, G.G.-L.; Jiang, L.; Kwok, H.-F.; Lee, J.K.-M.; Chan, K.-M.; Fung, K.-P.; Leung, P.-C.; Lau, C.B.-S. Turmeric ethanolic extract possesses stronger inhibitory activities on colon tumour growth than curcumin—The importance of turmerones. J. Funct. Foods 2016, 22, 565–577. [Google Scholar]
- Liu, R.H. Potential Synergy of Phytochemicals in Cancer Prevention: Mechanism of Action. J. Nutr. 2004, 134, 3479S–3485S. [Google Scholar] [PubMed]
- Murillo, G.; Naithani, R.; Mehta, R.G. Efficacy of Herbal Products in Colorectal Cancer Prevention. Curr. Colorectal Cancer Rep. 2008, 4, 34–42. [Google Scholar] [CrossRef]
- Xavier, C.P.R.; Lima, C.F.; Fernandes-Ferreira, M.; Pereira-Wilson, C. Salvia fruticosa, Salvia officinalis, and rosmarinic acid induce apoptosis and inhibit proliferation of human colorectal cell lines: The role in MAPK/ERK pathway. Nutr. Cancer 2009, 61, 564–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, L.; Abeywardena, M.; Burnard, S.; Forsyth, S.; Head, R.; King, K.; Patten, G.; Watkins, P.; Williams, R.; Zabaras, D.; et al. Molecular size fractions of bay leaf (Laurus nobilis) exhibit differentiated regulation of colorectal cancer cell growth in vitro. Nutr. Cancer 2013, 65, 746–764. [Google Scholar] [CrossRef] [PubMed]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Gawlik-Dziki, U. Dietary spices as a natural effectors of lipoxygenase, xanthine oxidase, peroxidase and antioxidant agents. LWT Food Sci. Technol. 2012, 47, 138–146. [Google Scholar] [CrossRef]
- Durak, A.; Gawlik-Dziki, U.; Kowlska, I. Coffee with ginger—Interactions of biologically active phytochemicals in the model system. Food Chem. 2015, 166, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shen, P.; Zhang, X.C.; Zhao, M.D.; Zhang, X.G.; Yang, L. Efficacy and safety profile of celecoxib for treating advanced cancers: A meta-analysis of 11 randomized clinical trials. Clin. Ther. 2014, 36, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Willenberg, I.; Meschede, A.K.; Gueler, F.; Jang, M.-S.; Shushakova, N.; Schebb, N.H. Food Polyphenols Fail to Cause a Biologically Relevant Reduction of COX-2 Activity. PLoS ONE 2015, 10, e0139147. [Google Scholar] [CrossRef] [PubMed]
- Puangsombat, K.; Smith, J.S. Inhibition of heterocyclic amine formation in beef patties by ethanolic extracts of rosemary. J. Food Sci. 2010, 75, T40–T47. [Google Scholar] [CrossRef] [PubMed]
- Generalić, I.; Skroza, D.; Surjak, J.; Možina, S.S.; Ljubenkov, I.; Katalinić, A.; Simat, V.; Katalinić, V. Seasonal Variations of Phenolic Compounds and Biological Properties in Sage (Salvia officinalis L.). Chem. Biodivers. 2012, 9, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Anandaraj, M.; Prasath, D.; Kandiannan, K.; Zachariah, T.J.; Srinivasan, V.; Jha, A.K.; Singh, B.K.; Singh, A.K.; Pandey, V.P.; Singh, S.P.; et al. Genotype by environment interaction effects on yield and curcumin in turmeric (Curcuma longa L.). Ind. Crops Prod. 2014, 53, 358–364. [Google Scholar] [CrossRef]
- Dvorackova, E.; Snoblova, M.; Chromcova, L.; Hrdlicka, P. Effects of Extraction Methods on the Phenolic Compounds Contents and Antioxidant Capacities of Cinnamon Extracts. Food Sci. Biotechnol. 2015, 24, 1201–1207. [Google Scholar] [CrossRef]
- Shemesh, N.; Arber, N. Curcumin Alone and in Combination for Prevention of Colorectal Cancer. Curr. Colorectal Cancer Rep. 2014, 10, 62–67. [Google Scholar] [CrossRef]
- Surh, Y.J.; Chun, K.S.; Cha, H.H.; Han, S.S.; Keum, Y.S.; Park, K.K.; Lee, S.S. Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: Down-regulation of COX-2 and iNOS through suppression of NF-kappa B activation. Mutat. Res. 2001, 480–481, 243–268. [Google Scholar] [CrossRef]
- Romier, B.; Van De Walle, J.; During, A.; Larondelle, Y.; Schneider, Y.-J. Modulation of signalling nuclear factor-kappaB activation pathway by polyphenols in human intestinal Caco-2 cells. Br. J. Nutr. 2008, 100, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Bose, M.; Ju, J.; Ryu, J.-H.; Chen, X.; Sang, S.; Lee, M.-J.; Yang, C.S. Modulation of arachidonic acid metabolism by curcumin and related beta-diketone derivatives: Effects on cytosolic phospholipase A(2), cyclooxygenases and 5-lipoxygenase. Carcinogenesis 2004, 25, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharan, N.V.; Simmons, D.L. Protein family review the cyclooxygenases. Genome Biol. 2004, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ricciotti, E.; FitzGerald, G.A. Prostaglandins and Inflammation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef] [PubMed]
- Koeberle, A.; Northoff, H.; Werz, O. Curcumin blocks prostaglandin E2 biosynthesis through direct inhibition of the microsomal prostaglandin E2 synthase-1. Mol. Cancer Ther. 2009, 8, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Yuan, W.; Li, S.; Gupta, S.C. Curcumin-free turmeric exhibits anti-inflammatory and anticancer activities: Identification of novel components of turmeric. Mol. Nutr. Food Res. 2013, 57, 1529–1542. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Hobiger, S.; Jungbauer, A. Anti-inflammatory activity of extracts from fruits, herbs and spices. Food Chem. 2010, 122, 987–996. [Google Scholar] [CrossRef]
- Guo, Y.; Sakulnarmrat, K.; Konczak, I. Anti-inflammatory potential of native Australian herbs polyphenols. Toxicol. Rep. 2014, 1, 385–390. [Google Scholar]
- Haghighi, A.; Jahromi, N.Z. Compare the effect of ginger extract and aspirin on COX-2 gene expression in colon cancer cell line HT-29. J. Herb. Drugs 2016, 7, 163–168. [Google Scholar]
- Pacifico, S.; D’Abrosca, B.; Scognamiglio, M.; Gallicchio, M.; Potenza, N.; Piccolella, S.; Russo, A.; Monaco, P.; Fiorentino, A. Metabolic profiling of strawberry grape (Vitis × labruscana cv. “Isabella”) components by nuclear magnetic resonance (NMR) and evaluation of their antioxidant and antiproliferative properties. J. Agric. Food Chem. 2011, 59, 7679–7687. [Google Scholar] [PubMed]
- Vallverdú-Queralt, A.; Regueiro, J.; Martínez-Huélamo, M.; Rinaldi Alvarenga, J.F.; Leal, L.N.; Lamuela-Raventos, R.M. A comprehensive study on the phenolic profile of widely used culinary herbs and spices: Rosemary, thyme, oregano, cinnamon, cumin and bay. Food Chem. 2014, 154, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Dufour, M.; Faes, S.; Dormond-Meuwly, A.; Demartines, N.; Dormond, O. PGE2-induced colon cancer growth is mediated by mTORC1. Biochem. Biophys. Res. Commun. 2014, 451, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Shukla, Y.; Singh, M. Cancer preventive properties of ginger: A brief review. Food Chem. Toxicol. 2007, 45, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Dugasani, S.; Pichika, M.R.; Nadarajah, V.D.; Balijepalli, M.K.; Tandra, S.; Korlakunta, J.N. Comparative antioxidant and anti-inflammatory effects of [6]-gingerol, [8]-gingerol, [10]-gingerol and [6]-shogaol. J. Ethnopharmacol. 2010, 127, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Mashhadi, N.S.; Ghiasvand, R.; Askari, G.; Hariri, M.; Darvishi, L. Anti-Oxidative and Anti-Inflammatory Effects of Ginger in Health and Physical Activity: Review of Current Evidence. Int. J. Prev. Med. 2013, 4, 4–6. [Google Scholar]
- Van Breemen, R.B.; Tao, Y.; Li, W. Cyclooxygenase-2 inhibitors in ginger (Zingiber officinale). Fitoterapia 2011, 82, 38–43. [Google Scholar] [CrossRef] [PubMed]
- De Kok, T.M.; Van Breda, S.G.; Manson, M.M. Mechanisms of combined action of different chemopreventive dietary compounds: A review. Eur. J. Nutr. 2008, 47, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Van Breda, S.G.; van Agen, E.; van Sanden, S.; Burzykowski, T.; Kleinjans, J.C.; Delft, J.H. Vegetables affect the expression of genes involved in carcinogenic and anticarcinogenic processes in the lungs of female C57BL/6 mice. J. Nutr. 2005, 135, 2546–2552. [Google Scholar] [PubMed]
- Sheng, H.; Shao, J.; Morrow, J.D.; Beauchamp, R.D.; DuBois, R.N. Modulation of apoptosis and Bcl-2 expression by prostaglandin E2 in human colon cancer cells. Cancer Res. 1998, 58, 362–366. [Google Scholar]
- Rezonja, R.; Knez, L.; Cufer, T.; Mrhar, A. Oral treatment with etoposide in small cell lung cancer—Dilemmas and solutions. Radiol. Oncol. 2013, 47, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Poirier, M.G.; Eroglu, S.; Marko, J.F. The bending rigidity of mitotic chromosomes. Mol. Biol. Cell 2002, 13, 2170–2179. [Google Scholar] [CrossRef]
- Chaitanya, G.V.; Steven, A.J.; Babu, P.P. PARP-1 cleavage fragments: Signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal. 2010, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Shehzad, A.; Lee, J.; Huh, T.-L.; Lee, Y.S. Curcumin induces apoptosis in human colorectal carcinoma (HCT-15) cells by regulating expression of Prp4 and p53. Mol. Cells 2013, 35, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.R.M.R.; Haque, A.; Rahman, M.A.; Chen, Z.G.; Khuri, F.R.; Shin, D.M. Curcumin induces apoptosis of upper aerodigestive tract cancer cells by targeting multiple pathways. PLoS ONE 2015, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rodd, A.L.; Ververis, K.; Sayakkarage, D.; Khan, A.W.; Rafehi, H.; Ziemann, M.; Loveridge, S.J.; Lazarus, R.; Kerr, C.; Lockett, T.; et al. RNA sequencing supports distinct reactive oxygen species-mediated pathways of apoptosis by high and low size mass fractions of Bay leaf (Lauris nobilis) in HT-29 cells. Food Funct. 2015, 6, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-T.; Kardosh, A.; Cooc, J.; Schönthal, A.H. Potential misidentification of cyclooxygenase-2 by Western blot analysis and prevention through the inclusion of appropriate controls. Mol. Biotechnol. 2006, 34, 329–335. [Google Scholar] [CrossRef]
- Thongrakard, V.; Titone, R.; Follo, C.; Morani, F.; Suksamrarn, A.; Tencomnao, T.; Isidoro, C. Turmeric toxicity in A431 epidermoid cancer cells associates with autophagy degradation of anti-apoptotic and anti-autophagic p53 mutant. Phyther. Res. 2014, 28, 1761–1769. [Google Scholar] [CrossRef]
- Agarwal, B.; Swaroop, P.; Protiva, P.; Raj, S.V.; Shirin, H.; Holt, P.R. Cox-2 is needed but not sufficient for apoptosis induced by Cox-2 selective inhibitors in colon cancer cells. Apoptosis 2003, 8, 649–654. [Google Scholar] [CrossRef] [PubMed]

| Herb/Spice/Combinations | IC50 (μg GAE/mL) | IC50 (μg/mL of DW) |
|---|---|---|
| TE | 3.0 (±0.3) | 300 |
| BLTE | 3.3 (±0.7) | 227 |
| BLE | 4.7 (±0.2) | 177 |
| GE | 5.5 (±0.3) | 417 |
| BLSE | 5.5 (±0.3) | 180 |
| RTE | 6.0 (±0.4) | 382 |
| SGE | 6.8 (±0.1) | 352 |
| SE | 12.5 (±0.9) | 347 |
| SAE | 15.7 (±0.6) | 414 |
| RE | 15.9 (±0.4) | 347 |
| RAE | 16.2 (±0.4) | 432 |
| RA | 17.1 (±0.1) | 442 |
| SA | >20 (n/a) | >442 |
| Combinations | HCA-7 |
|---|---|
| RAE | 0.98 |
| SAE | n/a |
| RTE | 1.20 |
| BLTE | 0.90 |
| SGE | 0.67 |
| SBLE | 0.80 |
| Herbs/Spices | 24 H | 48 H | 72 H | Extracts Removed from Media * |
|---|---|---|---|---|
| IC50 (μg GAE/mL) (±SEM) | IC50 (μg GAE/mL) (±SEM) | IC50 (μg GAE/mL) (±SEM) | IC50 (μg GAE/mL) (±SEM) | |
| TE | 6.0 (±0.1) | 2.1 (±0.3) | 2.5 (±0.2) | 2.5 (±0.4) |
| GE | 10.0 (±0.5) | 6.1 (±0.6) | 5.8 (±0.1) | 7.8 (±0.8) |
| BLE | 10.5 (±0.3) | 6.0 (±0.5) | 9.2 (±0.2) | 8.4 (±0.6) |
| BLTE | 11.1 (±0.9) | 4.9 (±0.5) | 3.6 (±0.6) | 3.7 (±0.6) |
| SGE | 11.1 (±0.8) | 10.7 (±0.5) | 10.9 (±0.8) | 11.3 (±0.9) |
| Herbs/Spices | Sub G1 (%) (±SEM) | G1 (%) (±SEM) | S (%) (±SEM) | G2 (%) (±SEM) | ||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| Untreated control | 10 (±1.3) | 7 (±1.4) | 40 (±1.5) | 46 (±1.0) | 26 (±1.2) | 25 (±2.3) | 23 (±1.2) | 20 (±2.9) |
| Vehicle control (ethanol) | 9 (±1.2) | 4 (±0.7) | 41 (±1.6) | 47 (±1.3) | 27 (±1.2) | 24 (±1.2) | 23 (±0.4) | 23 (±0.3) |
| Vehicle control (H2O) | 10 (±1.0) | 4 (±1.0) | 39 (±0.9) | 45 (±0.6) | 25 (±1.8) | 25 (±0.3) | 24 (±1.2) | 23 (±0.3) |
| TE (2 μg GAE/mL) | 23 (±4.0) * | 49 (±3.1) * | 41 (±3.2) | 21 (±4.7) | 20 (±0.3) | 19 (±1.2) | 15 (±0.9) | 9 (±0.6) |
| GE (8 μg GAE/mL) | 27 (±3.9) * | 49 (±3.1) * | 41 (±2.3) | 25 (±2.9) | 17 (±0.3) | 15 (±1.2) | 12 (±0.6) | 8 (±0.7) |
| BLE (6 μg GAE/mL) | 28 (±3.2) * | 43 (±2.5) * | 38 (±3.4) | 28 (±1.5) | 20 (±1.8) | 15 (±1.2) | 13 (±1.8) | 12 (±1.0) |
| SE 16 μg GAE/mL | 16 (±2.3) | 30 (±1.3) * | 42 (±0.3) | 31 (±4.2) | 23 (±1.2) | 21 (±3.5) | 18 (±1.5) | 17 (±0.3) |
| RE (20 μg GAE/mL) | 14 (±0.3) | 17 (±4.3) | 42 (±0.9) | 41 (±1.2) | 31 (±0.7) | 20 (±0.4) | 10 (±0.3) | 20 (±1.6) |
| BLTE (3 μg GAE/mL BL and 1 μg GAE/mL TE) | 33 (±0.9) * | 33 (±0.6) * | 34 (±0.3) | 35 (±1.3) | 19 (±0.6) | 16 (±0.7) | 11 (±0.7) | 14 (±1.5) |
| BLSE (3 μg GAE/mL BL and 8 μg GAE/mL SE) | 19 (±0.8) * | 26 (±1.8) * | 41 (±0.8) | 42 (±3.2) | 18 (±1.6) | 18 (±1.7) | 12 (±0.4) | 14 (±1.0) |
| RTE (10 μg GAE/mL RE and 1 μg GAE/mL TE) | 16 (±0.9) * | 35 (±0.3) * | 42 (±1.5) | 32 (±2.5) | 26 (±2.3) | 17 (±1.2) | 18 (±0.3) | 12 (±0.3 |
| SGE (8 μg GAE/mL SE and 4 μg GAE/mL GE) | 23 (±0.3) * | 22 (±1.2) | 37 (±0.7) | 45 (±0.7) | 27 (±0.3) | 20 (±0.6) | 11 (±0.6) | 12 (±1.0) |
| Celecoxib (50 μM) | 23 | - | 45 | - | 18 | - | 13 | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaksevicius, A.; Carew, M.; Mistry, C.; Modjtahedi, H.; Opara, E.I. Inhibitory Effects of Culinary Herbs and Spices on the Growth of HCA-7 Colorectal Cancer Cells and Their COX-2 Expression. Nutrients 2017, 9, 1051. https://doi.org/10.3390/nu9101051
Jaksevicius A, Carew M, Mistry C, Modjtahedi H, Opara EI. Inhibitory Effects of Culinary Herbs and Spices on the Growth of HCA-7 Colorectal Cancer Cells and Their COX-2 Expression. Nutrients. 2017; 9(10):1051. https://doi.org/10.3390/nu9101051
Chicago/Turabian StyleJaksevicius, Andrius, Mark Carew, Calli Mistry, Helmout Modjtahedi, and Elizabeth I. Opara. 2017. "Inhibitory Effects of Culinary Herbs and Spices on the Growth of HCA-7 Colorectal Cancer Cells and Their COX-2 Expression" Nutrients 9, no. 10: 1051. https://doi.org/10.3390/nu9101051





