Changes in Composition of Caecal Microbiota Associated with Increased Colon Inflammation in Interleukin-10 Gene-Deficient Mice Inoculated with Enterococcus Species
Abstract
:1. Introduction
2. Experimental Section
2.1. Animal Experiments
2.2. Sample Collection
2.3. Caecal Bacterial DNA Extraction
2.4. Pyrotag Sequencing Analysis of Caecal Bacterial Composition
2.5. DGGE Analysis of Caecal Bacterial composition
2.6. Real-Time PCR Quantification of Bacteria
3. Results
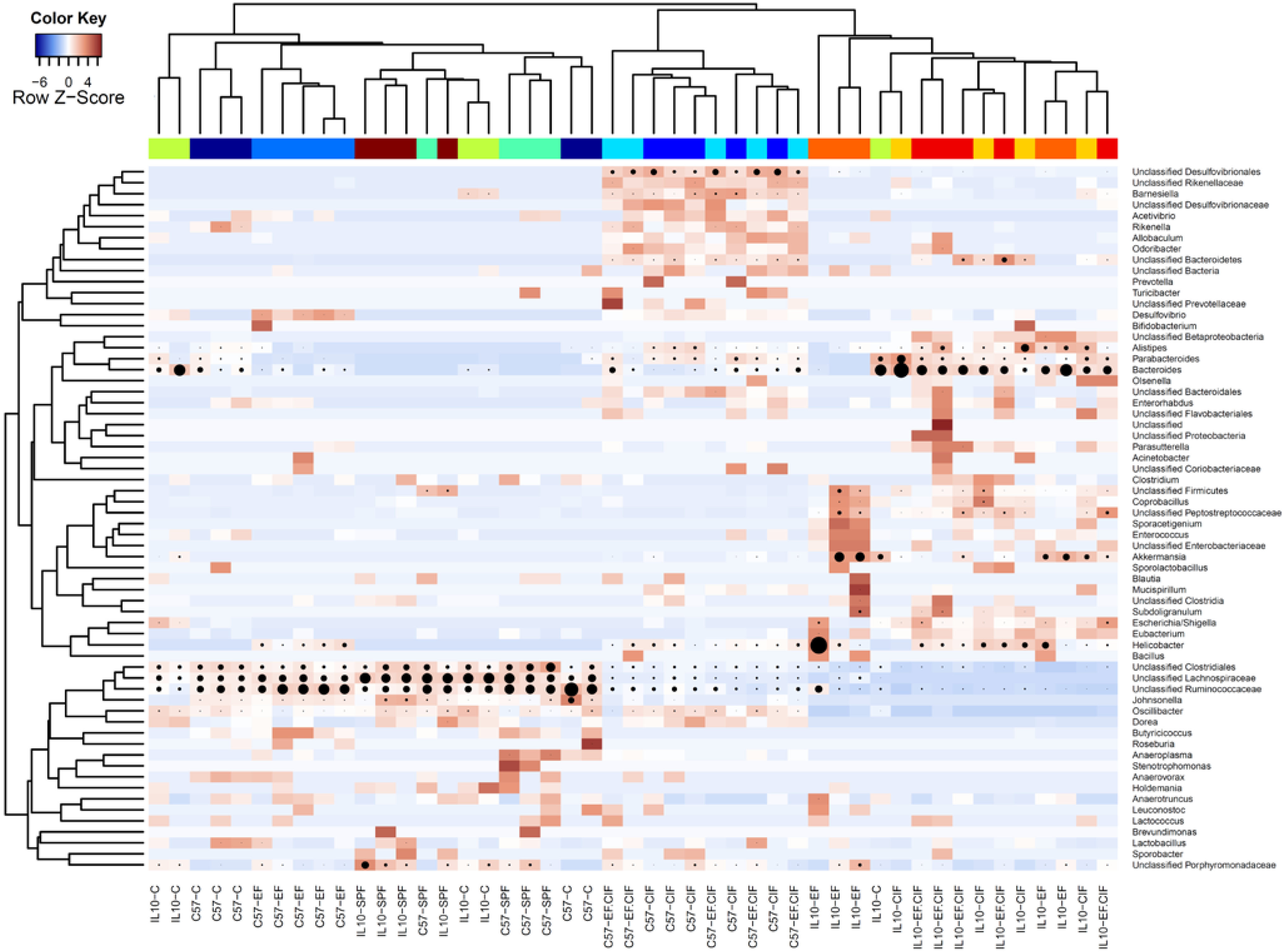
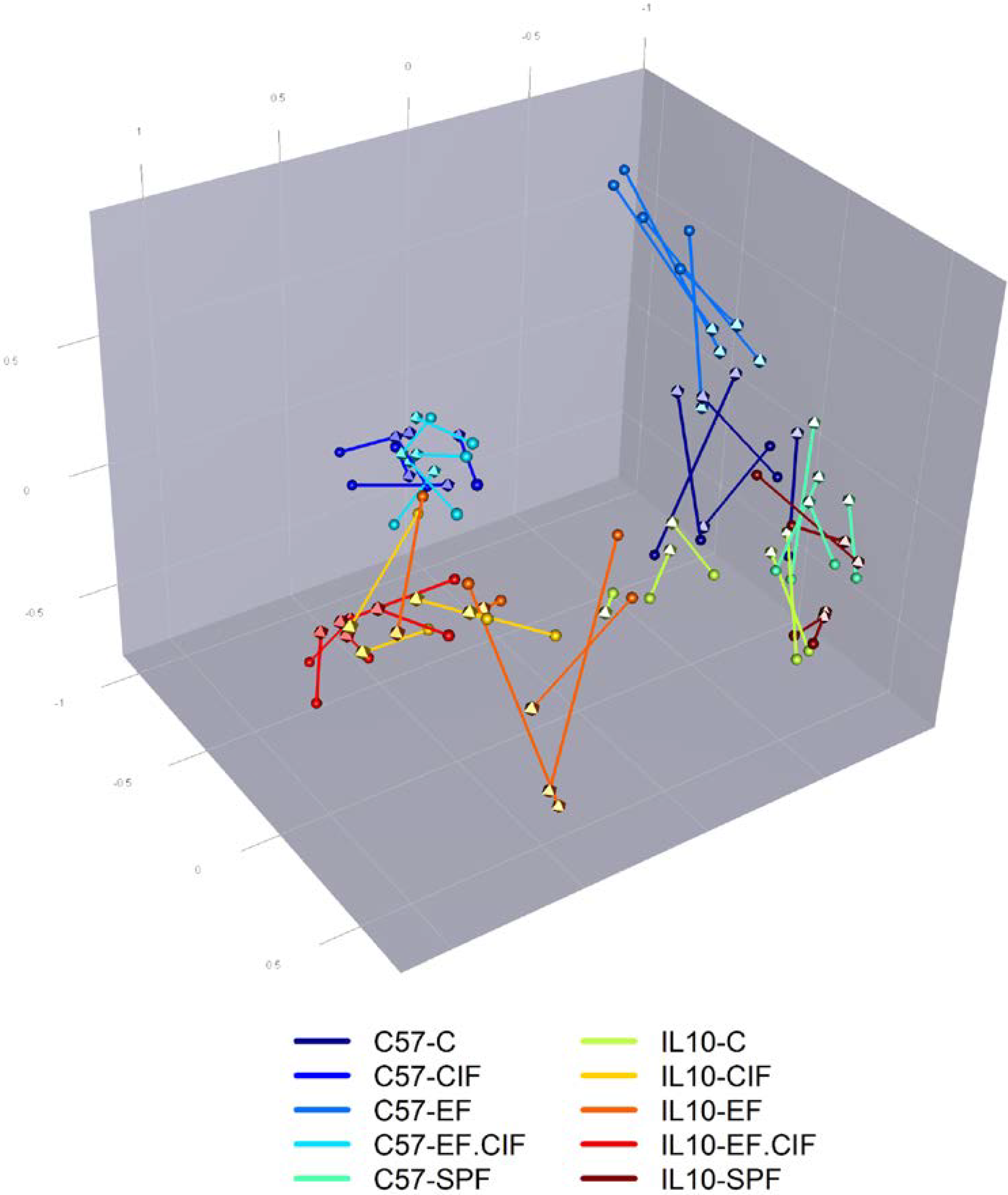
3.1. Inoculation Induced Dysbiosis of the Caecal Microbiota in Il10-/- Mice
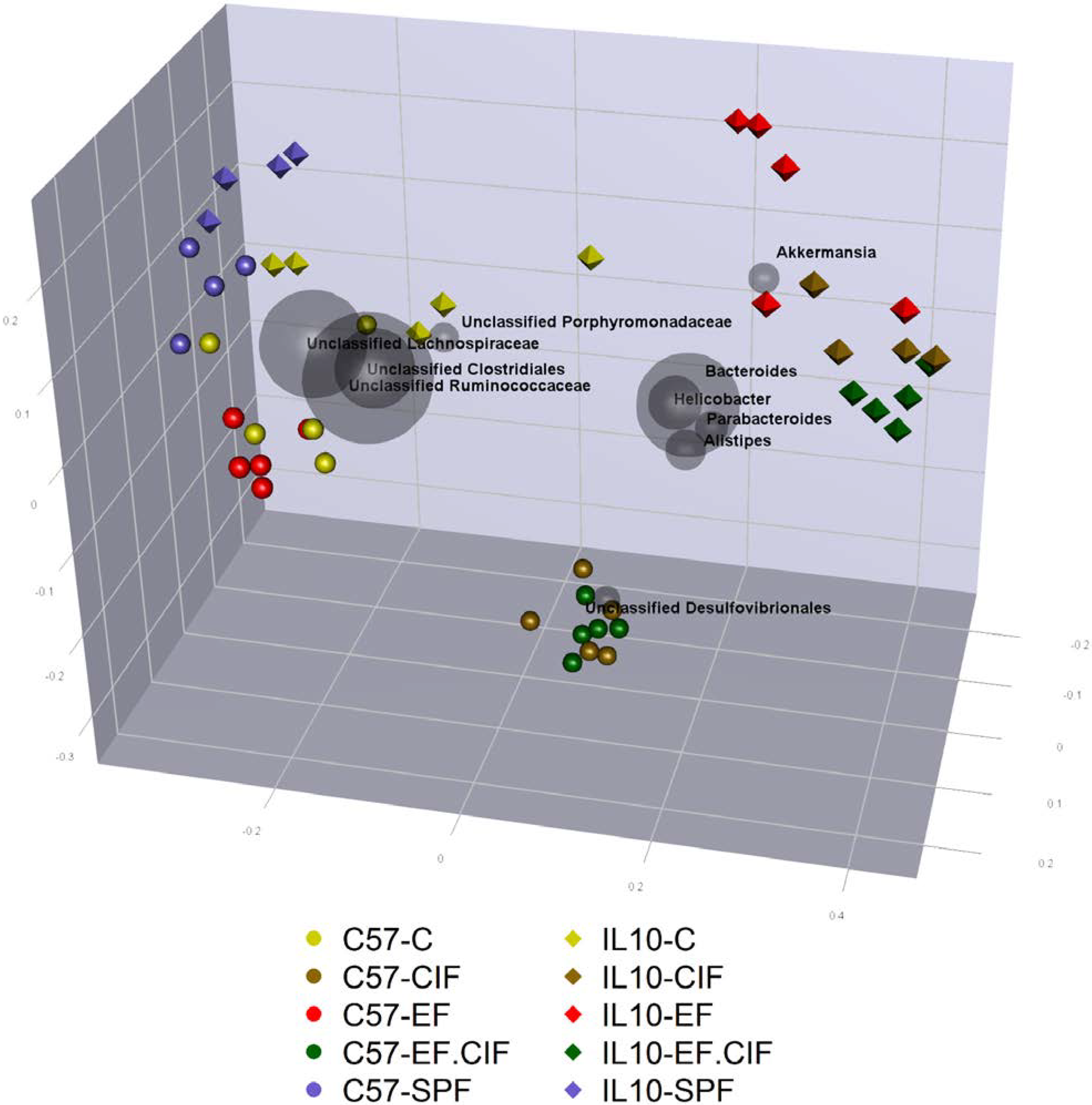

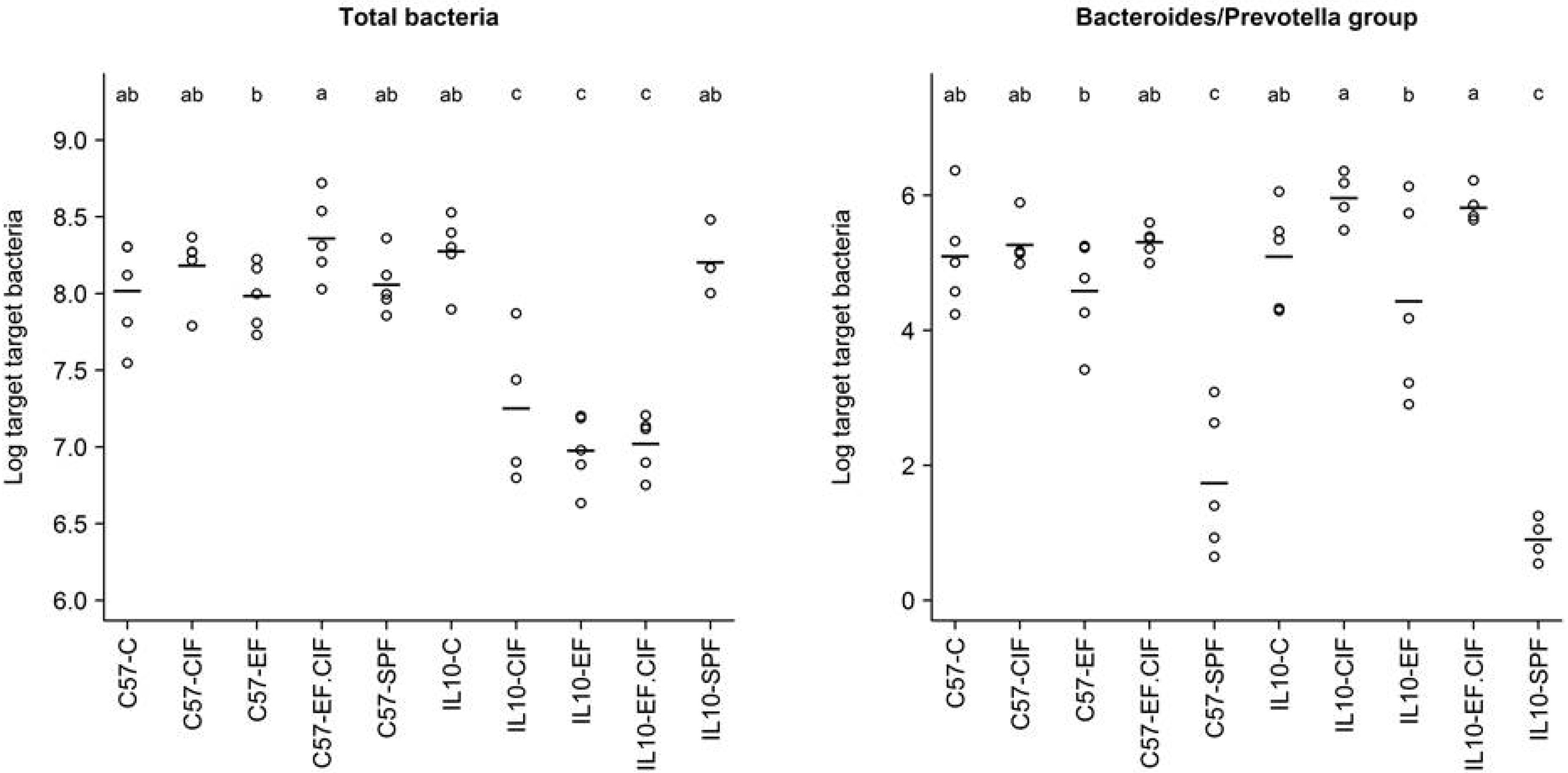
| Bacteroides-Prevotella Group | E. coli | Enterococcus | Total Bacteria | |
|---|---|---|---|---|
| Genotype | 0.56 | 6.33E-05 | 1.24E-07 | 3.59E-09 |
| Treatment | 1.80E-14 | 2.75E-07 | 2.70E-03 | 5.95E-06 |
| Genotype: Treatment | 0.25 | 1.13E-05 | 1.25E-03 | 2.69E-08 |
3.2. Inflammation-Associated Bacteria
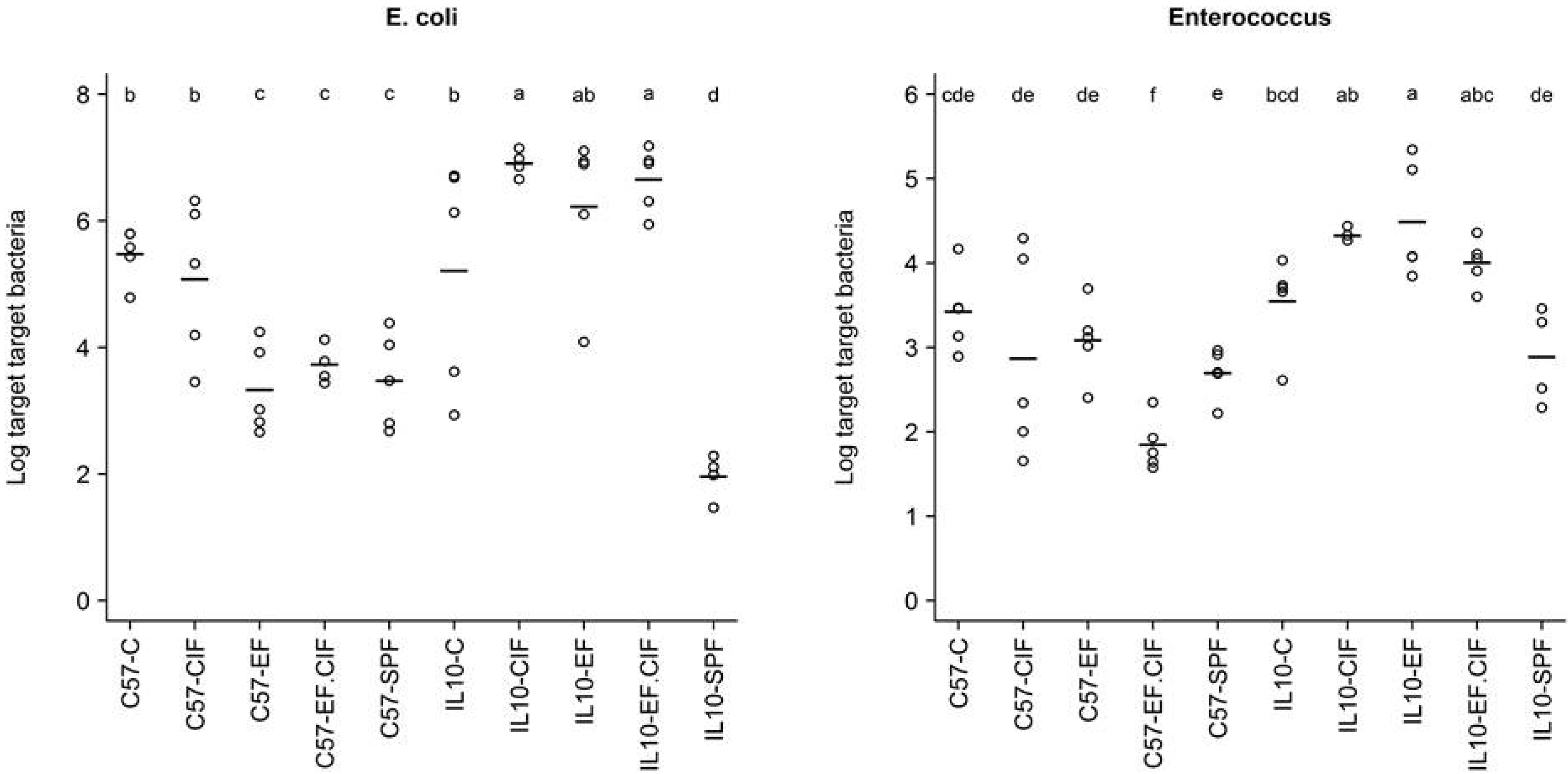

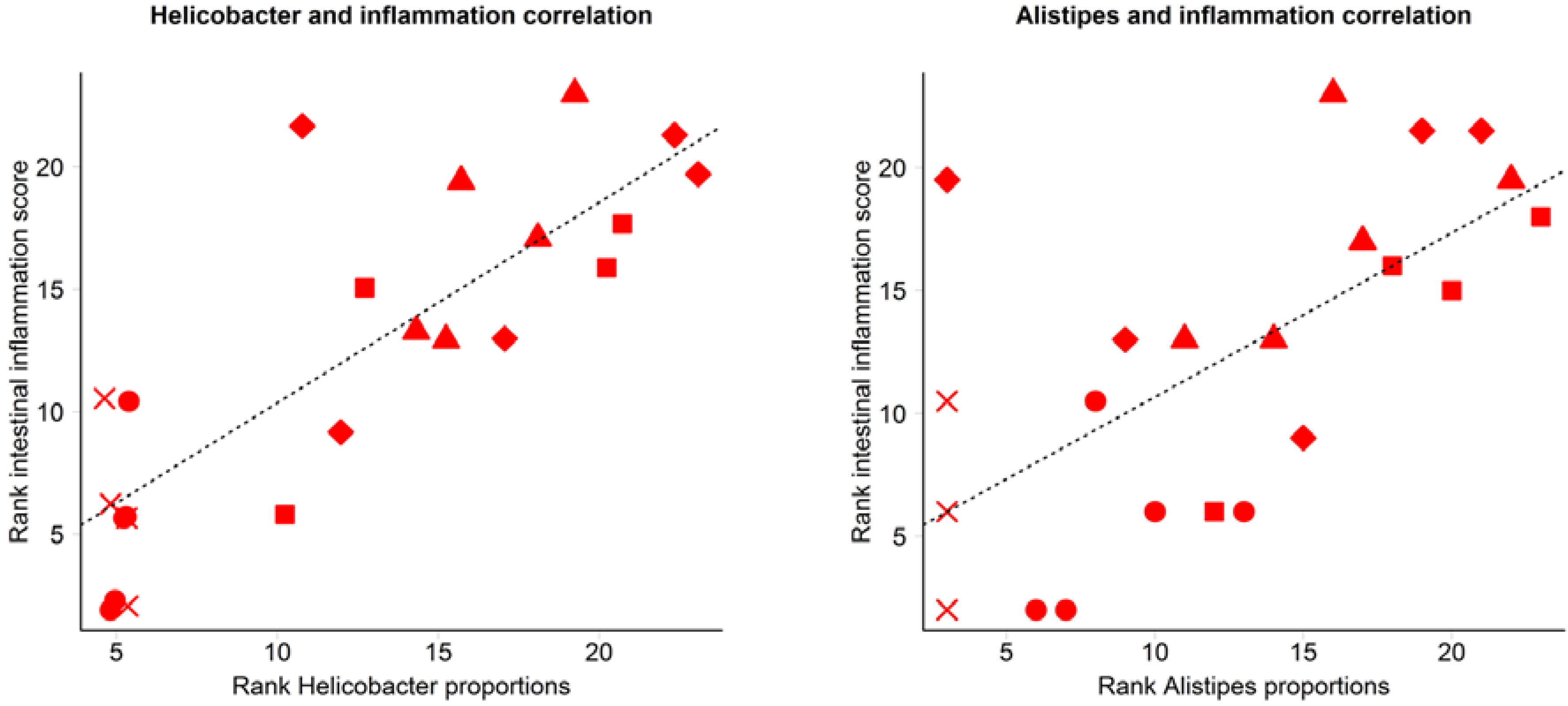
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Conte, M.P.; Schippa, S.; Zamboni, I.; Penta, M.; Chiarini, F.; Seganti, L.; Osborn, J.; Falconieri, P.; Borrelli, O.; Cucchiara, S. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006, 55, 1760–1767. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St. Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed]
- Gueimonde, M.; Ouwehand, A.; Huhtinen, H.; Salminen, E.; Salminen, S. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J. Gastroenterol. 2007, 13, 3985–3989. [Google Scholar] [CrossRef] [PubMed]
- Lepage, P.; Hösler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A.; et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011, 141, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Noor, S.O.; Ridgway, K.; Scovell, L.; Kemsley, E.K.; Lund, E.K.; Jamieson, C.; Johnson, I.T.; Narbad, A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010, 10, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, S.J.; Musfeldt, M.; Wenderoth, D.F.; Hampe, J.; Brant, O.; Fölsch, U.R.; Timmis, K.N.; Schreiber, S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Takaishi, H.; Matsuki, T.; Nakazawa, A.; Takada, T.; Kado, S.; Asahara, T.; Kamada, N.; Sakuraba, A.; Yajima, T.; Higuchi, H.; et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int. J. Med. Microbiol. 2008, 298, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.S.; Mishima, Y.; Wohlgemuth, S.; Liu, B.; Bower, M.; Carroll, I.M.; Sartor, R.B. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10-/- mice. Infect. Immun. 2014, 82, 2239–2246. [Google Scholar] [CrossRef] [PubMed]
- Sartor, R.B.; Mazmanian, S.K. Intestinal Microbes in Inflammatory Bowel Diseases. Am. J. Gastroenterol. Suppl. 2012, 1, 15–21. [Google Scholar] [CrossRef]
- Frank, D.; Robertson, C.; Hamm, C.; Kpadeh, Z.; Zhang, T.; Chen, H.; Zhu, W.; Sartor, R.; Boedeker, E.; Harpaz, N.; et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 2011, 17, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Sellon, R.K.; Tonkonogy, S.; Schultz, M.; Dieleman, L.A.; Grenther, W.; Balish, E.; Rennick, D.M.; Sartor, R.B. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 1998, 66, 5224–5231. [Google Scholar] [PubMed]
- Kuhn, R.; Lohler, J.; Rennick, D.; Rajewsky, K.; Muller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 1993, 75, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Balish, E.; Warner, T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am. J. Pathol. 2002, 160, 2253–2257. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Tonkonogy, S.L.; Albright, C.A.; Tsang, J.; Balish, E.J.; Braun, J.; Huycke, M.M.; Sartor, R.B. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 2005, 128, 891–906. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Tonkonogy, S.L.; Karrasch, T.; Jobin, C.; Sartor, R.B. Dual-association of gnotobiotic IL-10-/- mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm. Bowel Dis. 2007, 13, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Eaton, T.J.; Gasson, M.J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Jett, B.D.; Huycke, M.M.; Gilmore, M.S. Virulence of enterococci. Clin. Microbiol. Rev. 1994, 7, 462–478. [Google Scholar] [PubMed]
- Finegold, S.M.; Sutter, V.L.; Mathisen, G.E. Normal indigenous intestinal flora. In Human Intestinal Microflora in Health and Disease; Hentges, D.J., Ed.; Academic Press: New York, NY, USA, 1983; pp. 3–31. [Google Scholar]
- Tannock, G.W.; Cook, G. Enterococci as members of the intestinal microflora of humans. In The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance; Gilmore, M.S., Clewell, D.B., Courvalin, P.M., Dunny, G.M., Murray, B.E., Rice, L.B., Eds.; ASM Press: Washington DC, USA, 2002. [Google Scholar]
- Barnett, M.P.; McNabb, W.C.; Cookson, A.L.; Zhu, S.; Davy, M.; Knoch, B.; Nones, K.; Hodgkinson, A.J.; Roy, N.C. Changes in colon gene expression associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species. BMC Immunol. 2010, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.C.; Barnett, M.P.G.; Knoch, B.; Dommels, Y.E.M.; McNabb, W.C. Nutrigenomics applied to an animal model of Inflammatory Bowel Diseases: Transcriptomic analysis of the effects of eicosapentaenoic acid- and arachidonic acid-enriched diets. Mutat. Res. 2007, 622, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Bibiloni, R.; Simon, M.; Albright, C.A.; Sartor, B.; Tannock, G. Analysis of the large bowel microbiota of colitic mice using PCR/DGGE. Lett. Appl. Microbiol. 2005, 41, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Laboratory, T.J. Overview of the Jackson Laboratory Facility Barrier Levels. Available online: http://jaxmice.jax.org/health/barrier.html (accessed on 16 December 2014).
- Knoch, B.; Barnett, M.P.; Cooney, J.; McNabb, W.C.; Barraclough, D.; Laing, W.; Zhu, S.; Park, Z.A.; Maclean, P.; Knowles, S.O.; et al. Molecular characterization of the onset and progression of colitis in inoculated interleukin-10 gene-deficient mice: A role for PPARalpha. PPAR Res. 2010, 2010. [Google Scholar] [CrossRef]
- Knoch, B.; Barnett, M.P.; McNabb, W.C.; Zhu, S.; Park, Z.A.; Khan, A.; Roy, N.C. Dietary arachidonic acid-mediated effects on colon inflammation using transcriptome analysis. Mol. Nutr. Food. Res. 2010, 54 (Suppl 1), S62–74. [Google Scholar] [CrossRef] [PubMed]
- Knoch, B.; Barnett, M.P.G.; Zhu, S.T.; Park, Z.A.; Nones, K.; Dommels, Y.E.M.; Knowles, S.O.; McNabb, W.C.; Roy, N.C. Genome-wide analysis of dietary eicosapentaenoic acid- and oleic acid-induced modulation of colon inflammation in interleukin-10 gene-deficient mice. J. Nutrigenet. Nutrigenomics 2009, 2, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Russ, A.E.; Peters, J.S.; McNabb, W.C.; Barnett, M.P.G.; Anderson, R.C.; Park, Z.; Zhu, S.T.; Maclean, P.; Reynolds, G.W.; Roy, N.C. Gene expression changes in the colon epithelium are similar to those of intact colon in the interleukin-10 gene deficient mouse in late inflammation. PloS One 2013, 8, e63251. [Google Scholar] [CrossRef] [PubMed]
- Knoch, B.; Nones, K.; Barnett, M.P.; McNabb, W.C.; Roy, N.C. Diversity of caecal bacteria is altered in interleukin-10 gene-deficient mice before and after colitis onset and when fed polyunsaturated fatty acids. Microbiology 2010, 156, 3306–3316. [Google Scholar] [CrossRef] [PubMed]
- Claus, S.P.; Ellero, S.L.; Berger, B.; Krause, L.; Bruttin, A.; Molina, J.; Paris, A.; Want, E.J.; de Waziers, I.; Cloarec, O.; et al. Colonization-induced host-gut microbial metabolic interaction. MBio 2011, 2, e00271–00210. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. Available online: http://CRAN.R-project.org/package=agricolae (accessed on 3 February 2014).
- Tannock, G.W.; Munro, K.; Harmsen, H.J.; Welling, G.W.; Smart, J.; Gopal, P.K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 2000, 66, 2578–2588. [Google Scholar] [CrossRef] [PubMed]
- Nones, K.; Knoch, B.; Dommels, Y.E.; Paturi, G.; Butts, C.; McNabb, W.C.; Roy, N.C. Multidrug resistance gene deficient (mdr1a-/-) mice have an altered caecal microbiota that precedes the onset of intestinal inflammation. J. Appl. Microbiol. 2009, 107, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. Available online: http://CRAN.R-project.org/package=vegan (accessed on 3 February 2014).
- Bakken, J.S.; Borody, T.; Brandt, L.J.; Brill, J.V.; Demarco, D.C.; Franzos, M.A.; Kelly, C.; Khoruts, A.; Louie, T.; Martinelli, L.P.; et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Maharshak, N.; Packey, C.D.; Ellermann, M.; Manick, S.; Siddle, J.P.; Huh, E.Y.; Plevy, S.; Sartor, R.B.; Carroll, I.M. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes 2013, 4, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Winter, M.G.; Xavier, M.N.; Thiennimitr, P.; Poon, V.; Keestra, A.M.; Laughlin, R.C.; Gomez, G.; Wu, J.; Lawhon, S.D.; et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 2013, 339, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13. [Google Scholar] [CrossRef]
- Yang, I.; Eibach, D.; Kops, F.; Brenneke, B.; Woltemate, S.; Schulze, J.; Bleich, A.; Gruber, A.D.; Muthupalani, S.; Fox, J.G.; Josenhans, C.; et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One 2013, 8, e70783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Lapthorne, S.; Pereira-Fantini, P.M.; Fouhy, F.; Wilson, G.; Thomas, S.L.; Dellios, N.L.; Scurr, M.; O'Sullivan, O.; Ross, R.P.; Stanton, C.; et al. Gut microbial diversity is reduced and is associated with colonic inflammation in a piglet model of short bowel syndrome. Gut Microbes 2013, 4, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Vaneechoutte, M.; Doerffel, Y. Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 2008, 14, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.M.; Ferreyra, J.A.; Higginbottom, S.K.; Lynch, J.B.; Kashyap, P.C.; Gopinath, S.; Naidu, N.; Choudhury, B.; Weimer, B.C.; Monack, D.M.; et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502, 96–99. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bassett, S.A.; Young, W.; Barnett, M.P.G.; Cookson, A.L.; McNabb, W.C.; Roy, N.C. Changes in Composition of Caecal Microbiota Associated with Increased Colon Inflammation in Interleukin-10 Gene-Deficient Mice Inoculated with Enterococcus Species. Nutrients 2015, 7, 1798-1816. https://doi.org/10.3390/nu7031798
Bassett SA, Young W, Barnett MPG, Cookson AL, McNabb WC, Roy NC. Changes in Composition of Caecal Microbiota Associated with Increased Colon Inflammation in Interleukin-10 Gene-Deficient Mice Inoculated with Enterococcus Species. Nutrients. 2015; 7(3):1798-1816. https://doi.org/10.3390/nu7031798
Chicago/Turabian StyleBassett, Shalome A., Wayne Young, Matthew P. G. Barnett, Adrian L. Cookson, Warren C. McNabb, and Nicole C. Roy. 2015. "Changes in Composition of Caecal Microbiota Associated with Increased Colon Inflammation in Interleukin-10 Gene-Deficient Mice Inoculated with Enterococcus Species" Nutrients 7, no. 3: 1798-1816. https://doi.org/10.3390/nu7031798






