1. Introduction
Coronary heart disease (CHD) and ischemic stroke (IS) remain the major causes of morbidity and mortality worldwide [
1,
2]. The pathological basis of both diseases is atherosclerosis, a progressive inflammatory disorder and, therefore, CHD and IS may share common pathogenesis, as well as many risk factors [
2,
3]. Previous genome-wide association studies (GWASs) have identified many genes and loci in the predisposition to CHD [
4] or IS [
5] in different populations. Furthermore, some genetic variants originally shown to influence the risk of CHD were also subsequently found to be associated with IS [
6,
7].
The MAP-kinase activating death domain (MADD) and folate hydrolase 1 (FOLH1) gene (
MADD-FOLH1) is located on chromosome 11p. The MADD protein can modulate tumor necrosis factor-alpha and propagate the apoptotic signals [
8]. Folate is an essential nutrient that supports nucleotide synthesis and biological methylation reactions. Diminished folate status results in chromosome breakage and is associated with several diseases, including cardiovascular disease [
9,
10]. Genetic variants involved in folate metabolism may modify the effects of lifestyle (smoking and drinking) on plasma homocysteine concentrations—a risk factor for cardiovascular disease [
11]. A novel single nucleotide polymorphism (SNP) of rs7395662 in or near
MADD-FOLH1 has been found association with serum lipid levels [
12,
13] and the risk of CHD [
14] in a previous GWAS and two replication studies. Exactly as patterns of linkage disequilibrium (LD) vary between populations, genetic anomalies of ion channels or specific endothelial regulators may underlie coronary microvascular disease [
15,
16], the results are complicated and inconsistent across different populations [
17,
18]. In addition, there are still many variants have not been explored in the
MADD-FOLH1 cluster. Therefore, the purpose of the present study was to detect the association of six SNPs (rs7395662, rs326214, rs326217, rs1051006, rs3736101, and rs7120118) in or near
MADD-FOLH1 and their haplotypes with serum lipid traits and the risk of CHD and IS in a Chinese Han populations.
3. Results
3.1. General Characteristics of the Subjects
The general characteristics of the patients and healthy controls are summarized in
Table 1. The values of BMI, pulse pressure, and TG were higher but diastolic blood pressure, TC, HDL-C, ApoA1, the percentages of subjects who consumed alcohol, and the ratio of ApoA1 to ApoB were lower in CHD patients than in controls (
p < 0.001 for all). The values of BMI, systolic blood pressure, pulse pressure, and TG were higher but TC, HDL-C, ApoA1, the percentages of subjects who consumed alcohol, and the ratio of ApoA1 to ApoB were lower in IS patients than in controls (
p < 0.001 for all).
3.2. Genotypic and Allelic Frequencies in Controls and Patients
The genotypic and allelic frequencies of the 6
MADD-FOLH1 SNPs are presented in
Table 2. The genotype distribution of the 5 SNPs (not including rs1051006 in IS patients,
p = 0.026) was concordant with the Hardy–Weinberg equilibrium in patients and controls (
p > 0.05 for all). The genotypic and allelic frequencies of the rs7395662, but not the other five SNPs, were different between controls and patients (CHD and IS,
p < 0.01 for all), the rs7395662G allele and rs7395662GG genotype frequencies were lower in CHD (G, 36.6%; GG, 12.7%) or IS (G, 36.3%; GG, 14.4%) patients than in control subjects (G, 42.6%; GG, 18.8%;
p < 0.01 for all).
3.3. Genotypes of the Six MADD-FOLH1 SNPs and the Risk of CHD and IS
As shown in
Table 3, the genotypes of the rs7395662, but not the other five SNPs, were associated with the risk of CHD after the Bonferroni correction (a value of
p < 0.008 was considered statistically significant) in different genetic models: co-dominant model: GG
vs. AA (OR = 0.78, 95% CI = 0.66–0.92,
p = 0.0068); recessive model: GG
vs. AA/GA (OR = 0.63, 95% CI = 0.46–0.86,
p = 0.0038); and log-additive model: G
vs. A (OR = 0.78, 95% CI = 0.66–0.92,
p = 0.003).
The genotypes of the rs7395662 SNP were also associated with the risk of IS in different genetic models: dominant model: GA/GG vs. AA (OR = 0.70, 95% CI = 0.55–0.89, p = 0.0039); and log-additive model: G vs. A (OR = 0.78, 95% CI = 0.66–0.91, p = 0.0024).
3.4. Haplotypes and the Risk of CHD and IS
There was strong LD among the rs1051006, rs326214, rs326217, rs3736101, and rs7120118 SNPs in controls and patients (
D' = 0.8946–0.9983) but weak LD between the rs7395662 and the other five SNPs (
D' = 0.1127–0.3275). Thus, haplotype analyses among the five SNPs and the associations of their haplotypes and the risk of CHD and IS were performed. Six main haplotypes are shown in
Table 4. The haplotype of G-G-T-G-C (in the order of rs1051006, rs326214, rs326217, rs3736101 and rs7120118 SNPs) was associated with an increased risk for CHD (adjusted OR = 1.59, 95% CI = 1.06–2.38,
p = 0.026). The haplotype of G-A-T-G-T was associated with an increased risk for IS (adjusted OR = 1.95, 95% CI = 1.04–3.68,
p = 0.039).
3.5. Genotypes and Serum Lipid Levels
The association of the
MADD-FOLH1 SNPs and serum lipid levels in controls is presented in
Table 5. Serum TG and HDL-C levels were different among the three genotypes of the rs7395662 but not the other five SNPs (
p = 0.005 and 0.001; respectively), the subjects with rs7395662GG genotype had higher TG and lower HDL-C levels than the subjects with rs7395662AA and rs7395662AG genotypes. There was no difference in serum TC, LDL-C, ApoA1, ApoB levels, and the ApoA1/ApoB ratio among the three genotypes of the SNP.
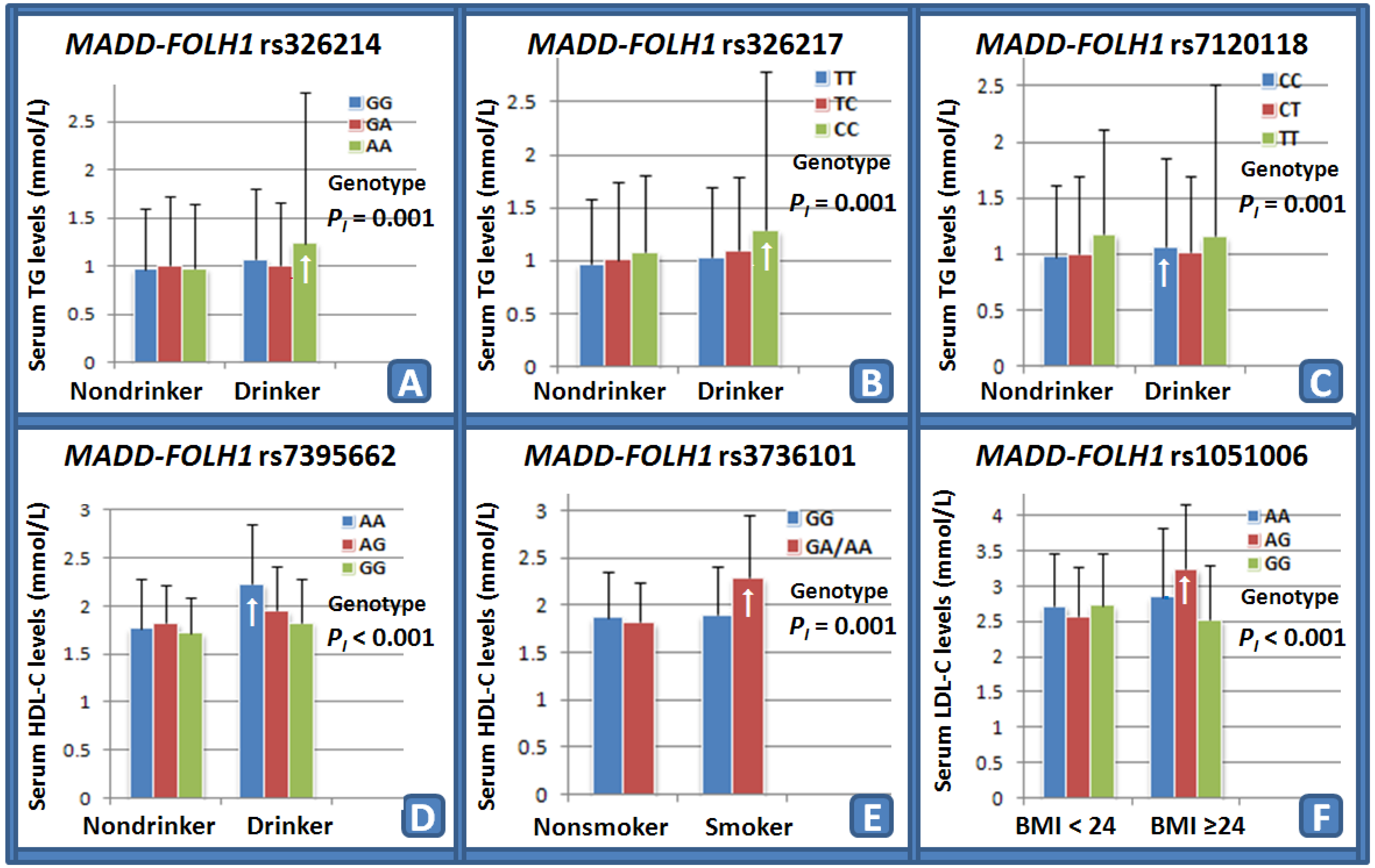
3.6. Interactions of the MADD-FOLH1 SNPs and Drinking, Smoking, BMI, Age, and Sex on Serum Lipid Levels and the Risk of CHD and IS
The interactions of the
MADD-FOLH1 SNPs and drinking, smoking, BMI, age, and sex on serum lipid levels and the risk of CHD and IS are shown in
Table 6. Several SNPs interacted with alcohol consumption to influence serum TG (rs326214,
Figure 1A; rs326217,
Figure 1B; and rs7120118,
Figure 1C) and HDL-C (rs7395662,
Figure 1D) levels. The SNP of rs3736101 interacted with cigarette smoking to modify serum HDL-C (
Figure 1E) levels. The SNP of rs1051006 interacted with BMI ≥ 24 kg/m
2 to modulate serum LDL-C (
Figure 1F) levels.
3.7. Interactions of the Genotypes and Drinking, Smoking, and BMI on the Risk of CHD and IS
The rs3736101GA/AA genotypes interacted with alcohol consumption to decrease the risk of CHD (OR = 0.33, 95% CI = 0.12–0.89, p = 0.0021). No interactions of the genotypes and smoking and BMI on the risk of CHD and IS was detected in our study population.
3.8. Interactions of the Haplotypes and Drinking on the Risk of CHD
The interactions of several haplotypes and alcohol consumption on the risk of CHD were noted in this study. As compared with the A-G-T-G-C haplotype in non-drinkers, the haplotypes of G-G-T-A-C (OR = 1.69, 95% CI = 1.02–2.81) and G-A-T-G-T (OR = 3.61, 95% CI = 1.29–10.15) in non-drinkers were associated with an increased risk for CHD, whereas the haplotypes of A-G-T-G-C (OR = 0.34, 95% CI = 0.22–0.51), G-A-C-G-T (OR = 0.39, 95% CI = 0.25–0.59), G-G-T-A-C (OR = 0.11, 95% CI = 0.04–0.30) and G-A-C-G-C (OR = 0.17, 95% CI = 0.07–0.40) in drinkers were associated with a decreased risk for CHD.
For the drinkers, as compared with the A-G-T-G-C haplotype, the haplotype of G-G-T-G-C (OR = 2.38, 95% CI = 1.25–4.56) was associated with an increased risk for CHD, whereas the haplotype of G-G-T-A-C (OR = 0.32, 95% CI = 0.12–0.87) was associated with a decreased risk for CHD.
As compared with the same haplotype in non-drinkers, the haplotypes of A-G-T-G-C (OR = 0.34, 95% CI = 0.22-0.51), G-A-C-G-T (OR = 0.36, 95% CI = 0.24–0.54), G-G-T-A-C (OR = 0.06, 95% CI = 0.02–0.19), G-A-C-G-C (OR = 0.18, 95% CI = 0.06–0.53), and G-A-T-G-T (OR = 0.07, 95% CI = 0.01–0.34) in drinkers were associated with a decreased risk for CHD.
3.9. Interactions of the Haplotypes and Drinking on the Risk of IS
The interactions of several haplotypes and alcohol consumption on the risk of IS were also noted in this study. As compared with the A-G-T-G-C haplotype in non-drinkers, the haplotype of G-A-T-G-T (OR = 4.05, 95% CI = 1.46–11.27) in non-drinkers was associated with an increased risk for IS, whereas the haplotypes of A-G-T-G-C (OR = 0.33, 95% CI = 0.22–0.50), G-A-C-G-T (OR = 0.39, 95% CI = 0.26–0.58), G-G-T-A-C (OR = 0.41, 95% CI = 0.22–0.79), G-A-C-G-C (OR = 0.25, 95% CI = 0.12–0.52), and G-A-T-G-T (OR = 0.28, 95% CI = 0.10–0.79) in drinkers were associated with a decreased risk for IS.
For the drinkers, as compared with the A-G-T-G-C haplotype, no significant association was detected between the haplotypes and IS.
As compared with the same haplotype in non-drinkers, the haplotypes of A-G-T-G-C (OR = 0.33, 95% CI = 0.22–0.50), G-A-C-G-T (OR = 0.48, 95% CI = 0.32–0.72), G-G-T-A-C (OR = 0.30, 95% CI = 0.13–0.67), G-A-C-G-C (OR = 0.37, 95% CI = 0.14–0.98), and G-A-T-G-T (OR = 0.07, 95% CI = 0.02–0.29) in drinkers were associated with a decreased risk for IS.
4. Discussion
In the present study, we showed that the genotypic and allelic frequencies of the rs7395662 SNP were different between controls and CHD or IS patients, and that the rs7395662 genotypes or alleles were associated with the risk of CHD and IS in different genetic models. The SNPs of rs1051006, rs326214, rs326217, rs3736101, and rs7120118 were strong LD in controls and patients. Six main haplotypes of the five SNPs were detected. The haplotype of G-G-T-G-C was associated with an increased risk for CHD, whereas the haplotype of G-A-T-G-T was associated with an increased risk for IS. The subjects with rs7395662GG genotype in controls had higher TG and lower HDL-C levels than the subjects with rs7395662AA and rs7395662AG genotypes. Several SNPs interacted with alcohol consumption to influence serum TG (rs326214, rs326217, and rs7120118) and HDL-C (rs7395662) levels. Two SNPs interacted with cigarette smoking to modify serum TG (rs7395662) and HDL-C (rs3736101) levels. The SNP of rs1051006 interacted with BMI ≥ 24 kg/m2 to modulate serum LDL-C levels. The rs3736101GA/AA genotypes interacted with alcohol consumption to decrease the risk of CHD. The interactions of several haplotypes and alcohol consumption on the risk of CHD and IS were also observed. To the best of our knowledge, this is the first report to evaluate the interaction between the six MADD-FOLH1 SNPs and their haplotypes and several environmental factors on serum lipid levels and the risk of CHD and IS.
We showed that the genotypic and allelic frequencies of the rs7395662, but not the other five SNPs were different between controls and CHD or IS patients, the patients with CHD (36.6%,
p = 0.003) or IS (36.3%,
p = 0.002) had lower frequencies of rs7395662G allele than the controls (42.6%). The genotypes of the rs7395662 SNP were also associated with the risk of CHD and IS after the Bonferroni correction in different genetic models. In a previous GWAS, the rs7395662G allele frequency was 61% in a total of 17,797–22,562 persons, aged 18-104 years and from geographic regions spanning from the Nordic countries to Southern Europe [
12]. In two recent studies, we and other researchers showed that the allelic frequency of rs7395662G was 44.9% in the healthy Mulao population [
13], 43.7% in Han Chinese [
13], 47.3% in CHD cases [
14], and 51.9% in non-CHD controls [
14]. The data in the International HapMap Project’s database have suggested that the frequency of rs7395662G allele was 62.7% in European, 58.9% in Han Chinese in Beijing, 48.9% in Japanese, and 44.2% in Sub-Saharan African [
13]. As compared with the other populations, we found that the frequency of rs7395662G allele in our study populations was lower than that in Han Chinese from Beijing, which may be caused by different sample sizes and Han Chinese from Beijing and Guangxi are different parts of Han. These results suggest that the prevalence of the rs7395662G allele variation may have racial/ethnic- and sex-specificity. The prevalence of the rs7395662G allele is higher in European than in Chinese. These findings may also partly explain why the prevalence of cardiovascular disease is different between European and Chinese.
The association of the rs1051006, rs326214, rs326217, rs3736101, and rs7120118 SNPs and the risk of CHD and IS has not been reported previously. In the present study, we showed that there were no associations of the five SNPs and the risk of CHD and IS. However, there was high LD among the rs1051006, rs326214, rs326217, rs3736101, and rs7120118 SNPs in controls and patients. Haplotype analyses of the five SNPs showed that the haplotype of G-G-T-G-C was associated with an increased risk for CHD, whereas the haplotype of G-A-T-G-T was associated with an increased risk for IS. However, these findings still need to be confirmed in the other populations with larger sample sizes.
The results of the present study showed that serum TG and HDL-C levels were different among the three genotypes of the rs7395662, but not the other five SNPs, the subjects with rs7395662GG genotype had higher TG and lower HDL-C levels than the subjects with rs7395662AA and rs7395662AG genotypes. There was no difference in serum TC, LDL-C, ApoA1, ApoB levels, and the ApoA1/ApoB ratio among the three genotypes of the SNP. The association between the rs7395662 SNP and serum or plasma lipid levels has been investigated in several previous studies [
12,
13,
14]. In a previous GWAS, Aulchenko
et al. [
12] showed significant association between the rs7395662 SNP and HDL-C levels in 16 population-based cohorts (
p = 6 × 10
−11). The coded allele (the allele for which effect was estimated) was the rs7395662G allele. In a previous comparative study of two ethnic groups, we found that there may be a sex-specific association of the rs7395662 SNP and serum lipid concentrations in the Mulao and Han populations [
13]. A recent study also showed that the rs7395662A allele was significantly associated with decreased HDL-C levels (
β = −0.024,
p = 0.007) in 1069 healthy control subjects [
14]. However, the association of the rs1051006, rs326214, rs326217, rs3736101, and rs7120118 SNPs and serum or plasma lipid phenotypes is not well-known. In a previous study, Akiyama
et al. [
8] investigated functional genes at homologous loci identified using human lipid GWASs that responded to a high-fat, high-cholesterol diet intervention in an animal model. They showed that the gene of
MADD rs7120118 SNP was potential target of lipid association (HDL-C).
MADD encodes the MADD protein, which interacts with tumor necrosis factor-alpha receptor 1 to activate mitogen-activated protein kinase and propagate apoptotic signals [
8]. In addition, G protein-coupled receptor kinase 5, which is also reported to interact with MADD [
45], has been demonstrated to exhibit a significant increase in its gene expression induced by high-fat, high-cholesterol diet intervention, supporting the potential involvement of both G protein-coupled receptor kinase 5 and MADD in lipid metabolism [
8].
The interactions of the MADD-FOLH1 SNPs and their haplotypes and some environmental factors on serum lipid levels and the risk of CHD and IS are not known. In the present study, we firstly showed that several MADD-FOLH1 SNPs interacted with alcohol consumption to influence serum TG (rs326214, rs326217, and rs7120118) and HDL-C (rs7395662) levels. The SNP of rs3736101 interacted with cigarette smoking to modify serum HDL-C levels. The SNP of rs1051006 interacted with BMI ≥ 24 kg/m2 to modulate serum LDL-C levels. The haplotypes of G-G-T-A-C and G-A-T-G-T in non-drinkers were associated with an increased risk for CHD, whereas the haplotypes of A-G-T-G-C, G-A-C-G-T, G-G-T-A-C, and G-A-C-G-C in drinkers were associated with a decreased risk for CHD. The haplotype of G-A-T-G-T in non-drinkers was associated with an increased risk for IS, whereas the haplotypes of A-G-T-G-C, G-A-C-G-T, G-G-T-A-C, G-A-C-G-C, and G-A-T-G-T in drinkers were associated with a decreased risk for IS. It is well known that heavy alcohol intake and cigarette smoking have a disadvantageous effect on lipid profiles. In the current study, however, we showed that the rs3736101 AG/GG genotypes interacted with cigarette smoking to increase serum HDL-C levels. The reason for this contradictory finding is not clear. In our study populations, most smokers also have drinking habits. Thus, these interactions still need to be determined.
There were several potential limitations in this study. Firstly, the sample size was relatively small compared to many GWASs and replication studies. Therefore, further studies with larger sample sizes are needed to confirm our results. Secondly, there were significant differences in the general characteristics between the control and patient groups. Although age, gender, BMI, cigarette smoking, and alcohol consumption have been adjusted for the statistical analysis, we could not completely eliminate the potential effects of these factors on serum lipid levels and the risk of CHD and IS. Thirdly, the association of the six SNPs and serum lipid levels in the CHD and IS groups was not analyzed because of the interference of lipid-lowering drugs. Finally, it is well known that both CHD and IS are the complex multifactorial disorders that are believed to result from an interaction between the genetic background of an individual and various environmental factors. Although we have detected the association between six MADD-FOLH1 SNPs and their haplotypes and the risk of CHD and IS, there are still many unmeasured environmental and genetic factors and their interactions.