Nutraceutical with Resveratrol and Omega-3 Fatty Acids Induces Autophagy in ARPE-19 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Treatments
2.3. Western Blot Analyses of Autophagy Markers p62 and LC3-I/LC3-II
2.4. Cell Viability Assays
2.5. Determination of Lysosome Fusion to the Autophagosome Using mCherry-GFP-LC3-Plasmid
2.6. Statistical Analyses
3. Results
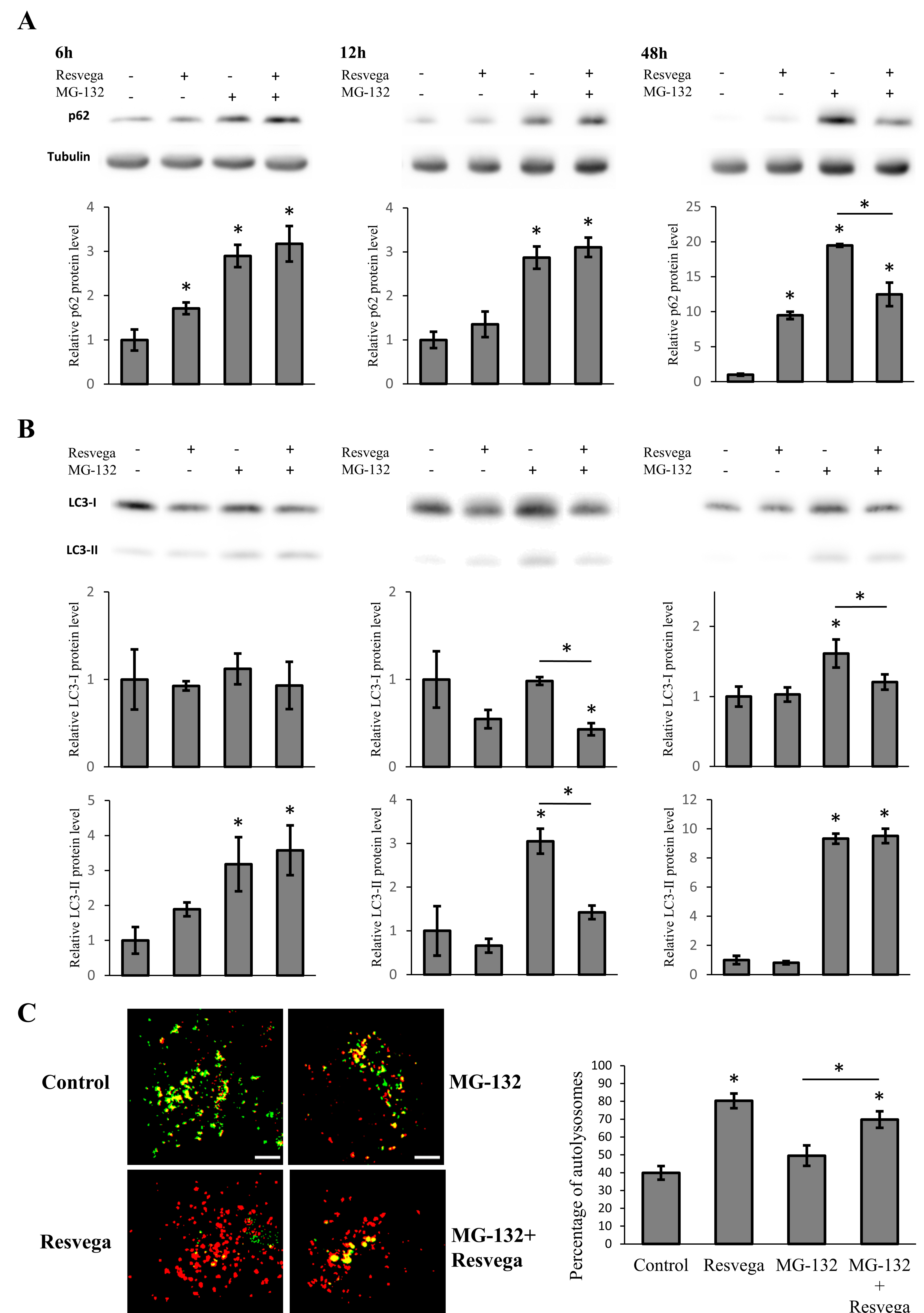
3.1. Resvega Alters the Level of Autophagy Markers p62 and LC3
3.2. Resvega Faciliates Protein Clearance by Autophagy in Proteasome Inhibition Model
3.3. Resvega Induces Autophagic Flux by Increasing the Number of Autolysosomes
3.4. Resvega Enhances Autophagy and Increases Cell Viability in the Starvation-Induced Autophagy Model
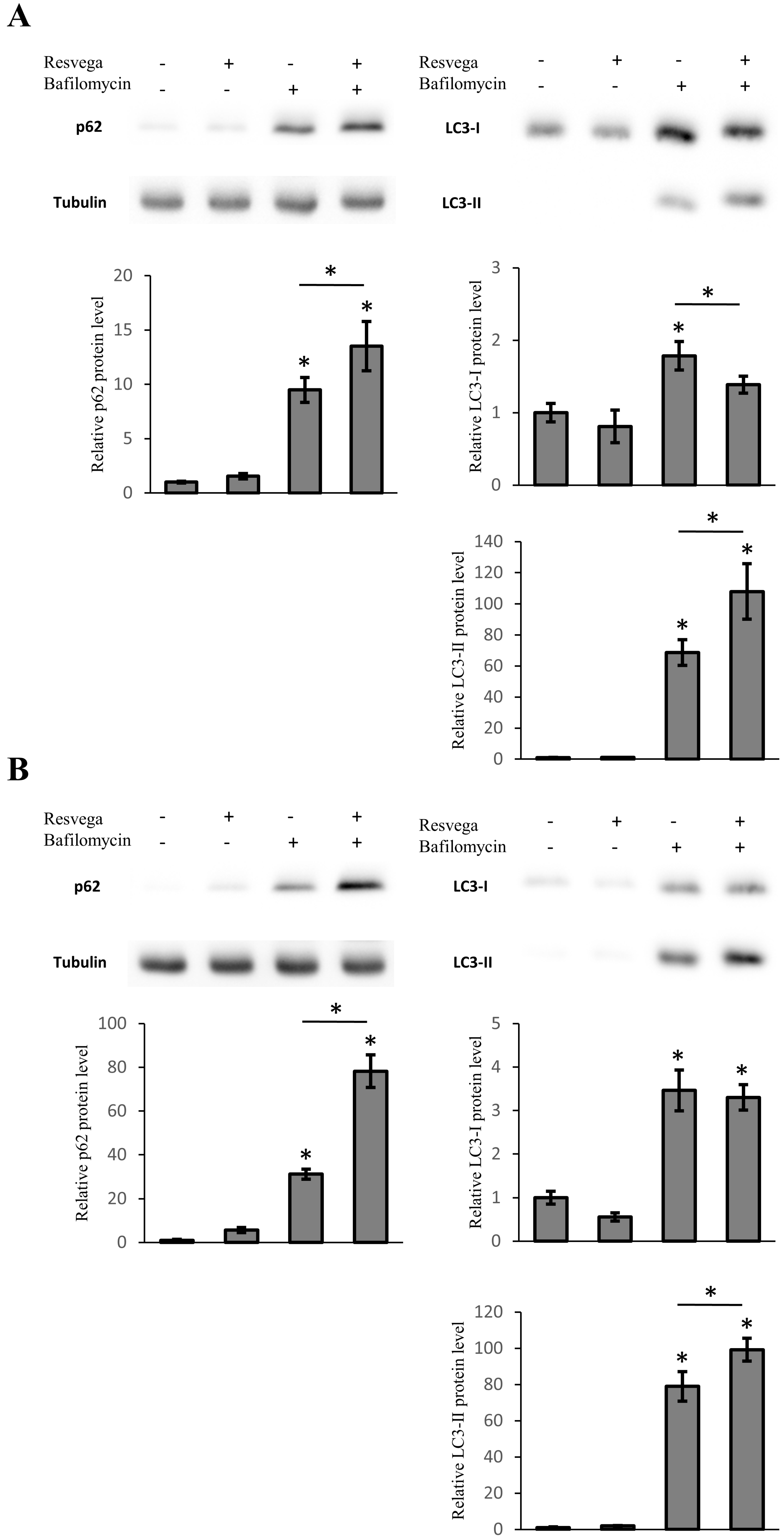
3.5. Resvega Induces Accumulation of LC3 and p62 under Autophagy Inhibition
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kaarniranta, K.; Sinha, D.; Blasiak, J.; Kauppinen, A.; Veréb, Z.; Salminen, A.; Boulton, M.E.; Petrovski, G. Autophagy and heterophagy dysregulation leads to retinal pigment epithelium dysfunction and development of age-related macular degeneration. Autophagy 2013, 9, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Gordois, A.; Cutler, H.; Pezzullo, L.; Gordon, K.; Cruess, A.; Winyard, S.; Hamilton, W.; Chua, K. An estimation of the worldwide economic and health burden of visual impairment. Glob. Public Health 2012, 7, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Kaarniranta, K.; Salminen, A.; Haapasalo, A.; Soininen, H.; Hiltunen, M. Age-related macular degeneration (AMD): Alzheimer’s disease in the eye? J. Alzheimers Dis. 2011, 24, 615–631. [Google Scholar] [PubMed]
- Ferrington, D.; Sinha, D.; Kaarniranta, K. Defects in retinal pigment epithelial cell proteolysis and the pathology associated with age-related macular degeneration. Prog. Retin. Eye Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kauppinen, A.; Paterno, J.J.; Blasiak, J.; Salminen, A.; Kaarniranta, K. Inflammation and its role in age-related macular degeneration. Cell. Mol. Life Sci. 2015, 73, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Stanhill, A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta 2014, 1843, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Lilienbaum, A. Relationship between the proteasomal system and autophagy. Int. J. Biochem. Mol. Biol. 2013, 31, 1–26. [Google Scholar]
- Kaarniranta, K.; Salminen, A.; Eskelinen, E.L.; Kopitz, J. Heat shock proteins as gatekeepers of proteolytic pathways–Implications for age-related macular degeneration (AMD). Ageing Res. Rev. 2009, 8, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Nowak, J.Z. Oxidative stress, polyunsaturated fatty acids-derived oxidation products and bisretinoids as potential inducers of CNS diseases; focus on age-related macular degeneration. Pharmacol. Rep. 2013, 65, 288–304. [Google Scholar] [CrossRef]
- Squier, T.C. Oxidative stress and protein aggregation during biological aging. Exp. Gerontol. 2001, 36, 1539–1550. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Pankiv, S.; Øvervatn, A.; Brech, A.; Johansen, T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009, 452, 181–197. [Google Scholar] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [PubMed]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 15, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Rhone, M.; Basu, A. Phytochemicals and age-related eye diseases. Nutr. Rev. 2008, 66, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.Y.; Lin, C.H.; Fang, J.Y. Natural compounds and aging: Between autophagy and inflammasome. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Singh, S. Antioxidants as a preventative therapeutic option for age related neurodegenerative diseases. Ther. Targets Neurol. Dis. 2016. [Google Scholar] [CrossRef]
- Spagnuolo, C.; Napolitano, M.; Tedesco, I.; Moccia, S.; Milito, A.; Russo, G.L. Neuroprotective Role of Natural Polyphenols. Available online: http://benthamscience.com/journals/current-topics-in-medicinal-chemistry/article/139246/ (accessed on 20 February 2015).
- Petrovski, G.; Gurusamy, N.; Das, D.K. Resveratrol in cardiovascular health and disease. Ann. N. Y. Acad. Sci. 2011, 1215, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Koskela, A.; Reinisalo, M.; Hyttinen, J.M.; Kaarniranta, K.; Karjalainen, R.O. Pinosylvin-mediated protection against oxidative stress in human retinal pigment epithelial cells. Mol. Vis. 2014, 20, 760–769. [Google Scholar] [PubMed]
- Reinisalo, M.; Kårlund, A.; Koskela, A.; Kaarniranta, K.; Karjalainen, R.O. Polyphenol stilbenes: Molecular mechanisms of defence against oxidative stress and aging-related diseases. Oxid. Med. Cell. Longev. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Wang, X.J.; Zhao, F.; Villeneuve, N.F.; Wu, T.; Jiang, T.; Sun, Z.; White, E.; Zhang, D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010, 30, 3275–3285. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Peyton, K.J.; Shebib, A.R.; Wang, H.; Korthuis, R.J.; Durante, W. Activation of AMPK stimulates heme oxygenase-1 gene expression and human endothelial cell survival. Am. J. Physiol. Heart Circ. Physiol. 2011, 300, H84–H93. [Google Scholar] [CrossRef] [PubMed]
- Richer, S.; Patel, S.; Sockanathan, S.; Ulanski, L.J., 2nd; Miller, L.; Pdella, C. Resveratrol based oral nutritional supplement produces long-term beneficial effects on structure and visual function in human patients. Nutrients 2014, 6, 4404–4420. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Huang, C.H.; Li, H.J.; Hsiao, C.Y.; Su, C.C.; Lee, P.L.; Hung, C.F. Protective effects of resveratrol against UVA damage in ARPE-19 cells. Int. J. Mol. Sci. 2015, 16, 5789–5802. [Google Scholar] [CrossRef] [PubMed]
- Nagineni, C.N.; Raju, R.; Nagineni, K.K.; Kommineni, V.K.; Cherukuri, A.; Kutty, R.K.; Hooks, J.J.; Detrick, B. Resveratrol suppresses expression of VEGF by human retinal retinal pigment epithelial cells: Potential nutraceutical for age-related macular degeneration. Aging Dis. 2014, 5, 88–100. [Google Scholar] [PubMed]
- Ivanescu, A.A.; Fernández-Robredo, P.; Heras-Mulero, H.; Sádaba-Echarri, L.M.; García-García, L.; Fernández-García, V.; Moreno-Orduna, M.; Redondo-Exposito, A.; Recalde, S.; García-Layana, A. Modifying choroidal neovascularization development with a nutritional supplement in mice. Nutrients 2015, 7, 5423–5442. [Google Scholar] [CrossRef] [PubMed]
- Johansson, I.; Monsen, V.T.; Pettersen, K.; Mildenberger, J.; Misund, K.; Kaarniranta, K.; Schønberg, S.; Bjørkøy, G. The marine n-3 PUFA DHA evokes cytoprotection against oxidative stress and protein misfolding by inducing autophagy and NFE2L2 in human retinal pigment epithelial cells. Autophagy 2015, 11, 1636–1651. [Google Scholar] [CrossRef] [PubMed]
- Jing, K.; Song, K.S.; Shin, S.; Kim, N.; Jeong, S.; Oh, H.R.; Park, J.H.; Seo, K.S.; Heo, J.Y.; Han, J.Y.; et al. Docosahexaenoic acid induces autophagy through p52/AMPK/mTOR signaling and promotes apoptosis in human cancer cell harboring wild-type p52. Autophagy 2011, 7, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Age-Related Eye Disease Study Group. A Randomized, placebo-controlled clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report No. 8. Arch. Ophthalmol. 2001, 119, 1417–1436. [Google Scholar]
- Chew, E.Y.; Clemons, T.E.; SanGiovanni, J.P.; Danis, R.; Ferris, F.L., 3rd; Elman, M.; Antoszyk, A.; Ruby, A.; Orth, D.; Bressler, S.; et al. Age-Related Eye Disease Study 2 Research Group. Lutein + zeaxanthin and omega-3 fatty acids for age-related macular degeneration: The Age-Related Eye Disease Study 2 (AREDS2) randomized clinical trial. JAMA 2013, 309, 2005–2015. [Google Scholar]
- Hansen, M.B.; Nielsen, S.E.; Berg, K. Re-examination and further development of a precise and rapid method for measuring cell growth/cell kill. J. Immunol. Methods 1989, 119, 203–210. [Google Scholar] [CrossRef]
- Viiri, J.; Amadio, M.; Marchesi, N.; Hyttinen, J.M.; Kivinen, N.; Sironen, R.; Rilla, K.; Akhtar, S.; Provenzani, A.; D’Agostino, V.G.; et al. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Lan, D.; Wang, W.; Zhuang, J.; Zhao, Z. Proteasome inhibitor-induced autophagy in PC12 cells overexpressing A53T mutant α-synuclein. Mol. Med. Rep. 2015, 11, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Valapala, M.; Wilson, C.; Hose, S.; Bhutto, I.A.; Grebe, R.; Dong, A.; Greenbaum, S.; Gu, L.; Sengupta, S.; Cano, M.; et al. Lysosomal-mediated waste clearance in retinal pigment epithelial cells is regulated by CRBA1/βA3/A1-crystallin cia V-ATPase-MTORC1 signalling. Autophagy 2014, 10, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.K.; Chen, S.D.; Chuang, Y.C.; Lin, H.Y.; Huang, C.R.; Chuang, J.H.; Wang, P.W.; Huang, S.T.; Tiao, M.M.; Chen, J.B.; et al. Resveratrol partially prevents rotenone-induced neurotoxicity in dopaminergic SH-SY5Y cells through induction of heme oxygenase-1 dependent autophagy. Int. J. Mol. Sci. 2014, 15, 1625–1646. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy, 3rd ed. Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Cano, M.; Handa, J.T. p62 provides dual cytoprotection against oxidative stress in the retinal pigment epithelium. Biochim. Biophys. Acta 2014, 1843, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.S.; Kim, Y.; Jung, J.Y.; Yang, S.H.; Lee, T.R.; Shin, D.W. Resveratrol induces autophagy through death-associated protein kinase 1 (DAPK1) in human dermal fibroblasts under normal culture conditions. Exp. Dermatol. 2013, 22, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, J.; Yu, X.; Shi, L.; Du, W.; Hu, L.; Liu, C.; Cao, Y. SIRT1 regulates accumulation of oxidized LDL in HUVEC via the autophagy-lysosomal pathway. Prostaglandins Other Lipid Mediat. 2015, 122, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Lamark, T.; Sjøttem, E.; Larsen, K.B.; Awuh, J.A.; Øvervatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 16, 22576–22591. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koskela, A.; Reinisalo, M.; Petrovski, G.; Sinha, D.; Olmiere, C.; Karjalainen, R.; Kaarniranta, K. Nutraceutical with Resveratrol and Omega-3 Fatty Acids Induces Autophagy in ARPE-19 Cells. Nutrients 2016, 8, 284. https://doi.org/10.3390/nu8050284
Koskela A, Reinisalo M, Petrovski G, Sinha D, Olmiere C, Karjalainen R, Kaarniranta K. Nutraceutical with Resveratrol and Omega-3 Fatty Acids Induces Autophagy in ARPE-19 Cells. Nutrients. 2016; 8(5):284. https://doi.org/10.3390/nu8050284
Chicago/Turabian StyleKoskela, Ali, Mika Reinisalo, Goran Petrovski, Debasish Sinha, Céline Olmiere, Reijo Karjalainen, and Kai Kaarniranta. 2016. "Nutraceutical with Resveratrol and Omega-3 Fatty Acids Induces Autophagy in ARPE-19 Cells" Nutrients 8, no. 5: 284. https://doi.org/10.3390/nu8050284




