Effect of Green Tea Extract on Systemic Metabolic Homeostasis in Diet-Induced Obese Mice Determined via RNA-Seq Transcriptome Profiles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Measurement of Energy Expenditures
2.3. Analysis of Plasma and Hepatic Lipids
2.4. Levels of Plasma Aspartate Aminotransferase (AST) and Alanine Aminotransferase (ALT)
2.5. Plasma Glucose and Insulin Resistance Index
2.6. Plasma Hormones, Adipokines, and Proinflammatory Cytokines
2.7. Hepatic Enzyme Activities and Glycogen Concentration
2.8. Histological Analysis of eWAT and the Liver
2.9. RNA Preparation, Library Preparation, and RNA-Seq
2.10. Preprocessing of the RNA-Seq Data
2.11. Differential Transcriptome and Functional Analysis
2.12. Molecular Pathway and Function Analysis
2.13. Statistical Analysis
3. Results
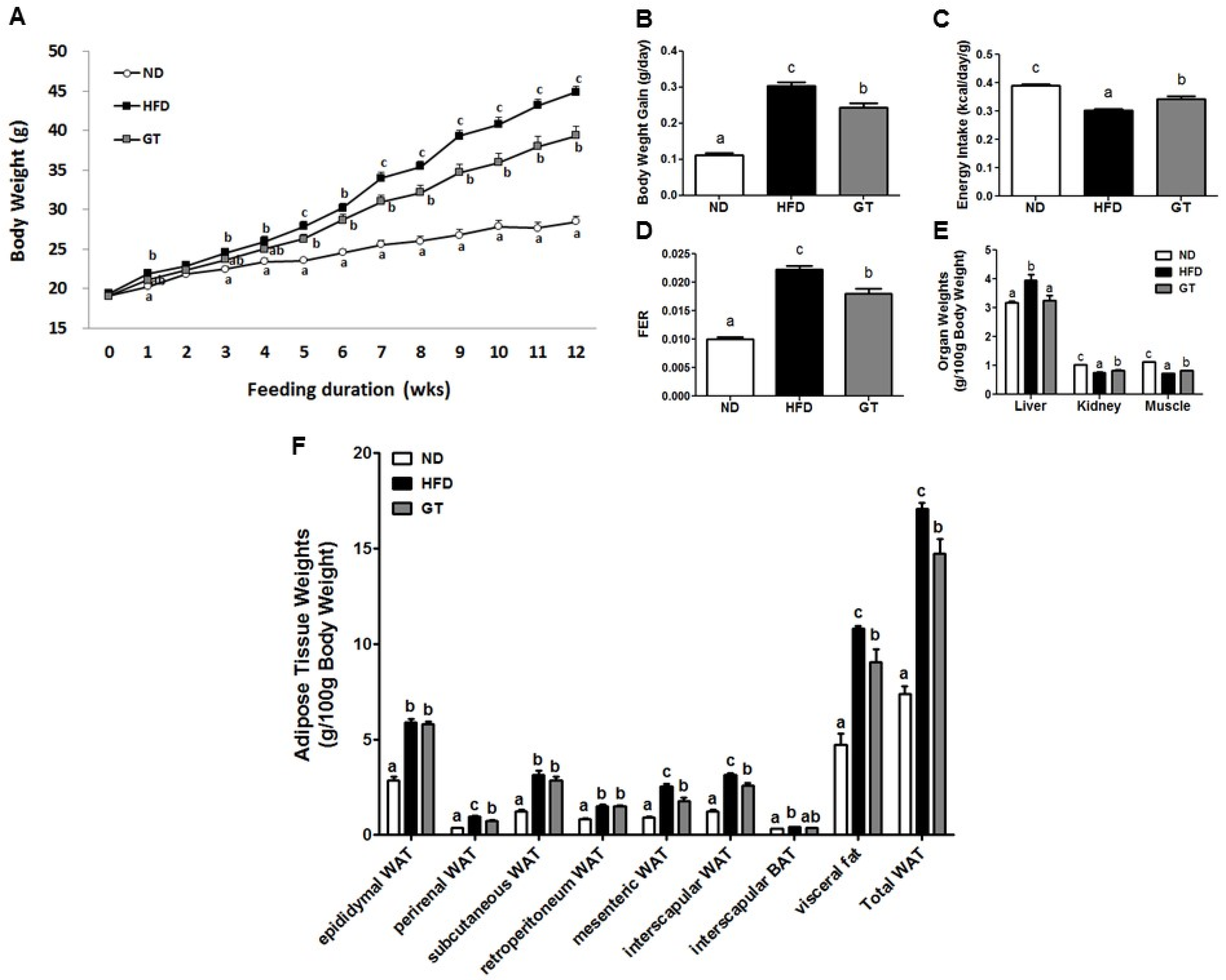
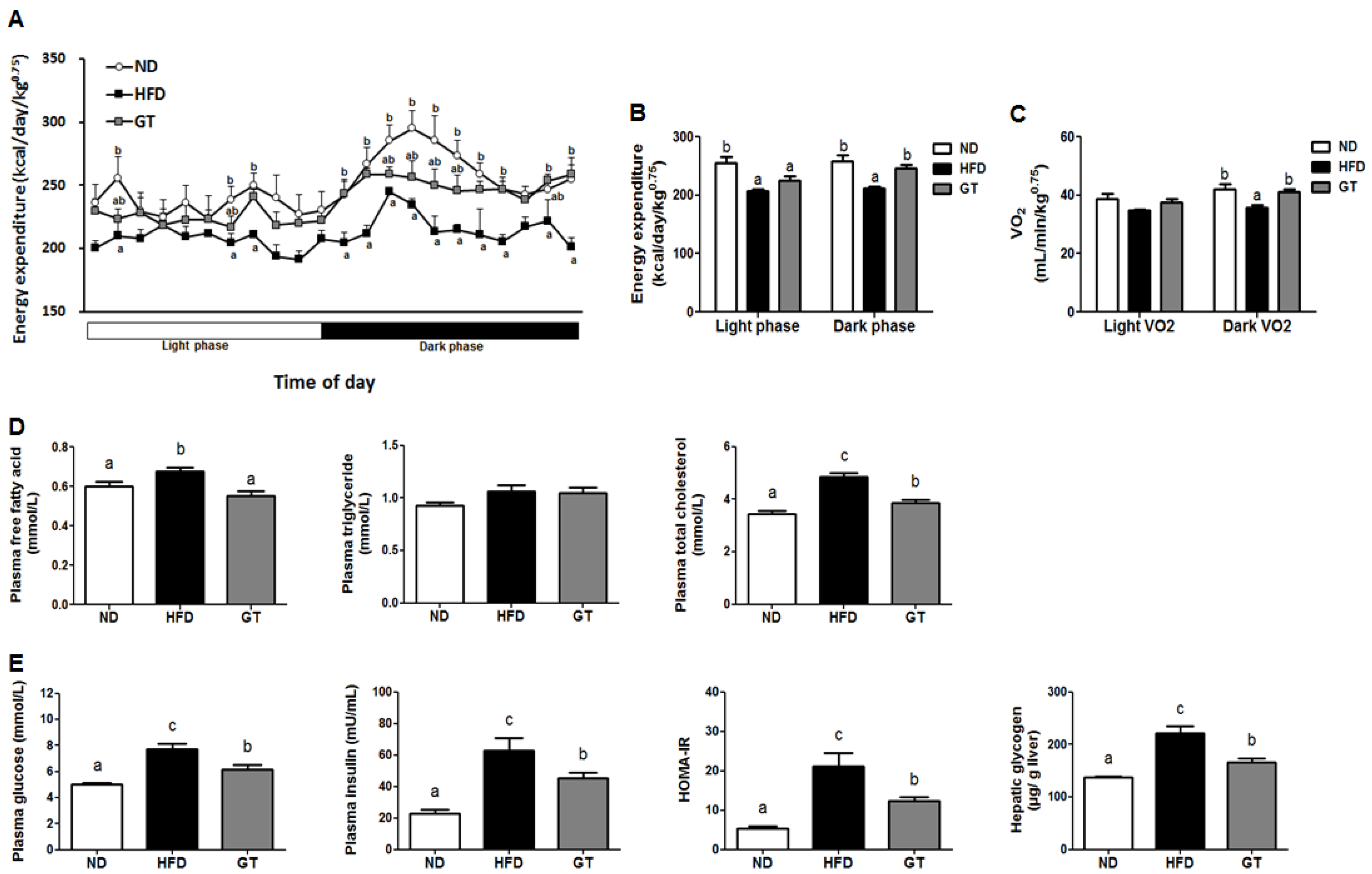
3.1. Supplementation with GT Ethanol Extract Reduced Body Weight Gain and Body Fat Mass with Enhanced Energy Expenditure and Plasma Lipid and Glucose Profiles in Diet-Induce Obesity (DIO) Mice
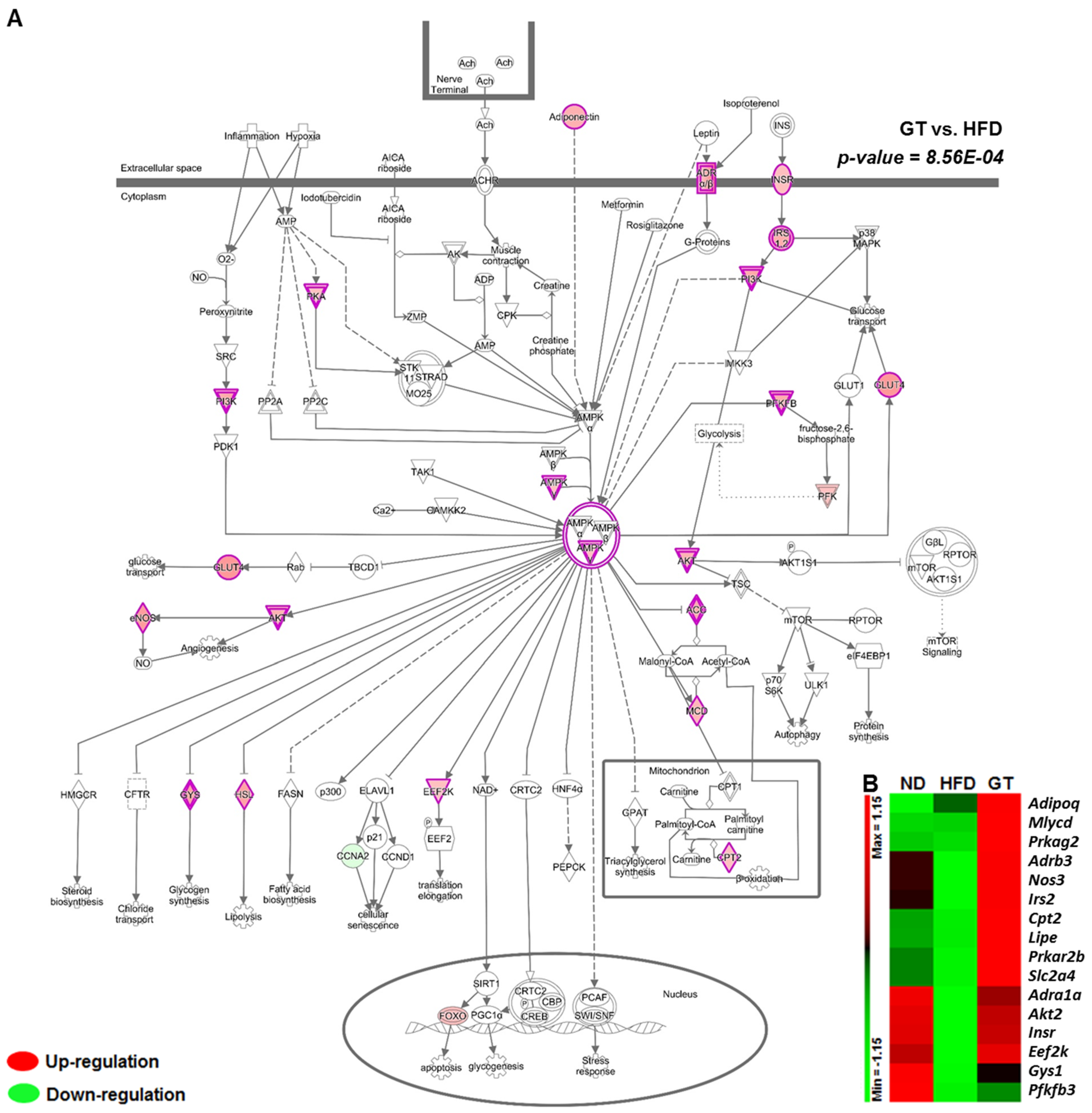
3.2. GT Ethanol Extract Attenuated the Level of Plasma Adipokines in DIO Mice and Modulated Transcriptional Responses to a HFD in eWAT
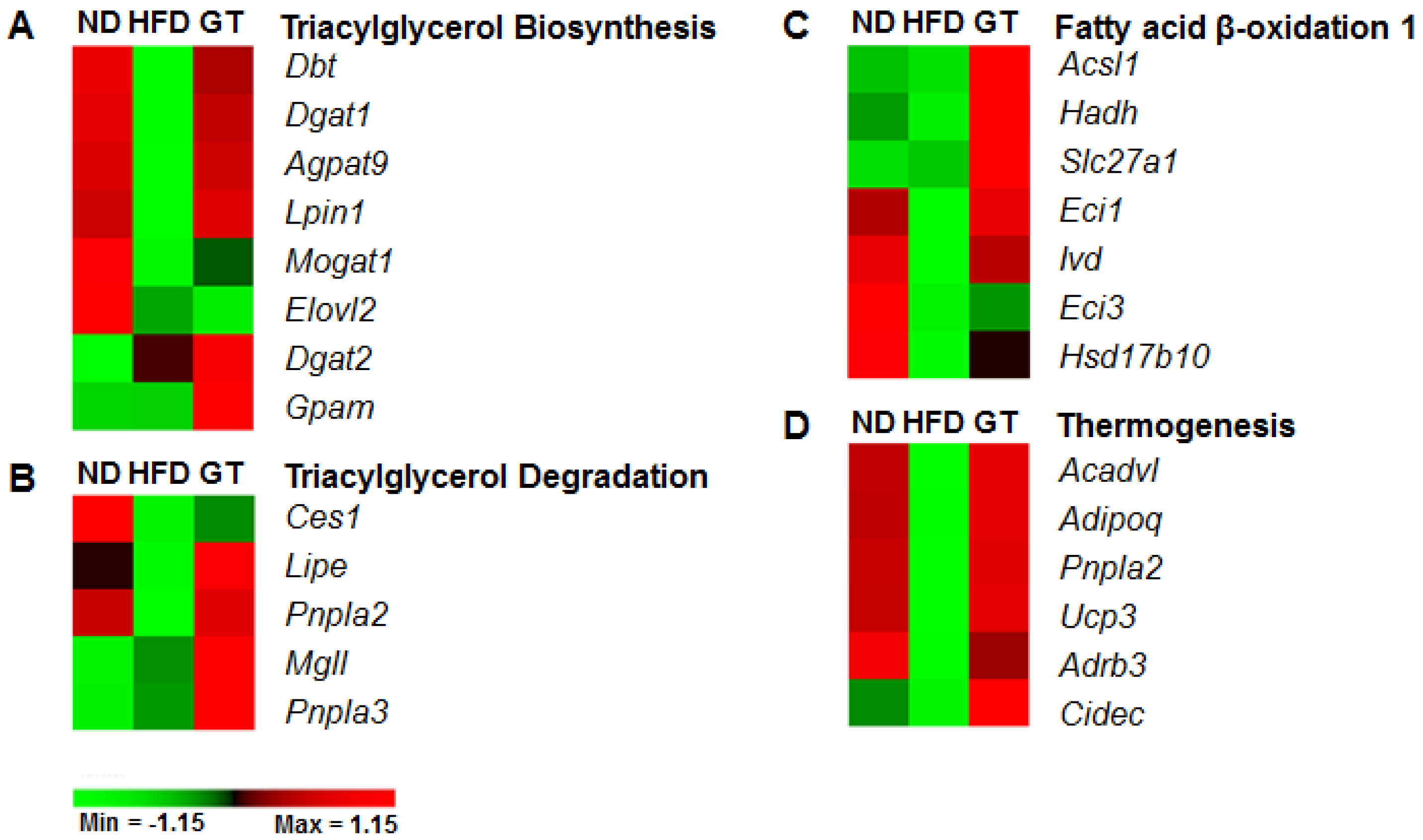
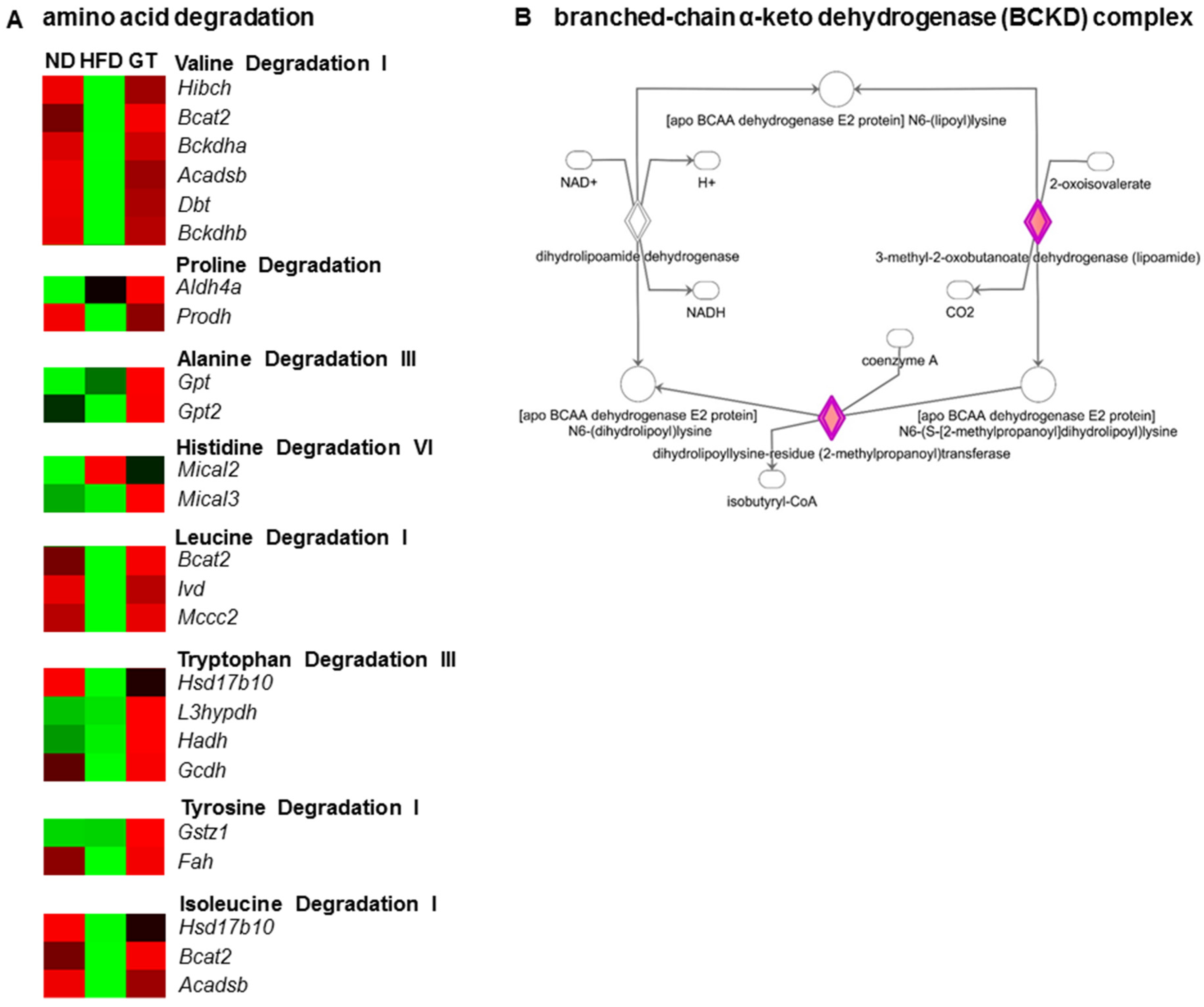
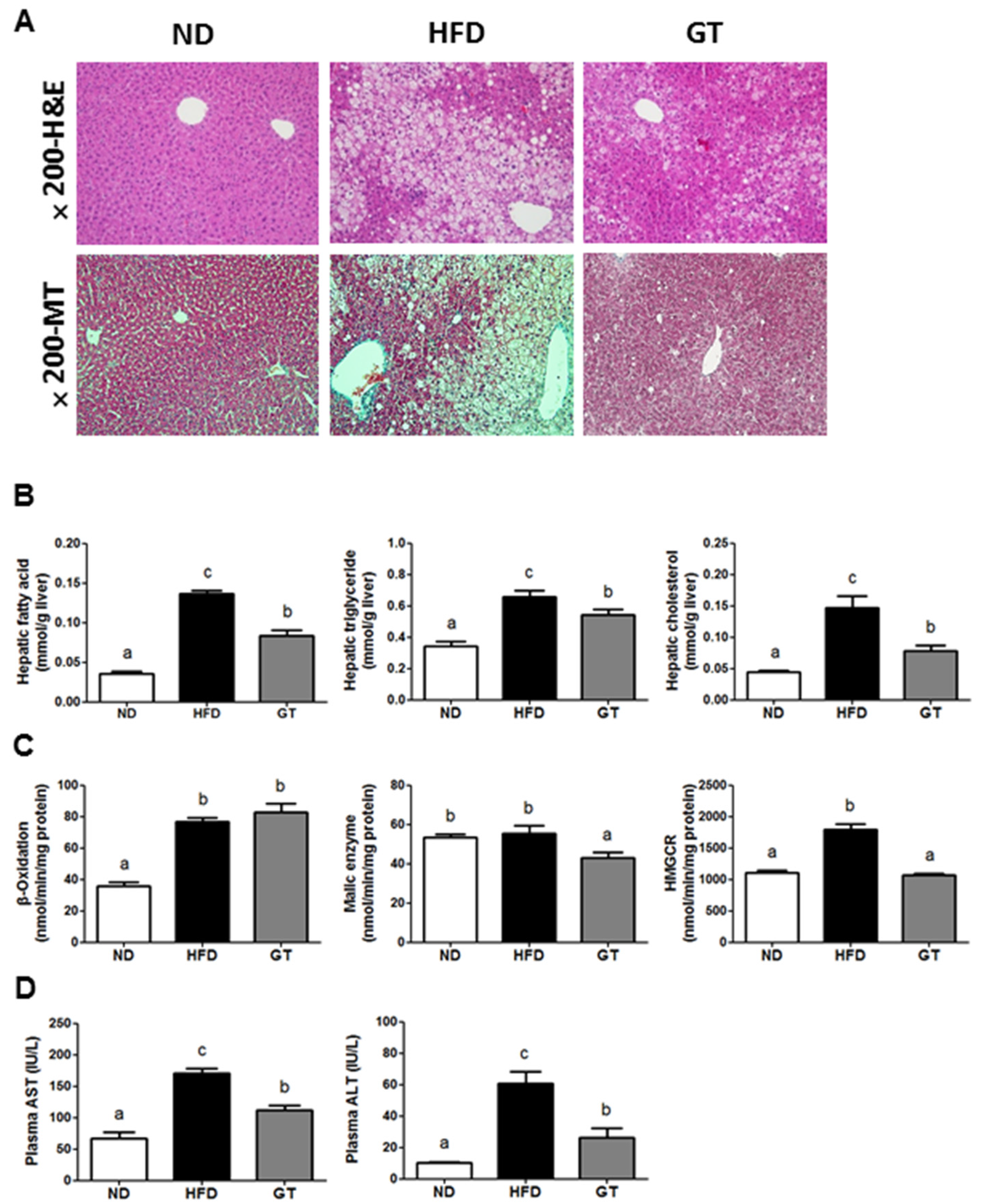
3.3. GT Extract Ameliorated Hepatic Steatosis via the Metabolic and Transcriptional Responses in the Livers of DIO Mice
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ALT | alanine aminotransferase |
| AMPK | AMP-activated protein kinase |
| AST | aspartate aminotransferase |
| BCAAs | branched chain amino acids |
| BCKD | branched-chain α-keto dehydrogenase |
| DEGs | differentially expressed genes |
| DIO | diet-induced obesity |
| EC | epicatechin |
| ECG | epicatechin gallate |
| EGC | epigallocatechin |
| EGCG | epigallocatechin gallate |
| eWAT | epididymal white adipose tissue |
| FER | food efficiency ratio |
| GT | green tea |
| HFD | high-fat diet |
| HMGCR | HMG-CoA reductase |
| HOMA-IR | homeostasis model assessment for insulin resistance |
| IFN-γ | interferon γ |
| IPA | ingenuity pathway analysis |
| MCP-1 | monocyte chemoattractant protein 1 |
| ND | normal diet |
| PAI-1 | plasminogen activator inhibitor 1 |
| TNF-α | tumor necrosis factor α |
References
- Kopelman, P.G. Obesity as a medical problem. Nature 2000, 404, 635–643. [Google Scholar] [PubMed]
- Gesta, S.; Tseng, Y.H.; Kahn, R.C. Developmental origin of fat: Tracking obesity to its source. Cell 2007, 131, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Barnes, G.T.; Yang, Q.; Tan, G.; Yang, D.; Chou, C.J.; Sole, J.; Nichols, A.; Ross, J.S.; Tartaglia, L.A.; et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Investig. 2003, 112, 1821–1830. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Flier, J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Hauner, H. Secretory factors from human adipose tissue and their functional role. Proc. Nutr. Soc. 2005, 64, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Lackey, D.E.; Lynch, C.J.; Olson, K.C.; Mostaedi, R.; Ali, M.; Smith, W.H.; Karpe, F.; Humphreys, S.; Bedinger, D.H.; Dunn, T.N.; et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E1175–E1187. [Google Scholar] [CrossRef] [PubMed]
- Herman, M.A.; She, P.; Peroni, O.D.; Lynch, C.J.; Kahn, B.B. Adipose Tissue Branched Chain Amino Acid (BCAA) Metabolism Modulates Circulating BCAA Levels. J. Biol. Chem. 2010, 285, 11348–11356. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.J.; Adams, S.H.; Christopher, J.; Lynch, S.H.A. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef] [PubMed]
- McCormack, S.E.; Shaham, O.; McCarthy, M.A.; Deik, A.A.; Wang, T.J.; Gerszten, R.E.; Clish, C.B.; Mootha, V.K.; Grinspoon, S.K.; Fleischman, A. Circulating Branched-chain Amino Acid Concentrations Are Associated with Obesity and Future Insulin Resistance in Children and Adolescents. Pediatr. Obes. 2013, 8, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Freudenberg, A.; Petzke, K.J.; Klaus, S. Dietary l-leucine and l-alanine supplementation have similar acute effects in the prevention of high-fat diet-induced obesity. Amino Acids 2013, 44, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Cavener, D.R. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007, 5, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Wolfram, S.; Wang, Y.; Thielecke, F. Anti-obesity effects of green tea: From bedside to bench. Mol. Nutr. Food Res. 2006, 50, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Bose, M.; Lambert, J.D.; Ju, J.; Reuhl, K.R.; Shapses, S.A.; Yang, C.S. The Major Green Tea Polyphenol, (−)-Epigallocatechin-3-Gallate, Inhibits Obesity, Metabolic Syndrome, and Fatty Liver Disease in High-Fat-Fed Mice. J. Nutr. 2008, 138, 1677–1683. [Google Scholar] [PubMed]
- Park, H.J.; DiNatale, D.A.; Chung, M.Y.; Park, Y.K.; Lee, J.Y.; Koo, S.I.; O’Connor, M.; Manautou, J.E.; Bruno, R.S. Green tea extract attenuates hepatic steatosis by decreasing adipose lipogenesis and enhancing hepatic antioxidant defenses in ob/ob mice. J. Nutr. Biochem. 2011, 22, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Forester, S.C.; Lambert, J.D. The role of antioxidant versus pro-oxidant effects of green tea polyphenols in cancer prevention. Mol. Nutr. Food Res. 2011, 55, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Betts, N.M.; Mulugeta, A.; Tong, C.; Newman, E.; Lyons, T.J. Green tea supplementation increases glutathione and plasma antioxidant capacity in adults with the metabolic syndrome. Nutr. Res. 2013, 33, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhou, R.; Wang, B.; Yu, X.; Yang, X.; Liu, K.; Mi, M. Effect of green tea consumption on blood pressure: A meta-analysis of 13 randomized controlled trials. Sci. Rep. 2014, 1, 6251. [Google Scholar] [CrossRef] [PubMed]
- Dulloo, A.G.; Duret, C.; Rohrer, D.; Girardier, L.; Mensi, N.; Fathi, M.; Chantre, P.; Vandermander, J. Efficacy of a green tea extract rich in catechin polyphenols and caffeine in increasing 24-h energy expenditure and fat oxidation in humans. Am. J. Clin. Nutr. 1999, 70, 1040–1045. [Google Scholar] [PubMed]
- Diepvens, K.; Westerterp, K.R.; Westerterp-Plantenga, M.S. Obesity and thermogenesis related to the consumption of caffeine, ephedrine, capsaicin, and green tea. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R77–R85. [Google Scholar] [CrossRef] [PubMed]
- Bérubé-Parent, S.; Pelletier, C.; Doré, J.; Tremblay, A. Effects of encapsulated green tea and Guarana extracts containing a mixture of epigallocatechin-3-gallate and caffeine on 24 h energy expenditure and fat oxidation in men. Br. J. Nutr. 2005, 94, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Rocha, A.; Bolin, A.P.; Cardoso, C.A.; Otton, R. Green tea extract activates AMPK and ameliorates white adipose tissue metabolic dysfunction induced by obesity. Eur. J. Nutr. 2015, 55, 2231–2244. [Google Scholar] [CrossRef] [PubMed]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. 2008, 22, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Sloane-Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [PubMed]
- Lazarow, P.B. Assay of peroxisomal β-oxidation of fatty acids. Methods Enzymol. 1981, 72, 315–319. [Google Scholar] [PubMed]
- Markwell, M.A.; McGroarty, E.J.; Bieber, L.L.; Tolbert, N.E. The Subcellular Distribution of Carnitine Acyltransferases in Mammalian Liver and Kidney A new peroxisomal enzyme. J. Biol. Chem. 1973, 248, 3426–3432. [Google Scholar] [PubMed]
- Ochoa, S. Malic dehydrogenase from pig heart: l-Malate + DPN+ ⇆ Oxalacetate + DPNH + H+. Methods Enzymol. 1955, 1, 735–739. [Google Scholar]
- Shapiro, D.J.; Nordstrom, J.L.; Mitschelen, J.J.; Rodwell, V.W.; Schimke, R.T. Micro assay for 3-hdyroxy-3-methylglutaryl-CoA reductase in rat liver and in L-cell fibroblasts. Biochim. Biophys. Acta 1974, 70, 369–377. [Google Scholar] [CrossRef]
- Seifter, S.; Dayton, S.; Novic, B.; Muntwyler, E. The estimation of glycogen with the anthrone reagent. Arch. Biochem. 1950, 25, 191–200. [Google Scholar] [PubMed]
- Do, G.M.; Jung, U.J.; Park, H.J.; Kwon, E.Y.; Jeon, S.M.; McGregor, R.A.; Choi, M.S. Resveratrol ameliorates diabetes-related metabolic changes via activation of AMP-activated protein kinase and its downstream targets in db/db mice. Mol. Nutr. Food Res. 2012, 56, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 8, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinf. 2013, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.C.; Zierath, J.R. AMP-activated protein kinase signaling in metabolic regulation. J. Clin. Investig. 2006, 116, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.T.; Park, I.J.; Shin, J.I.; Lee, Y.K.; Lee, S.K.; Baik, H.W.; Ha, J.; Park, O.J. Genistein, EGCG, and capsaicin inhibit adipocyte differentiation process via activating AMP-activated protein kinase. Biochem. Biophys. Res. Commun. 2005, 338, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.S.; Chung, C.S.; Lee, H.G.; Kim, T.G.; Choi, Y.J.; Cho, C.S. Inhibitory effect of (−)-epigallocatechin-3-gallate on lipid accumulation of 3T3-L1 cells. Obesity 2007, 15, 2571–2582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.F.; Li, Q.; Liang, J.; Dai, X.Q.; Ding, Y.; Wang, J.B.; Li, Y. Epigallocatechin-3-O-gallate (EGCG) protects the insulin sensitivity in rat L6 muscle cells exposed to dexamethasone condition. Phytomedicine 2010, 17, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Santamarina, A.B.; Oliveira, J.L.; Silva, F.P.; Carnier, J.; Mennitti, L.V.; Santana, A.A.; de Souza, G.H.; Ribeiro, E.B.; Oller do Nascimento, C.M.; Lira, F.S.; et al. Green Tea Extract Rich in Epigallocatechin-3-Gallate Prevents Fatty Liver by AMPK Activation via LKB1 in Mice Fed a High-Fat Diet. PLoS ONE 2015, 10, e0141227. [Google Scholar] [CrossRef] [PubMed]
- She, P.; Van Horn, C.; Reid, T.; Hutson, S.M.; Cooney, R.N.; Lynch, C.J. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1552–E1563. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Sun, H.; She, P.; Youn, J.Y.; Warburton, S.; Ping, P.; Vondriska, T.M.; Cai, H.; Lynch, C.J.; Wang, Y. Protein phosphatase 2Cm is a critical regulator of branched-chain amino acid catabolism in mice and cultured cells. J. Clin. Investig. 2009, 119, 1678–1687. [Google Scholar] [CrossRef] [PubMed]
- Lian, K.; Du, C.; Liu, Y.; Zhu, D.; Yan, W.; Zhang, H.; Hong, Z.; Liu, P.; Zhang, L.; Pei, H.; et al. Impaired adiponectin signaling contributes to disturbed catabolism of branched-chain amino acids in diabetic mice. Diabetes 2015, 64, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.A.; Smart, D.E. Transcriptional regulation of hepatic stellate cell activation. Gut 2002, 50, 891–896. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, J.-Y.; Kim, Y.J.; Ryu, R.; Cho, S.-J.; Kwon, E.-Y.; Choi, M.-S. Effect of Green Tea Extract on Systemic Metabolic Homeostasis in Diet-Induced Obese Mice Determined via RNA-Seq Transcriptome Profiles. Nutrients 2016, 8, 640. https://doi.org/10.3390/nu8100640
Choi J-Y, Kim YJ, Ryu R, Cho S-J, Kwon E-Y, Choi M-S. Effect of Green Tea Extract on Systemic Metabolic Homeostasis in Diet-Induced Obese Mice Determined via RNA-Seq Transcriptome Profiles. Nutrients. 2016; 8(10):640. https://doi.org/10.3390/nu8100640
Chicago/Turabian StyleChoi, Ji-Young, Ye Jin Kim, Ri Ryu, Su-Jung Cho, Eun-Young Kwon, and Myung-Sook Choi. 2016. "Effect of Green Tea Extract on Systemic Metabolic Homeostasis in Diet-Induced Obese Mice Determined via RNA-Seq Transcriptome Profiles" Nutrients 8, no. 10: 640. https://doi.org/10.3390/nu8100640




