Biomarkers of Nutrition and Health: New Tools for New Approaches
Abstract
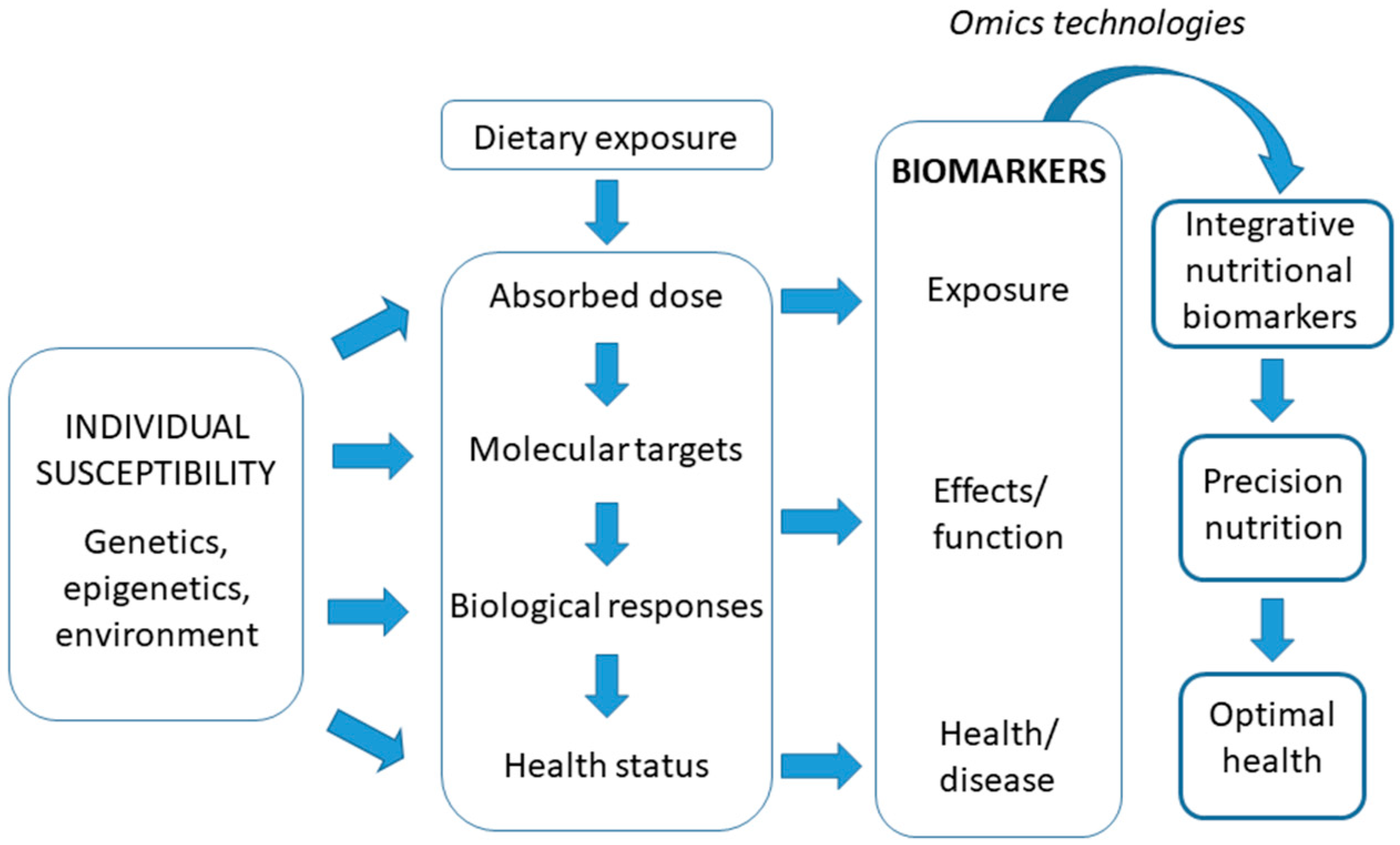
:1. Introduction
2. Biomarkers of Nutritional Status
| Proposed Biomarker | Sample Type | Intended Use (As Nutritional Biomarker) | References |
|---|---|---|---|
| Alkylresorcinols | Plasma | Whole-grain food consumption | Original research [14,15] Reviewed in Reference [16] |
| Allyl methyl sulfoxide (AMSO) or allyl methyl sulfone (AMSO2) | Urine | Intake of garlic | Original research [17] BFIRev ** [18] |
| Allyl methyl sulphide (AMS) | Urine/breath | Intake of garlic | Original research [17,19,20] BFIRev [18] |
| Arbutin | Plasma | Pear intake | Original research [21] BFIRev [22] |
| Carotenoids | Plasma | Fruit and vegetable intake | Systematic review and meta-analysis [23] |
| Carotenoids with Vitamin C | Plasma/serum | Fruit and vegetable intake Combined marker (suggested as better biomarker than carotenoids or vitamin C alone) | Reviewed in Reference [24] |
| Creatine | Serum | Intake of meat and fish | Reviewed in Reference [25] |
| Creatinine | Urine | Intake of meat and fish | Reviewed in Reference [25] |
| Daidzein | Urine/plasma | Intake of soy or soy-based products | Systematic review [26] |
| Dyhydrocaffeic acid derivatives | Urine | Acute and habitual exposure to coffee | Original research [27,28,29] Reviewed in Reference [30] |
| Erythronic acid, alone or with fructose and/or sucrose | Urine | Sugar intake Combined marker | Original research [31] |
| Genistein | Urine/plasma | Intake of soy or soy-based products | Systematic review [26] |
| Homocysteine | Plasma | One carbon metabolism and folate status | Reviewed in References [32,33] |
| Hydroxylated and sulfonated metabolites of esculeogenin B | Urine | Intake of tomato juice | Original research [34] |
| 1-Methylhistidine | Urine | Meat and oily fish consumption | Original research [27,35,36] Reviewed in References [30,37] |
| n-3 fatty acids: docosahexaenoic acid (DHA) | Blood: erythrocytes or platelets | DHA status | Systematic review [38] |
| n-3 fatty acids: DHA (as phospholipid) | Plasma | DHA status | Systematic review [38] |
| n-3 fatty acids: eicosapentaenoic acid (EPA as phospholipid) | Plasma | EPA status | Systematic review [38] |
| N-acetyl-S-(2carboxypropyl)cysteine (CPMA) | Urine | Intake of onion and garlic | Original research [39] BFIRev [18] |
| Nitrogen* | Urine (24h) | Protein intake | Reviewed in Reference [40] |
| O-acetylcarnitine | Urine | Red-meat consumption | Original research [41] Reviewed in Reference [42] |
| Pentadecanoic acid (C15:0) | Plasma/serum | Total dairy fat intake | Reviewed in Reference [43] |
| Phenylacetylglutamine | Urine | Vegetable intake | Original research [41] Reviewed in Reference [30] |
| Phloretin | Urine | Apple intake | Original research [44,45] BFIRev [22] |
| Phloretin glucuronide | Urine | Apple intake | Original research [46,47] BFIRev [22] |
| Proline betaine | Urine | Acute and habitual citrus exposure | Original research [27,48,49] Reviewed in Reference [30] |
| S-allylcysteine (SAC) | Plasma | Intake of garlic | Original research [19] BFIRev [18] |
| S-allylmercapturic acid (ALMA) | Urine | Intake of garlic | Original research [50] BFIRev [18] |
| Urolithin B | Urine | Intake of ellagitannins (present in fruits as strawberries, raspberries and walnuts and oak-aged red wine, among others) | Original research [51] |
3. Current Challenges in the Development of Health Biomarkers
The New Concept of Integrative Nutritional Biomarkers
4. Sources of Biomarkers in Nutritional Studies
5. Types of Analysis
6. Nutrigenomic Approach in the Identification of Biomarkers
6.1. Genetic Biomarkers
6.2. Epigenetic Markers
6.3. Transcriptome Markers
6.3.1. Non-coding RNAs
6.4. Proteomic Markers
6.5. Metabolomic and Lipidomic Markers
7. Empowering Citizens to Monitor and Follow a Healthy Diet
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thompson, F.E.; Subar, A.F.; Loria, C.M.; Reedy, J.L.; Baranowski, T. Need for technological innovation in dietary assessment. J. Am. Diet. Assoc. 2010, 110, 48–51. [Google Scholar] [CrossRef] [PubMed]
- Frobisher, C.; Maxwell, S.M. The estimation of food portion sizes: A comparison between using descriptions of portion sizes and a photographic food atlas by children and adults. J. Hum. Nutr. Diet. 2003, 16, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Maurer, J.; Taren, D.L.; Teixeira, P.J.; Thomson, C.A.; Lohman, T.G.; Going, S.B.; Houtkooper, L.B. The psychosocial and behavioral characteristics related to energy misreporting. Nutr. Rev. 2006, 64, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.K.; Soultanakis, R.P.; Matthews, D.E. Literacy and body fatness are associated with underreporting of energy intake in US low-income women using the multiple-pass 24-hour recall: A doubly labeled water study. J. Am. Diet. Assoc. 1998, 98, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Elmadfa, I.; Meyer, A.L. Importance of food composition data to nutrition and public health. Eur. J. Clin. Nutr. 2010, 64 (Suppl. 3), S4–S7. [Google Scholar] [CrossRef] [PubMed]
- Potischman, N. Biologic and methodologic issues for nutritional biomarkers. J. Nutr. 2003, 133 (Suppl. 3), 875S–880S. [Google Scholar] [CrossRef]
- Riedl, J.; Linseisen, J.; Hoffmann, J.; Wolfram, G. Some dietary fibres reduce the absorption of carotenoids in women. J. Nutr. 1999, 129, 2170–2176. [Google Scholar] [CrossRef]
- Forbes, A.L.; Arnaud, M.J.; Chichester, C.O.; Cook, J.D.; Harrison, B.N.; Hurrell, R.F.; Kahn, S.G.; Morris, E.R.; Tanner, J.T.; Whittaker, P.; et al. Comparison of in vitro, animal and clinical determinations of iron bioavailability: International Nutritional Anemia Consultative Group Task Force report on iron bioavailability. Am. J. Clin. Nutr. 1989, 49, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, A.J.; Wallace, O.A.M.; Kruger, M.C.; Prosser, C.G. Effect of the dietary delivery matrix on vitamin D3 bioavailability and bone mineralisation in vitamin-D3-deficient growing male rats. Br. J. Nutr. 2018, 119, 143–152. [Google Scholar] [CrossRef]
- McKinnon, H.; Kruger, M.; Prosser, C.; Lowry, D. The effect of formulated goats’ milk on calcium bioavailability in male growing rats. J. Sci. Food Agric. 2010, 90, 112–116. [Google Scholar] [CrossRef]
- Wang, C.; Li, B.; Wang, B.; Xie, N. Degradation and antioxidant activities of peptides and zinc-peptide complexes during in vitro gastrointestinal digestion. Food Chem. 2015, 173, 733–740. [Google Scholar] [CrossRef]
- Zielinska-Dawidziak, M. Plant ferritin—A source of iron to prevent its deficiency. Nutrients 2015, 7, 1184–1201. [Google Scholar] [CrossRef]
- Chan, L.N.; Mike, L.A. The science and practice of micronutrient supplementations in nutritional anemia: An evidence-based review. J. Parenter. Enter. Nutr. 2014, 38, 656–672. [Google Scholar] [CrossRef]
- Ross, A.B.; Bourgeois, A.; Macharia, H.N.; Kochhar, S.; Jebb, S.A.; Brownlee, I.A.; Seal, C.J. Plasma alkylresorcinols as a biomarker of whole-grain food consumption in a large population: Results from the WHOLEheart Intervention Study. Am. J. Clin. Nutr. 2012, 95, 204–211. [Google Scholar] [CrossRef]
- Ma, J.; Ross, A.B.; Shea, M.K.; Bruce, S.J.; Jacques, P.F.; Saltzman, E.; Lichtenstein, A.H.; Booth, S.L.; McKeown, N.M. Plasma alkylresorcinols, biomarkers of whole-grain intake, are related to lower BMI in older adults. J. Nutr. 2012, 142, 1859–1864. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.B. Present status and perspectives on the use of alkylresorcinols as biomarkers of wholegrain wheat and rye intake. J. Nutr. Metab. 2012, 2012, 462967. [Google Scholar] [CrossRef]
- Scheffler, L.; Sauermann, Y.; Heinlein, A.; Sharapa, C.; Buettner, A. Detection of Volatile Metabolites Derived from Garlic (Allium sativum) in Human Urine. Metabolites 2016, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Pratico, G.; Gao, Q.; Manach, C.; Dragsted, L.O. Biomarkers of food intake for Allium vegetables. Genes Nutr. 2018, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Rosen, R.T.; Hiserodt, R.D.; Fukuda, E.K.; Ruiz, R.J.; Zhou, Z.; Lech, J.; Rosen, S.L.; Hartman, T.G. Determination of allicin, S-allylcysteine and volatile metabolites of garlic in breath, plasma or simulated gastric fluids. J. Nutr. 2001, 131, 968S–971S. [Google Scholar] [CrossRef] [PubMed]
- Taucher, J.; Hansel, A.; Jordan, A.; Lindinger, W. Analysis of compounds in human breath after ingestion of garlic using proton-tranfer-reaction mass spectrometry. J. Agric. Food Chem. 1996, 44, 3778–3782. [Google Scholar] [CrossRef]
- Nieman, D.C.; Gillitt, N.D.; Sha, W.; Meaney, M.P.; John, C.; Pappan, K.L.; Kinchen, J.M. Metabolomics-Based Analysis of Banana and Pear Ingestion on Exercise Performance and Recovery. J. Proteome Res. 2015, 14, 5367–5377. [Google Scholar] [CrossRef]
- Ulaszewska, M.; Vazquez-Manjarrez, N.; Garcia-Aloy, M.; Llorach, R.; Mattivi, F.; Dragsted, L.O.; Pratico, G.; Manach, C. Food intake biomarkers for apple, pear and stone fruit. Genes Nutr. 2018, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Pennant, M.; Steur, M.; Moore, C.; Butterworth, A.; Johnson, L. Comparative validity of vitamin C and carotenoids as indicators of fruit and vegetable intake: A systematic review and meta-analysis of randomised controlled trials. Br. J. Nutr. 2015, 114, 1331–1340. [Google Scholar] [CrossRef]
- Woodside, J.V.; Draper, J.; Lloyd, A.; McKinley, M.C. Use of biomarkers to assess fruit and vegetable intake. Proc. Nutr. Soc. 2017, 76, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Dragsted, L.O. Biomarkers of meat intake and the application of nutrigenomics. Meat Sci. 2010, 84, 301–307. [Google Scholar] [CrossRef]
- Sri Harsha, P.S.C.; Wahab, R.A.; Garcia-Aloy, M.; Madrid-Gambin, F.; Estruel-Amades, S.; Watzl, B.; Andrés-Lacueva, C.; Brennan, L. Biomarkers of legume intake in human intervention and observational studies: A systematic review. Genes Nutr. 2018, 13, 25. [Google Scholar] [CrossRef]
- Lloyd, A.J.; Beckmann, M.; Haldar, S.; Seal, C.; Brandt, K.; Draper, J. Data-driven strategy for the discovery of potential urinary biomarkers of habitual dietary exposure. Am. J. Clin. Nutr. 2013, 97, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Renouf, M.; Guy, P.A.; Marmet, C.; Fraering, A.L.; Longet, K.; Moulin, J.; Enslen, M.; Barron, D.; Dionisi, F.; Cavin, C.; et al. Measurement of caffeic and ferulic acid equivalents in plasma after coffee consumption: Small intestine and colon are key sites for coffee metabolism. Mol. Nutr. Food Res. 2010, 54, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Stalmach, A.; Mullen, W.; Barron, D.; Uchida, K.; Yokota, T.; Cavin, C.; Steiling, H.; Williamson, G.; Crozier, A. Metabolite profiling of hydroxycinnamate derivatives in plasma and urine after the ingestion of coffee by humans: Identification of biomarkers of coffee consumption. Drug Metab. Dispos. 2009, 37, 1749–1758. [Google Scholar] [CrossRef] [PubMed]
- Astarita, G.; Langridge, J. An emerging role for metabolomics in nutrition science. J. Nutrigenet. Nutrigenomics 2013, 6, 181–200. [Google Scholar] [CrossRef]
- Beckmann, M.; Joosen, A.M.; Clarke, M.M.; Mugridge, O.; Frost, G.; Engel, B.; Taillart, K.; Lloyd, A.J.; Draper, J.; Lodge, J.K. Changes in the human plasma and urinary metabolome associated with acute dietary exposure to sucrose and the identification of potential biomarkers of sucrose intake. Mol. Nutr. Food Res. 2016, 60, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Pratico, G.; Scalbert, A.; Vergeres, G.; Kolehmainen, M.; Manach, C.; Brennan, L.; Afman, L.A.; Wishart, D.S.; Andres-Lacueva, C.; et al. A scheme for a flexible classification of dietary and health biomarkers. Genes Nutr. 2017, 12, 34. [Google Scholar] [CrossRef]
- Mason, J.B. Biomarkers of nutrient exposure and status in one-carbon (methyl) metabolism. J. Nutr. 2003, 133 (Suppl. 3), 941S–947S. [Google Scholar] [CrossRef]
- Hovelmann, Y.; Jagels, A.; Schmid, R.; Hubner, F.; Humpf, H.U. Identification of potential human urinary biomarkers for tomato juice intake by mass spectrometry-based metabolomics. Eur. J. Nutr. 2019. [Google Scholar] [CrossRef]
- Myint, T.; Fraser, G.E.; Lindsted, K.D.; Knutsen, S.F.; Hubbard, R.W.; Bennett, H.W. Urinary 1-methylhistidine is a marker of meat consumption in Black and in White California Seventh-day Adventists. Am. J. Epidemiol. 2000, 152, 752–755. [Google Scholar] [CrossRef]
- Lloyd, A.J.; Fave, G.; Beckmann, M.; Lin, W.; Tailliart, K.; Xie, L.; Mathers, J.C.; Draper, J. Use of mass spectrometry fingerprinting to identify urinary metabolites after consumption of specific foods. Am. J. Clin. Nutr. 2011, 94, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; van der Hooft, J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin. Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef] [PubMed]
- Serra-Majem, L.; Nissensohn, M.; Overby, N.C.; Fekete, K. Dietary methods and biomarkers of omega 3 fatty acids: A systematic review. Br. J. Nutr. 2012, 107 (Suppl. 2), S64–S76. [Google Scholar] [CrossRef]
- Jandke, J.; Spiteller, G. Unusual conjugates in biological profiles originating from consumption of onions and garlic. J. Chromatogr. 1987, 421, 1–8. [Google Scholar] [CrossRef]
- Bingham, S.A. Biomarkers in nutritional epidemiology. Public Health Nutr. 2002, 5, 821–827. [Google Scholar] [CrossRef]
- O’Sullivan, A.; Gibney, M.J.; Brennan, L. Dietary intake patterns are reflected in metabolomic profiles: Potential role in dietary assessment studies. Am. J. Clin. Nutr. 2011, 93, 314–321. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, A.; Gibbons, H.; Brennan, L. Metabolomics in the identification of biomarkers of dietary intake. Comput. Struct. Biotechnol. J. 2013, 4, e201301004. [Google Scholar] [CrossRef] [PubMed]
- Riserus, U.; Marklund, M. Milk fat biomarkers and cardiometabolic disease. Curr. Opin. Lipidol. 2017, 28, 46–51. [Google Scholar] [CrossRef]
- Mennen, L.I.; Sapinho, D.; Ito, H.; Bertrais, S.; Galan, P.; Hercberg, S.; Scalbert, A. Urinary flavonoids and phenolic acids as biomarkers of intake for polyphenol-rich foods. Br. J. Nutr. 2006, 96, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Saenger, T.; Hubner, F.; Humpf, H.U. Short-term biomarkers of apple consumption. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef]
- Edmands, W.M.; Ferrari, P.; Rothwell, J.A.; Rinaldi, S.; Slimani, N.; Barupal, D.K.; Biessy, C.; Jenab, M.; Clavel-Chapelon, F.; Fagherazzi, G.; et al. Polyphenol metabolome in human urine and its association with intake of polyphenol-rich foods across European countries. Am. J. Clin. Nutr. 2015, 102, 905–913. [Google Scholar] [CrossRef]
- Makarova, E.; Gornas, P.; Konrade, I.; Tirzite, D.; Cirule, H.; Gulbe, A.; Pugajeva, I.; Seglina, D.; Dambrova, M. Acute anti-hyperglycaemic effects of an unripe apple preparation containing phlorizin in healthy volunteers: A preliminary study. J. Sci. Food Agric. 2015, 95, 560–568. [Google Scholar] [CrossRef]
- Fave, G.; Beckmann, M.; Lloyd, A.J.; Zhou, S.; Harold, G.; Lin, W.; Tailliart, K.; Xie, L.; Draper, J.; Mathers, J.C. Development and validation of a standardized protocol to monitor human dietary exposure by metabolite fingerprinting of urine samples. Metabolomics 2011, 7, 469–484. [Google Scholar] [CrossRef]
- Lloyd, A.J.; Beckmann, M.; Fave, G.; Mathers, J.C.; Draper, J. Proline betaine and its biotransformation products in fasting urine samples are potential biomarkers of habitual citrus fruit consumption. Br. J. Nutr. 2011, 106, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Verhagen, H.; Hageman, G.J.; Rauma, A.L.; Versluis-de Haan, G.; van Herwijnen, M.H.; de Groot, J.; Torronen, R.; Mykkanen, H. Biomonitoring the intake of garlic via urinary excretion of allyl mercapturic acid. Br. J. Nutr. 2001, 86 (Suppl. 1), S111–S114. [Google Scholar] [CrossRef]
- Cerda, B.; Tomas-Barberan, F.A.; Espin, J.C. Metabolism of antioxidant and chemopreventive ellagitannins from strawberries, raspberries, walnuts and oak-aged wine in humans: Identification of biomarkers and individual variability. J. Agric. Food Chem. 2005, 53, 227–235. [Google Scholar] [CrossRef]
- Pratico, G.; Gao, Q.; Scalbert, A.; Vergeres, G.; Kolehmainen, M.; Manach, C.; Brennan, L.; Pedapati, S.H.; Afman, L.A.; Wishart, D.S.; et al. Guidelines for Biomarker of Food Intake Reviews (BFIRev): How to conduct an extensive literature search for biomarker of food intake discovery. Genes Nutr. 2018, 13, 3. [Google Scholar] [CrossRef]
- Corella, D.; Ordovas, J.M. Biomarkers: Background, classification and guidelines for applications in nutritional epidemiology. Nutr. Hosp. 2015, 31 (Suppl. 3), 177–188. [Google Scholar]
- Combs, G.F., Jr. Biomarkers of selenium status. Nutrients 2015, 7, 2209–2236. [Google Scholar] [CrossRef] [PubMed]
- Van Ommen, B.; Keijer, J.; Heil, S.G.; Kaput, J. Challenging homeostasis to define biomarkers for nutrition related health. Mol. Nutr. Food Res. 2009, 53, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Van Ommen, B.; van der Greef, J.; Ordovas, J.M.; Daniel, H. Phenotypic flexibility as key factor in the human nutrition and health relationship. Genes Nutr. 2014, 9, 423. [Google Scholar] [CrossRef]
- Van der Heijden, R.A.; Sheedfar, F.; Morrison, M.C.; Hommelberg, P.P.; Kor, D.; Kloosterhuis, N.J.; Gruben, N.; Youssef, S.A.; de Bruin, A.; Hofker, M.H.; et al. High-fat diet induced obesity primes inflammation in adipose tissue prior to liver in C57BL/6j mice. Aging 2015, 7, 256–268. [Google Scholar] [CrossRef]
- Arab, L. Biomarkers of fat and fatty acid intake. J. Nutr. 2003, 133 (Suppl. 3), 925S–932S. [Google Scholar] [CrossRef]
- Ramos-Lopez, O.; Milagro, F.I.; Allayee, H.; Chmurzynska, A.; Choi, M.S.; Curi, R.; De Caterina, R.; Ferguson, L.R.; Goni, L.; Kang, J.X.; et al. Guide for Current Nutrigenetic, Nutrigenomic and Nutriepigenetic Approaches for Precision Nutrition Involving the Prevention and Management of Chronic Diseases Associated with Obesity. J. Nutrigenet. Nutrigenomics 2017, 10, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Raiten, D.J.; Namaste, S.; Brabin, B.; Combs, G., Jr.; L’Abbe, M.R.; Wasantwisut, E.; Darnton-Hill, I. Executive summary--Biomarkers of Nutrition for Development: Building a Consensus. Am. J. Clin. Nutr. 2011, 94, 633S–650S. [Google Scholar] [CrossRef] [PubMed]
- De Vries, J.; Antoine, J.M.; Burzykowski, T.; Chiodini, A.; Gibney, M.; Kuhnle, G.; Meheust, A.; Pijls, L.; Rowland, I. Markers for nutrition studies: Review of criteria for the evaluation of markers. Eur. J. Nutr. 2013, 52, 1685–1699. [Google Scholar] [CrossRef]
- Marshall, W.J.; Bangert, S.K. Clinical Biochemistry: Metabolic and Clinical Aspects; Churchill Livingstone/Elsevier: London, UK, 2008. [Google Scholar]
- Raghavan, R.; Ashour, F.S.; Bailey, R. A Review of Cutoffs for Nutritional Biomarkers. Adv. Nutr. 2016, 7, 112–120. [Google Scholar] [CrossRef]
- Hossein-nezhad, A.; Holick, M.F. Vitamin D for health: A global perspective. Mayo Clin. Proc. 2013, 88, 720–755. [Google Scholar] [CrossRef]
- O’Connor, M.Y.; Thoreson, C.K.; Ramsey, N.L.; Ricks, M.; Sumner, A.E. The uncertain significance of low vitamin D levels in African descent populations: A review of the bone and cardiometabolic literature. Prog. Cardiovasc. Dis. 2013, 56, 261–269. [Google Scholar] [CrossRef] [PubMed]
- De Mello, V.D.; Kolehmanien, M.; Schwab, U.; Pulkkinen, L.; Uusitupa, M. Gene expression of peripheral blood mononuclear cells as a tool in dietary intervention studies: What do we know so far? Mol. Nutr. Food Res. 2012, 56, 1160–1172. [Google Scholar] [CrossRef]
- Sanchez, J.; Bonet, M.L.; Keijer, J.; van Schothorst, E.M.; Molller, I.; Chetrit, C.; Martinez-Puig, D.; Palou, A. Blood cells transcriptomics as source of potential biomarkers of articular health improvement: Effects of oral intake of a rooster combs extract rich in hyaluronic acid. Genes Nutr. 2014, 9, 417. [Google Scholar] [CrossRef] [PubMed]
- Caimari, A.; Oliver, P.; Rodenburg, W.; Keijer, J.; Palou, A. Feeding conditions control the expression of genes involved in sterol metabolism in peripheral blood mononuclear cells of normoweight and diet-induced (cafeteria) obese rats. J. Nutr. Biochem. 2010, 21, 1127–1133. [Google Scholar] [CrossRef]
- Konieczna, J.; Sanchez, J.; van Schothorst, E.M.; Torrens, J.M.; Bunschoten, A.; Palou, M.; Pico, C.; Keijer, J.; Palou, A. Identification of early transcriptome-based biomarkers related to lipid metabolism in peripheral blood mononuclear cells of rats nutritionally programmed for improved metabolic health. Genes Nutr. 2014, 9, 366. [Google Scholar] [CrossRef]
- Diaz-Rua, R.; Keijer, J.; Caimari, A.; van Schothorst, E.M.; Palou, A.; Oliver, P. Peripheral blood mononuclear cells as a source to detect markers of homeostatic alterations caused by the intake of diets with an unbalanced macronutrient composition. J. Nutr. Biochem. 2015, 26, 398–407. [Google Scholar] [CrossRef]
- Reynes, B.; Klein Hazebroek, M.; Garcia-Ruiz, E.; Keijer, J.; Oliver, P.; Palou, A. Specific Features of the Hypothalamic Leptin Signaling Response to Cold Exposure Are Reflected in Peripheral Blood Mononuclear Cells in Rats and Ferrets. Front. Physiol. 2017, 8, 581. [Google Scholar] [CrossRef] [PubMed]
- Rainen, L.; Oelmueller, U.; Jurgensen, S.; Wyrich, R.; Ballas, C.; Schram, J.; Herdman, C.; Bankaitis-Davis, D.; Nicholls, N.; Trollinger, D.; et al. Stabilization of mRNA expression in whole blood samples. Clin. Chem. 2002, 48, 1883–1890. [Google Scholar]
- Sanchez, J.; Priego, T.; Pico, C.; Ahrens, W.; De Henauw, S.; Fraterman, A.; Marild, S.; Molnar, D.; Moreno, L.A.; Peplies, J.; et al. Blood cells as a source of transcriptional biomarkers of childhood obesity and its related metabolic alterations: Results of the IDEFICS study. J. Clin. Endocrinol. Metab. 2012, 97, E648–E652. [Google Scholar] [CrossRef] [PubMed]
- Min, J.L.; Barrett, A.; Watts, T.; Pettersson, F.H.; Lockstone, H.E.; Lindgren, C.M.; Taylor, J.M.; Allen, M.; Zondervan, K.T.; McCarthy, M.I. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genom. 2010, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Joehanes, R.; Johnson, A.D.; Barb, J.J.; Raghavachari, N.; Liu, P.; Woodhouse, K.A.; O’Donnell, C.J.; Munson, P.J.; Levy, D. Gene expression analysis of whole blood, peripheral blood mononuclear cells and lymphoblastoid cell lines from the Framingham Heart Study. Physiol. Genom. 2012, 44, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Denes, J.; Szabo, E.; Robinette, S.L.; Szatmari, I.; Szonyi, L.; Kreuder, J.G.; Rauterberg, E.W.; Takats, Z. Metabonomics of newborn screening dried blood spot samples: A novel approach in the screening and diagnostics of inborn errors of metabolism. Anal. Chem. 2012, 84, 10113–10120. [Google Scholar] [CrossRef]
- Yuste, S.; Macia, A.; Ludwig, I.A.; Romero, M.P.; Fernandez-Castillejo, S.; Catalan, U.; Motilva, M.J.; Rubio, L. Validation of Dried Blood Spot Cards to Determine Apple Phenolic Metabolites in Human Blood and Plasma After an Acute Intake of Red-Fleshed Apple Snack. Mol. Nutr. Food Res. 2018, 62, e1800623. [Google Scholar] [CrossRef] [PubMed]
- Hewawasam, E.; Liu, G.; Jeffery, D.W.; Muhlhausler, B.S.; Gibson, R.A. A stable method for routine analysis of oxylipins from dried blood spots using ultra-high performance liquid chromatography-tandem mass spectrometry. Prostaglandins Leukot. Essent. Fatty Acids 2018, 137, 12–18. [Google Scholar] [CrossRef]
- Yu, T.; Han, Y.; Pu, X.; Duan, H.; Zhi, L.; Zhang, Y.; Lei, L.; Zhao, Y.; Cao, Q.; Xu, H. Performance of LC/MS/MS in Analyzing Multiple Amino Acids and Acylcarnitines in Dried Blood Spot of Very Low Birth Weight, Low Birth Weight and Normal Weight Neonates. Clin. Lab. 2018, 64, 1333–1339. [Google Scholar] [CrossRef]
- Maleska, A.; Hirtz, C.; Casteleyn, E.; Villard, O.; Ducos, J.; Avignon, A.; Sultan, A.; Lehmann, S. Comparison of HbA1c detection in whole blood and dried blood spots using an automated ion-exchange HPLC system. Bioanalysis 2017, 9, 427–434. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Scalia, F.; Marino Gammazza, A.; Carlisi, D.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C. Extracellular Vesicle-Mediated Cell(-)Cell Communication in the Nervous System: Focus on Neurological Diseases. Int. J. Mol. Sci. 2019, 20, 434. [Google Scholar] [CrossRef]
- Shi, M.; Sheng, L.; Stewart, T.; Zabetian, C.P.; Zhang, J. New windows into the brain: Central nervous system-derived extracellular vesicles in blood. Prog. Neurobiol. 2019, 175, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Torrano, V.; Royo, F.; Peinado, H.; Loizaga-Iriarte, A.; Unda, M.; Falcon-Perez, J.M.; Carracedo, A. Vesicle-MaNiA: Extracellular vesicles in liquid biopsy and cancer. Curr. Opin. Pharmacol. 2016, 29, 47–53. [Google Scholar] [CrossRef]
- Jansen, F.; Nickenig, G.; Werner, N. Extracellular Vesicles in Cardiovascular Disease: Potential Applications in Diagnosis, Prognosis and Epidemiology. Circ. Res. 2017, 120, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Cesare Marincola, F.; Dessì, A.; Corbu, S.; Reali, A.; Fanos, V. Clinical impact of human breast milk metabolomics. Clin. Chim. Acta 2015, 451 (Pt A), 103–106. [Google Scholar] [CrossRef]
- De Vries, J.Y.; Pundir, S.; Mckenzie, E.; Keijer, J.; Kussmann, M. Maternal Circulating Vitamin Status and Colostrum Vitamin Composition in Healthy Lactating Women-A Systematic Approach. Nutrients 2018, 10, 687. [Google Scholar] [CrossRef] [PubMed]
- Leghi, G.E.; Middleton, P.F.; Muhlhausler, B.S. A methodological approach to identify the most reliable human milk collection method for compositional analysis: A systematic review protocol. Syst. Rev. 2018, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Palou, M.; Picó, C.; Palou, A. Leptin as a breast milk component for the prevention of obesity. Nutr. Rev. 2018, 76, 875–892. [Google Scholar] [CrossRef]
- Servera, M.; López, N.; Zamanillo, R.; Picó, C.; Palou, A.; Serra, F. Nutrigenomics and Breast Milk, Perspectives in Obesity. In Lactation: Natural Processes, Physiological Responses and Role in Maternity; Lisa, M., Cruz, R., Ortiz, D.C., Eds.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2012; ISBN 978-1-62257-243-4. [Google Scholar]
- Bravi, F.; Wiens, F.; Decarli, A.; Dal Pont, A.; Agostoni, C.; Ferraroni, M. Impact of maternal nutrition on breast-milk composition: A systematic review. Am. J. Clin. Nutr. 2016, 104, 646–662. [Google Scholar] [CrossRef]
- Patel, M.S.; Srinivasan, M. Metabolic programming in the immediate postnatal life. Ann. Nutr. Metab. 2011, 58 (Suppl. 2), 18–28. [Google Scholar] [CrossRef] [PubMed]
- Palou, M.; Torrens, J.M.; Castillo, P.; Sánchez, J.; Palou, A.; Picó, C. Metabolomic approach in milk from calorie-restricted rats during lactation: A potential link to the programming of a healthy phenotype in offspring. Eur. J. Nutr. 2019. [Google Scholar] [CrossRef]
- Rechner, A.R.; Kuhnle, G.; Hu, H.; Roedig-Penman, A.; van den Braak, M.H.; Moore, K.P.; Rice-Evans, C.A. The metabolism of dietary polyphenols and the relevance to circulating levels of conjugated metabolites. Free Radic. Res. 2002, 36, 1229–1241. [Google Scholar] [CrossRef]
- Cerda, B.; Periago, P.; Espin, J.C.; Tomas-Barberan, F.A. Identification of urolithin a as a metabolite produced by human colon microflora from ellagic acid and related compounds. J. Agric. Food Chem. 2005, 53, 5571–5576. [Google Scholar] [CrossRef]
- Cogswell, M.E.; Maalouf, J.; Elliott, P.; Loria, C.M.; Patel, S.; Bowman, B.A. Use of Urine Biomarkers to Assess Sodium Intake: Challenges and Opportunities. Annu. Rev. Nutr. 2015, 35, 349–387. [Google Scholar] [CrossRef]
- Khamis, M.M.; Adamko, D.J.; El-Aneed, A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom. Rev. 2017, 36, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Wierdsma, N.J.; Peters, J.H.; van Bokhorst-de van der Schueren, M.A.; Mulder, C.J.; Metgod, I.; van Bodegraven, A.A. Bomb calorimetry, the gold standard for assessment of intestinal absorption capacity: Normative values in healthy ambulant adults. J. Hum. Nutr. Diet. 2014, 27 (Suppl. 2), 57–64. [Google Scholar] [CrossRef]
- Rivero-Marcotegui, A.; Olivera-Olmedo, J.E.; Valverde-Visus, F.S.; Palacios-Sarrasqueta, M.; Grijalba-Uche, A.; Garcia-Merlo, S. Water, fat, nitrogen and sugar content in feces: Reference intervals in children. Clin. Chem. 1998, 44, 1540–1544. [Google Scholar]
- Bjorkman, L.; Sandborgh-Englund, G.; Ekstrand, J. Mercury in saliva and feces after removal of amalgam fillings. Toxicol. Appl. Pharmacol. 1997, 144, 156–162. [Google Scholar] [CrossRef]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on Gut Microbiota Associated to Mediterranean Diet Adherence and Specific Dietary Intakes on General Adult Population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef]
- De la Cuesta-Zuluaga, J.; Mueller, N.T.; Alvarez-Quintero, R.; Velasquez-Mejia, E.P.; Sierra, J.A.; Corrales-Agudelo, V.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Higher Fecal Short-Chain Fatty Acid Levels Are Associated with Gut Microbiome Dysbiosis, Obesity, Hypertension and Cardiometabolic Disease Risk Factors. Nutrients 2018, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Gart, E.; Souto Lima, E.; Schuren, F.; de Ruiter, C.G.F.; Attema, J.; Verschuren, L.; Keijer, J.; Salic, K.; Morrison, M.C.; Kleemann, R. Diet-Independent Correlations between Bacteria and Dysfunction of Gut, Adipose Tissue and Liver: A Comprehensive Microbiota Analysis in Feces and Mucosa of the Ileum and Colon in Obese Mice with NAFLD. Int. J. Mol. Sci. 2018, 20, 1. [Google Scholar] [CrossRef]
- Colston, J.M.; Penataro Yori, P.; Colantuoni, E.; Moulton, L.H.; Ambikapathi, R.; Lee, G.; Rengifo Trigoso, D.; Siguas Salas, M.; Kosek, M.N. A methodologic framework for modeling and assessing biomarkers of environmental enteropathy as predictors of growth in infants: An example from a Peruvian birth cohort. Am. J. Clin. Nutr. 2017, 106, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Matysika, S.; Le Royb, C.I.; Liebischa, G.; Claus, S.P. Metabolomics of fecal samples: A practical consideration. Trends Food Sci. Technol. 2016, 57, 244–255. [Google Scholar] [CrossRef]
- Kolk, A.H.; van Berkel, J.J.; Claassens, M.M.; Walters, E.; Kuijper, S.; Dallinga, J.W.; van Schooten, F.J. Breath analysis as a potential diagnostic tool for tuberculosis. Int. J. Tuberc. Lung Dis. 2012, 16, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Smolinska, A.; Bodelier, A.G.; Dallinga, J.W.; Masclee, A.A.; Jonkers, D.M.; van Schooten, F.J.; Pierik, M.J. The potential of volatile organic compounds for the detection of active disease in patients with ulcerative colitis. Aliment. Pharmacol. Ther. 2017, 45, 1244–1254. [Google Scholar] [CrossRef] [PubMed]
- Montuschi, P.; Santini, G.; Mores, N.; Vignoli, A.; Macagno, F.; Shoreh, R.; Tenori, L.; Zini, G.; Fuso, L.; Mondino, C.; et al. Breathomics for Assessing the Effects of Treatment and Withdrawal with Inhaled Beclomethasone/Formoterol in Patients With COPD. Front. Pharmacol. 2018, 9, 258. [Google Scholar] [CrossRef]
- Smolinska, A.; Baranska, A.; Dallinga, J.W.; Mensink, R.P.; Baumgartner, S.; van de Heijning, B.J.M.; van Schooten, F.J. Comparing patterns of volatile organic compounds exhaled in breath after consumption of two infant formulae with a different lipid structure: A randomized trial. Sci. Rep. 2019, 9, 554. [Google Scholar] [CrossRef]
- Hageman, J.H.J.; Nieuwenhuizen, A.G.; van Ruth, S.M.; Hageman, J.A.; Keijer, J. Application of volatile organic compound (VOC) analysis in a nutritional intervention study: differential responses during five hours following consumption of a high- and a low-fat dairy drink. 2019; Submitted for publication. [Google Scholar]
- Desai, G.S.; Mathews, S.T. Saliva as a non-invasive diagnostic tool for inflammation and insulin-resistance. World J. Diabetes 2014, 5, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Azkargorta, M.; Soria, J.; Acera, A.; Iloro, I.; Elortza, F. Human tear proteomics and peptidomics in ophthalmology: Toward the translation of proteomic biomarkers into clinical practice. J. Proteom. 2017, 150, 359–367. [Google Scholar] [CrossRef]
- Balne, P.K.; Au, V.B.; Tong, L.; Ghosh, A.; Agrawal, M.; Connolly, J.; Agrawal, R. Bead Based Multiplex Assay for Analysis of Tear Cytokine Profiles. J. Vis. Exp. 2017, 128. [Google Scholar] [CrossRef] [PubMed]
- Walter, S.D.; Gronert, K.; McClellan, A.L.; Levitt, R.C.; Sarantopoulos, K.D.; Galor, A. ω-3 Tear Film Lipids Correlate with Clinical Measures of Dry Eye. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2472–2478. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Di Ilio, C.; Sacchetta, P.; Del Boccio, P. Unraveling the molecular repertoire of tears as a source of biomarkers: Beyond ocular diseases. Proteomics Clin. Appl. 2015, 9, 169–186. [Google Scholar] [CrossRef]
- Cicalini, I.; Rossi, C.; Pieragostino, D.; Agnifili, L.; Mastropasqua, L.; di Ioia, M.; De Luca, G.; Onofrj, M.; Federici, L.; Del Boccio, P. Integrated Lipidomics and Metabolomics Analysis of Tears in Multiple Sclerosis: An Insight into Diagnostic Potential of Lacrimal Fluid. Int. J. Mol. Sci. 2019, 20, 1265. [Google Scholar] [CrossRef] [PubMed]
- Hagan, S.; Martin, E.; Enriquez-de-Salamanca, A. Tear fluid biomarkers in ocular and systemic disease: Potential use for predictive, preventive and personalised medicine. EPMA J. 2016, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. 2018, 15, 68. [Google Scholar] [CrossRef] [PubMed]
- Cappelle, D.; Neels, H.; De Keukeleire, S.; Fransen, E.; Dom, G.; Vermassen, A.; Covaci, A.; Crunelle, C.L.; van Nuijs, A.L.N. Ethyl glucuronide in keratinous matrices as biomarker of alcohol use: A correlation study between hair and nails. Forensic Sci. Int. 2017, 279, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Katz, S.A.; Katz, R.B. Use of hair analysis for evaluating mercury intoxication of the human body: A review. J. Appl. Toxicol. 1992, 12, 79–84. [Google Scholar] [CrossRef]
- Golasik, M.; Przybylowicz, A.; Wozniak, A.; Herman, M.; Gawecki, W.; Golusinski, W.; Walas, S.; Krejpcio, Z.; Szyfter, K.; Florek, E.; et al. Essential metals profile of the hair and nails of patients with laryngeal cancer. J. Trace Elem. Med. Biol. 2015, 31, 67–73. [Google Scholar] [CrossRef]
- King, J.C.; Brown, K.H.; Gibson, R.S.; Krebs, N.F.; Lowe, N.M.; Siekmann, J.H.; Raiten, D.J. Biomarkers of Nutrition for Development (BOND)-Zinc Review. J. Nutr. 2016. [Google Scholar] [CrossRef]
- Nowak, B.; Chmielnicka, J. Relationship of lead and cadmium to essential elements in hair, teeth and nails of environmentally exposed people. Ecotoxicol. Environ. Saf. 2000, 46, 265–274. [Google Scholar] [CrossRef]
- Ryynanen, J.; Neme, A.; Tuomainen, T.P.; Virtanen, J.K.; Voutilainen, S.; Nurmi, T.; de Mello, V.D.; Uusitupa, M.; Carlberg, C. Changes in vitamin D target gene expression in adipose tissue monitor the vitamin D response of human individuals. Mol. Nutr. Food Res. 2014, 58, 2036–2045. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.; Portune, K.J.; Steuer, N.; Lan, A.; Cerrudo, V.; Audebert, M.; Dumont, F.; Mancano, G.; Khodorova, N.; Andriamihaja, M.; et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: A randomized, parallel, double-blind trial in overweight humans. Am. J. Clin. Nutr. 2017, 106, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Schrezenmeir, J.; Fenselau, S.; Keppler, I.; Abel, J.; Orth, B.; Laue, C.; Sturmer, W.; Fauth, U.; Halmagyi, M.; Marz, W. Postprandial triglyceride high response and the metabolic syndrome. Ann. N. Y. Acad. Sci. 1997, 827, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Horakova, O.; Medrikova, D.; van Schothorst, E.M.; Bunschoten, A.; Flachs, P.; Kus, V.; Kuda, O.; Bardova, K.; Janovska, P.; Hensler, M.; et al. Preservation of metabolic flexibility in skeletal muscle by a combined use of n-3 PUFA and rosiglitazone in dietary obese mice. PLoS ONE 2012, 7, e43764. [Google Scholar] [CrossRef] [PubMed]
- Duivenvoorde, L.P.; van Schothorst, E.M.; Swarts, H.M.; Kuda, O.; Steenbergh, E.; Termeulen, S.; Kopecky, J.; Keijer, J. A Difference in Fatty Acid Composition of Isocaloric High-Fat Diets Alters Metabolic Flexibility in Male C57BL/6JOlaHsd Mice. PLoS ONE 2015, 10, e0128515. [Google Scholar] [CrossRef] [PubMed]
- Wopereis, S.; Stroeve, J.H.M.; Stafleu, A.; Bakker, G.C.M.; Burggraaf, J.; van Erk, M.J.; Pellis, L.; Boessen, R.; Kardinaal, A.A.F.; van Ommen, B. Multi-parameter comparison of a standardized mixed meal tolerance test in healthy and type 2 diabetic subjects: The PhenFlex challenge. Genes Nutr. 2017, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Duivenvoorde, L.P.; van Schothorst, E.M.; Derous, D.; van der Stelt, I.; Masania, J.; Rabbani, N.; Thornalley, P.J.; Keijer, J. Oxygen restriction as challenge test reveals early high-fat-diet-induced changes in glucose and lipid metabolism. Pflug. Arch. 2015, 467, 1179–1193. [Google Scholar] [CrossRef]
- Duivenvoorde, L.P.; van Schothorst, E.M.; Swarts, H.J.; Keijer, J. Assessment of metabolic flexibility of old and adult mice using three noninvasive, indirect calorimetry-based treatments. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 282–293. [Google Scholar] [CrossRef]
- Oliver, P.; Reynes, B.; Caimari, A.; Palou, A. Peripheral blood mononuclear cells: A potential source of homeostatic imbalance markers associated with obesity development. Pflug. Arch. 2013, 465, 459–468. [Google Scholar] [CrossRef]
- Mathers, J.C. Nutrigenomics in the modern era. Proc. Nutr. Soc. 2017, 76, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Keijer, J.; van Helden, Y.G.; Bunschoten, A.; van Schothorst, E.M. Transcriptome analysis in benefit-risk assessment of micronutrients and bioactive food components. Mol. Nutr. Food Res. 2010, 54, 240–248. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, A.; Brennan, L. The role of metabolomics in determination of new dietary biomarkers. Proc. Nutr. Soc. 2017, 76, 295–302. [Google Scholar] [CrossRef]
- Van Duynhoven, J.P.M.; Jacobs, D.M. Assessment of dietary exposure and effect in humans: The role of NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 96, 58–72. [Google Scholar] [CrossRef]
- Qiu, C.; Kaplan, C.D. Functional assays for transcription mechanisms in high-throughput. Methods 2019. [Google Scholar] [CrossRef]
- Carneiro, G.; Radcenco, A.L.; Evaristo, J.; Monnerat, G. Novel strategies for clinical investigation and biomarker discovery: A guide to applied metabolomics. Horm. Mol. Biol. Clin. Investig. 2019. [Google Scholar] [CrossRef]
- Stuart, T.; Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 2019. [Google Scholar] [CrossRef]
- Munoz Garcia, A.; Eijssen, L.M.; Kutmon, M.; Sarathy, C.; Cengo, A.; Hewison, M.; Evelo, C.T.; Lenz, M.; Coort, S.L. A bioinformatics workflow to decipher transcriptomic data from vitamin D studies. J. Steroid Biochem. Mol. Biol. 2019, 189, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Fito, M.; Melander, O.; Martinez, J.A.; Toledo, E.; Carpene, C.; Corella, D. Advances in Integrating Traditional and Omic Biomarkers When Analyzing the Effects of the Mediterranean Diet Intervention in Cardiovascular Prevention. Int. J. Mol. Sci. 2016, 17, 1469. [Google Scholar] [CrossRef]
- Odriozola, L.; Corrales, F.J. Discovery of nutritional biomarkers: Future directions based on omics technologies. Int. J. Food Sci. Nutr. 2015, 66 (Suppl. 1), S31–S40. [Google Scholar] [CrossRef]
- Enattah, N.S.; Sahi, T.; Savilahti, E.; Terwilliger, J.D.; Peltonen, L.; Jarvela, I. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 2002, 30, 233–237. [Google Scholar] [CrossRef]
- Almon, R.; Patterson, E.; Nilsson, T.K.; Engfeldt, P.; Sjostrom, M. Body fat and dairy product intake in lactase persistent and non-persistent children and adolescents. Food Nutr. Res. 2010, 54. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; Byrne, E.M.; Esko, T.; Nalls, M.A.; Ganna, A.; Paynter, N.; Monda, K.L.; Amin, N.; Fischer, K.; Renstrom, F.; et al. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol. Psychiatry 2015, 20, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Ngwa, J.S.; van Rooij, F.J.; Zillikens, M.C.; Wojczynski, M.K.; Frazier-Wood, A.C.; Houston, D.K.; Kanoni, S.; Lemaitre, R.N.; Luan, J.; et al. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am. J. Clin. Nutr. 2013, 97, 1395–1402. [Google Scholar] [CrossRef]
- Dashti, H.S.; Shea, M.K.; Smith, C.E.; Tanaka, T.; Hruby, A.; Richardson, K.; Wang, T.J.; Nalls, M.A.; Guo, X.; Liu, Y.; et al. Meta-analysis of genome-wide association studies for circulating phylloquinone concentrations. Am. J. Clin. Nutr. 2014, 100, 1462–1469. [Google Scholar] [CrossRef]
- Erkkila, A.T.; Booth, S.L.; Hu, F.B.; Jacques, P.F.; Lichtenstein, A.H. Phylloquinone intake and risk of cardiovascular diseases in men. Nutr. Metab. Cardiovasc. Dis. 2007, 17, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M. Genotype-phenotype associations: Modulation by diet and obesity. Obesity 2008, 16 (Suppl. 3), S40–S46. [Google Scholar] [CrossRef]
- Pena-Romero, A.C.; Navas-Carrillo, D.; Marin, F.; Orenes-Pinero, E. The future of nutrition: Nutrigenomics and nutrigenetics in obesity and cardiovascular diseases. Crit. Rev. Food Sci. Nutr. 2018, 58, 3030–3041. [Google Scholar] [CrossRef]
- Day, K.J.; Adamski, M.M.; Dordevic, A.L.; Murgia, C. Genetic Variations as Modifying Factors to Dietary Zinc Requirements-A Systematic Review. Nutrients 2017, 9, 148. [Google Scholar] [CrossRef]
- Galmes, S.; Serra, F.; Palou, A. Vitamin E Metabolic Effects and Genetic Variants: A Challenge for Precision Nutrition in Obesity and Associated Disturbances. Nutrients 2018, 10, 1919. [Google Scholar] [CrossRef]
- Meplan, C. Selenium and chronic diseases: A nutritional genomics perspective. Nutrients 2015, 7, 3621–3651. [Google Scholar] [CrossRef]
- Steinbrenner, H.; Speckmann, B.; Klotz, L.O. Selenoproteins: Antioxidant selenoenzymes and beyond. Arch. Biochem. Biophys. 2016, 595, 113–119. [Google Scholar] [CrossRef]
- Kipp, A.; Banning, A.; van Schothorst, E.M.; Meplan, C.; Schomburg, L.; Evelo, C.; Coort, S.; Gaj, S.; Keijer, J.; Hesketh, J.; et al. Four selenoproteins, protein biosynthesis and Wnt signalling are particularly sensitive to limited selenium intake in mouse colon. Mol. Nutr. Food Res. 2009, 53, 1561–1572. [Google Scholar] [CrossRef]
- Combs, G.F., Jr.; Watts, J.C.; Jackson, M.I.; Johnson, L.K.; Zeng, H.; Scheett, A.J.; Uthus, E.O.; Schomburg, L.; Hoeg, A.; Hoefig, C.S.; et al. Determinants of selenium status in healthy adults. Nutr. J. 2011, 10, 75. [Google Scholar] [CrossRef]
- Portha, B.; Fournier, A.; Kioon, M.D.; Mezger, V.; Movassat, J. Early environmental factors, alteration of epigenetic marks and metabolic disease susceptibility. Biochimie 2014, 97, 1–15. [Google Scholar] [CrossRef]
- De Mello, V.D.; Pulkkinen, L.; Lalli, M.; Kolehmainen, M.; Pihlajamaki, J.; Uusitupa, M. DNA methylation in obesity and type 2 diabetes. Ann. Med. 2014, 46, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Palou, M.; Pico, C.; McKay, J.A.; Sanchez, J.; Priego, T.; Mathers, J.C.; Palou, A. Protective effects of leptin during the suckling period against later obesity may be associated with changes in promoter methylation of the hypothalamic pro-opiomelanocortin gene. Br. J. Nutr. 2011, 106, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Chaplin, A.; Palou, A.; Serra, F. Methylation analysis in fatty-acid-related genes reveals their plasticity associated with conjugated linoleic acid and calcium supplementation in adult mice. Eur. J. Nutr. 2017, 56, 879–891. [Google Scholar] [CrossRef]
- Park, J.H.; Stoffers, D.A.; Nicholls, R.D.; Simmons, R.A. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J. Clin. Investig. 2008, 118, 2316–2324. [Google Scholar] [CrossRef]
- Kasch, J.; Kanzleiter, I.; Saussenthaler, S.; Schurmann, A.; Keijer, J.; van Schothorst, E.; Klaus, S.; Schumann, S. Insulin sensitivity linked skeletal muscle Nr4a1 DNA methylation is programmed by the maternal diet and modulated by voluntary exercise in mice. J. Nutr. Biochem. 2018, 57, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Waterland, R.A.; Kellermayer, R.; Rached, M.T.; Tatevian, N.; Gomes, M.V.; Zhang, J.; Zhang, L.; Chakravarty, A.; Zhu, W.; Laritsky, E.; et al. Epigenomic profiling indicates a role for DNA methylation in early postnatal liver development. Hum. Mol. Genet. 2009, 18, 3026–3038. [Google Scholar] [CrossRef]
- Burton, M.A.; Lillycrop, K.A. Nutritional modulation of the epigenome and its implication for future health. Proc. Nutr. Soc. 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Waterland, R.A.; Kellermayer, R.; Laritsky, E.; Rayco-Solon, P.; Harris, R.A.; Travisano, M.; Zhang, W.; Torskaya, M.S.; Zhang, J.; Shen, L.; et al. Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 2010, 6, e1001252. [Google Scholar] [CrossRef]
- Dominguez-Salas, P.; Moore, S.E.; Baker, M.S.; Bergen, A.W.; Cox, S.E.; Dyer, R.A.; Fulford, A.J.; Guan, Y.; Laritsky, E.; Silver, M.J.; et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat. Commun. 2014, 5, 3746. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef]
- Tobi, E.W.; Lumey, L.H.; Talens, R.P.; Kremer, D.; Putter, H.; Stein, A.D.; Slagboom, P.E.; Heijmans, B.T. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet. 2009, 18, 4046–4053. [Google Scholar] [CrossRef]
- Gillberg, L.; Orskov, A.D.; Liu, M.; Harslof, L.B.S.; Jones, P.A.; Gronbaek, K. Vitamin C - A new player in regulation of the cancer epigenome. Semin. Cancer Biol. 2018, 51, 59–67. [Google Scholar] [CrossRef]
- Janssen, J.J.E.; Grefte, S.; Keijer, J.; de Boer, V.C.J. Mito-Nuclear Communication by Mitochondrial Metabolites and Its Regulation by B-Vitamins. Front. Physiol. 2019, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, R.; Kupkova, K.; Shetty, S.J.; Linford, A.S.; Pray-Grant, M.G.; Wagar, L.E.; Davis, M.M.; Haque, R.; Gaultier, A.; Mayo, M.W.; et al. Histone H3 lysine 4 methylation signature associated with human undernutrition. Proc. Natl. Acad. Sci. USA 2018, 115, E11264–E11273. [Google Scholar] [CrossRef]
- McDonnell, E.; Crown, S.B.; Fox, D.B.; Kitir, B.; Ilkayeva, O.R.; Olsen, C.A.; Grimsrud, P.A.; Hirschey, M.D. Lipids Reprogram Metabolism to Become a Major Carbon Source for Histone Acetylation. Cell Rep. 2016, 17, 1463–1472. [Google Scholar] [CrossRef]
- Denu, J.M. Vitamin B3 and sirtuin function. Trends Biochem. Sci. 2005, 30, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Timmers, S.; Konings, E.; Bilet, L.; Houtkooper, R.H.; van de Weijer, T.; Goossens, G.H.; Hoeks, J.; van der Krieken, S.; Ryu, D.; Kersten, S.; et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011, 14, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Bo, S.; Togliatto, G.; Gambino, R.; Ponzo, V.; Lombardo, G.; Rosato, R.; Cassader, M.; Brizzi, M.F. Impact of sirtuin-1 expression on H3K56 acetylation and oxidative stress: A double-blind randomized controlled trial with resveratrol supplementation. Acta Diabetol. 2018, 55, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Platt, A.R.; Jones, D.R.; Reifenberger, J.G.; Sass, L.E.; McInerney, P.; Thompson, J.F.; Bowers, J.; Jarosz, M.; Milos, P.M. Direct RNA sequencing. Nature 2009, 461, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Bodian, D.L.; Kothiyal, P.; Hauser, N.S. Pitfalls of clinical exome and gene panel testing: Alternative transcripts. Genet. Med. 2018. [Google Scholar] [CrossRef]
- Koch, C.M.; Chiu, S.F.; Akbarpour, M.; Bharat, A.; Ridge, K.M.; Bartom, E.T.; Winter, D.R. A Beginner’s Guide to Analysis of RNA Sequencing Data. Am. J. Respir. Cell Mol. Biol. 2018, 59, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.C.; Yao, J.; Ho, K.S.; Lambowitz, A.M.; Wilke, C.O. Limitations of alignment-free tools in total RNA-seq quantification. BMC Genomics 2018, 19, 510. [Google Scholar] [CrossRef]
- Dal Molin, A.; Di Camillo, B. How to design a single-cell RNA-sequencing experiment: Pitfalls, challenges and perspectives. Brief. Bioinform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kussmann, M.; Raymond, F.; Affolter, M. OMICS-driven biomarker discovery in nutrition and health. J. Biotechnol. 2006, 124, 758–787. [Google Scholar] [CrossRef]
- Corella, D.; Coltell, O.; Macian, F.; Ordovas, J.M. Advances in Understanding the Molecular Basis of the Mediterranean Diet Effect. Annu. Rev. Food Sci. Technol. 2018, 9, 227–249. [Google Scholar] [CrossRef]
- Mohr, S.; Liew, C.C. The peripheral-blood transcriptome: New insights into disease and risk assessment. Trends Mol. Med. 2007, 13, 422–432. [Google Scholar] [CrossRef]
- Sanchez, J.; Pico, C.; Ahrens, W.; Foraita, R.; Fraterman, A.; Moreno, L.A.; Russo, P.; Siani, A.; Palou, A. Transcriptome analysis in blood cells from children reveals potential early biomarkers of metabolic alterations. Int. J. Obes. 2017, 41, 1481–1488. [Google Scholar] [CrossRef]
- Bouchard-Mercier, A.; Paradis, A.M.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.C. Associations between dietary patterns and gene expression profiles of healthy men and women: A cross-sectional study. Nutr. J. 2013, 12, 24. [Google Scholar] [CrossRef]
- Afman, L.; Milenkovic, D.; Roche, H.M. Nutritional aspects of metabolic inflammation in relation to health--insights from transcriptomic biomarkers in PBMC of fatty acids and polyphenols. Mol. Nutr. Food Res. 2014, 58, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Priego, T.; Sanchez, J.; Pico, C.; Ahrens, W.; De Henauw, S.; Kourides, Y.; Lissner, L.; Molnar, D.; Moreno, L.A.; Russo, P.; et al. TAS1R3 and UCN2 Transcript Levels in Blood Cells Are Associated with Sugary and Fatty Food Consumption in Children. J. Clin. Endocrinol. Metab. 2015, 100, 3556–3564. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Rando, O.J. Metabolic Inputs into the Epigenome. Cell Metab. 2017, 25, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Lorente-Cebrian, S.; Gonzalez-Muniesa, P.; Milagro, F.I.; Martinez, J.A. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: Emerging roles as biomarkers and therapeutic targets. Clin. Sci. 2019, 133, 23–40. [Google Scholar] [CrossRef]
- Sun, W.; Shi, Y.; Wang, Z.; Zhang, J.; Cai, H.; Zhang, J.; Huang, D. Interaction of long-chain non-coding RNAs and important signaling pathways on human cancers (Review). Int. J. Oncol. 2018, 53, 2343–2355. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen Martinez-Jimenez, V.; Mendez-Mancilla, A.; Patricia Portales-Perez, D. miRNAs in nutrition, obesity and cancer: The biology of miRNAs in metabolic disorders and its relationship with cancer development. Mol. Nutr. Food Res. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.A.; Davis, C.D. The emerging role of microRNAs and nutrition in modulating health and disease. Annu. Rev. Nutr. 2014, 34, 305–336. [Google Scholar] [CrossRef]
- Cui, J.; Zhou, B.; Ross, S.A.; Zempleni, J. Nutrition, microRNAs and Human Health. Adv. Nutr. 2017, 8, 105–112. [Google Scholar] [CrossRef]
- Parra, P.; Serra, F.; Palou, A. Expression of adipose microRNAs is sensitive to dietary conjugated linoleic acid treatment in mice. PLoS ONE 2010, 5, e13005. [Google Scholar] [CrossRef]
- Ortega, F.J.; Cardona-Alvarado, M.I.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramirez-Chavez, E.; Ricart, W.; Molina-Torres, J.; et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef]
- Zempleni, J.; Aguilar-Lozano, A.; Sadri, M.; Sukreet, S.; Manca, S.; Wu, D.; Zhou, F.; Mutai, E. Biological Activities of Extracellular Vesicles and Their Cargos from Bovine and Human Milk in Humans and Implications for Infants. J. Nutr. 2017, 147, 3–10. [Google Scholar] [CrossRef]
- Pomar, C.A.; Castro, H.; Picó, C.; Serra, F.; Palou, A.; Sánchez, J. Cafeteria Diet Consumption during Lactation in Rats, Rather than Obesity Per Se, alters miR-222, miR-200a and miR-26a Levels in Milk. Mol. Nutr. Food Res. 2019, 63, e1800928. [Google Scholar] [CrossRef]
- Barderas, M.G.; Vivanco, F.; Alvarez-Llamas, G. Vascular proteomics. Methods Mol. Biol. 2013, 1000, 1–20. [Google Scholar] [CrossRef]
- De Roos, B.; Duivenvoorden, I.; Rucklidge, G.; Reid, M.; Ross, K.; Lamers, R.J.; Voshol, P.J.; Havekes, L.M.; Teusink, B. Response of apolipoprotein E*3-Leiden transgenic mice to dietary fatty acids: Combining liver proteomics with physiological data. FASEB J. 2005, 19, 813–815. [Google Scholar] [CrossRef]
- Zhang, X.; Yap, Y.; Wei, D.; Chen, G.; Chen, F. Novel omics technologies in nutrition research. Biotechnol. Adv. 2008, 26, 169–176. [Google Scholar] [CrossRef]
- Cominetti, O.; Nunez Galindo, A.; Corthesy, J.; Oller Moreno, S.; Irincheeva, I.; Valsesia, A.; Astrup, A.; Saris, W.H.; Hager, J.; Kussmann, M.; et al. Proteomic Biomarker Discovery in 1000 Human Plasma Samples with Mass Spectrometry. J. Proteome Res. 2016, 15, 389–399. [Google Scholar] [CrossRef]
- Curran, A.M.; Fogarty Draper, C.; Scott-Boyer, M.P.; Valsesia, A.; Roche, H.M.; Ryan, M.F.; Gibney, M.J.; Kutmon, M.; Evelo, C.T.; Coort, S.L.; et al. Sexual Dimorphism, Age and Fat Mass Are Key Phenotypic Drivers of Proteomic Signatures. J. Proteome Res. 2017, 16, 4122–4133. [Google Scholar] [CrossRef]
- Chiu, C.J.; Rabbani, N.; Rowan, S.; Chang, M.L.; Sawyer, S.; Hu, F.B.; Willett, W.; Thornalley, P.J.; Anwar, A.; Bar, L.; et al. Studies of advanced glycation end products and oxidation biomarkers for type 2 diabetes. Biofactors 2018, 44, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Schroll, M.M.; Hummon, A.B. Employing proteomics to understand the effects of nutritional intervention in cancer treatment. Anal. Bioanal. Chem. 2018, 410, 6371–6386. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, H.M.; McDonald, M.R.; Sullivan, J.A.; Tsao, R.; Meckling, K.A. Proteomic Profiles of Adipose and Liver Tissues from an Animal Model of Metabolic Syndrome Fed Purple Vegetables. Nutrients 2018, 10, 456. [Google Scholar] [CrossRef]
- Senechal, S.; Kussmann, M. Nutriproteomics: Technologies and applications for identification and quantification of biomarkers and ingredients. Proc. Nutr. Soc. 2011, 70, 351–364. [Google Scholar] [CrossRef]
- Marshall, J.; Bowden, P.; Schmit, J.C.; Betsou, F. Creation of a federated database of blood proteins: A powerful new tool for finding and characterizing biomarkers in serum. Clin. Proteomics 2014, 11, 3. [Google Scholar] [CrossRef]
- Newgard, C.B. Metabolomics and Metabolic Diseases: Where Do We Stand? Cell Metab. 2017, 25, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Shaham, O.; Wei, R.; Wang, T.J.; Ricciardi, C.; Lewis, G.D.; Vasan, R.S.; Carr, S.A.; Thadhani, R.; Gerszten, R.E.; Mootha, V.K. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol. Syst. Biol. 2008, 4, 214. [Google Scholar] [CrossRef]
- Bertram, H.C.; Jakobsen, L.M.A. Nutrimetabolomics: Integrating metabolomics in nutrition to disentangle intake of animal-based foods. Metabolomics 2018, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.; McNamara, A.E.; Brennan, L. Role of metabolomics in identification of biomarkers related to food intake. Proc. Nutr. Soc. 2019, 1–8. [Google Scholar] [CrossRef]
- Brennan, L.; Hu, F.B. Metabolomics-Based Dietary Biomarkers in Nutritional Epidemiology-Current Status and Future Opportunities. Mol. Nutr. Food Res. 2019, 63, e1701064. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.; Keski-Rahkonen, P.; Assi, N.; Ferrari, P.; Freisling, H.; Rinaldi, S.; Slimani, N.; Zamora-Ros, R.; Rundle, M.; Frost, G.; et al. A metabolomic study of biomarkers of meat and fish intake. Am. J. Clin. Nutr. 2017, 105, 600–608. [Google Scholar] [CrossRef]
- Madrid-Gambin, F.; Llorach, R.; Vazquez-Fresno, R.; Urpi-Sarda, M.; Almanza-Aguilera, E.; Garcia-Aloy, M.; Estruch, R.; Corella, D.; Andres-Lacueva, C. Urinary (1)H Nuclear Magnetic Resonance Metabolomic Fingerprinting Reveals Biomarkers of Pulse Consumption Related to Energy-Metabolism Modulation in a Subcohort from the PREDIMED study. J. Proteome Res. 2017, 16, 1483–1491. [Google Scholar] [CrossRef]
- Wang, Y.; Gapstur, S.M.; Carter, B.D.; Hartman, T.J.; Stevens, V.L.; Gaudet, M.M.; McCullough, M.L. Untargeted Metabolomics Identifies Novel Potential Biomarkers of Habitual Food Intake in a Cross-Sectional Study of Postmenopausal Women. J. Nutr. 2018, 148, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, A.; Barbas, C. Chronic Diseases and Lifestyle Biomarkers Identification by Metabolomics. Adv. Exp. Med. Biol. 2017, 965, 235–263. [Google Scholar] [CrossRef]
- Savolainen, O.; Lind, M.V.; Bergstrom, G.; Fagerberg, B.; Sandberg, A.S.; Ross, A. Biomarkers of food intake and nutrient status are associated with glucose tolerance status and development of type 2 diabetes in older Swedish women. Am. J. Clin. Nutr. 2017, 106, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Bondia-Pons, I.; Martinez, J.A.; de la Iglesia, R.; Lopez-Legarrea, P.; Poutanen, K.; Hanhineva, K.; Zulet Mde, L. Effects of short- and long-term Mediterranean-based dietary treatment on plasma LC-QTOF/MS metabolic profiling of subjects with metabolic syndrome features: The Metabolic Syndrome Reduction in Navarra (RESMENA) randomized controlled trial. Mol. Nutr. Food Res. 2015, 59, 711–728. [Google Scholar] [CrossRef]
- Vazquez-Fresno, R.; Llorach, R.; Urpi-Sarda, M.; Lupianez-Barbero, A.; Estruch, R.; Corella, D.; Fito, M.; Aros, F.; Ruiz-Canela, M.; Salas-Salvado, J.; et al. Metabolomic pattern analysis after mediterranean diet intervention in a nondiabetic population: A 1- and 3-year follow-up in the PREDIMED study. J. Proteome Res. 2015, 14, 531–540. [Google Scholar] [CrossRef]
- Del Bas, J.M.; Caimari, A.; Rodriguez-Naranjo, M.I.; Childs, C.E.; Paras Chavez, C.; West, A.L.; Miles, E.A.; Arola, L.; Calder, P.C. Impairment of lysophospholipid metabolism in obesity: Altered plasma profile and desensitization to the modulatory properties of n-3 polyunsaturated fatty acids in a randomized controlled trial. Am. J. Clin. Nutr. 2016, 104, 266–279. [Google Scholar] [CrossRef]
- Holmes, E.; Li, J.V.; Marchesi, J.R.; Nicholson, J.K. Gut Microbiota Composition and Activity in Relation to Host Metabolic Phenotype and Disease Risk. Cell Metab. 2012, 16, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Black, A.; Kales, S.N.; Vattem, D.; Ruiz-Canela, M.; Sotos-Prieto, M. Metabolomics and Microbiomes as Potential Tools to Evaluate the Effects of the Mediterranean Diet. Nutrients 2019, 11, 207. [Google Scholar] [CrossRef]
- Shaffer, M.; Armstrong, A.J.S.; Phelan, V.V.; Reisdorph, N.; Lozupone, C.A. Microbiome and metabolome data integration provides insight into health and disease. Transl. Res. 2017, 189, 51–64. [Google Scholar] [CrossRef]
- Mills, S.; Stanton, C.; Lane, J.A.; Smith, G.J.; Ross, R.P. Precision Nutrition and the Microbiome, Part I: Current State of the Science. Nutrients 2019, 11, 923. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calleja, J.M.S.; Konstanti, P.; Swarts, H.J.M.; Bouwman, L.M.S.; Garcia-Campayo, V.; Billecke, N.; Oosting, A.; Smidt, H.; Keijer, J.; van Schothorst, E.M. Non-invasive continuous real-time in vivo analysis of microbial hydrogen production shows adaptation to fermentable carbohydrates in mice. Sci. Rep. 2018, 8, 15351. [Google Scholar] [CrossRef]
- Bashiardes, S.; Godneva, A.; Elinav, E.; Segal, E. Towards utilization of the human genome and microbiome for personalized nutrition. Curr. Opin. Biotechnol. 2018, 51, 57–63. [Google Scholar] [CrossRef]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef]
- Houten, S.M.; Violante, S.; Ventura, F.V.; Wanders, R.J. The Biochemistry and Physiology of Mitochondrial Fatty Acid beta-Oxidation and Its Genetic Disorders. Annu. Rev. Physiol. 2016, 78, 23–44. [Google Scholar] [CrossRef]
- Rombaldova, M.; Janovska, P.; Kopecky, J.; Kuda, O. Omega-3 fatty acids promote fatty acid utilization and production of pro-resolving lipid mediators in alternatively activated adipose tissue macrophages. Biochem. Biophys. Res. Commun. 2017, 490, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Oseeva, M.; Paluchova, V.; Zacek, P.; Janovska, P.; Mracek, T.; Rossmeisl, M.; Hamplova, D.; Cadova, N.; Stohanzlova, I.; Flachs, P.; et al. Omega-3 index in the Czech Republic: No difference between urban and rural populations. Chem. Phys. Lipids 2019, 220, 23–27. [Google Scholar] [CrossRef]
- O’Gorman, A.; Gibbons, H.; Ryan, M.F.; Gibney, E.R.; Gibney, M.J.; Frost, G.S.; Roche, H.M.; Brennan, L. Exploring the Links between Diet and Health in an Irish Cohort: A Lipidomic Approach. J. Proteome Res. 2017, 16, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Kuang, A.; Erlund, I.; Herder, C.; Westerhuis, J.A.; Tuomilehto, J.; Cornelis, M.C. Lipidomic Response to Coffee Consumption. Nutrients 2018, 10, 1851. [Google Scholar] [CrossRef]
- Guertin, K.A.; Moore, S.C.; Sampson, J.N.; Huang, W.Y.; Xiao, Q.; Stolzenberg-Solomon, R.Z.; Sinha, R.; Cross, A.J. Metabolomics in nutritional epidemiology: Identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am. J. Clin. Nutr. 2014, 100, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Mias, G.I.; Li-Pook-Than, J.; Jiang, L.; Lam, H.Y.; Chen, R.; Miriami, E.; Karczewski, K.J.; Hariharan, M.; Dewey, F.E.; et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012, 148, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picó, C.; Serra, F.; Rodríguez, A.M.; Keijer, J.; Palou, A. Biomarkers of Nutrition and Health: New Tools for New Approaches. Nutrients 2019, 11, 1092. https://doi.org/10.3390/nu11051092
Picó C, Serra F, Rodríguez AM, Keijer J, Palou A. Biomarkers of Nutrition and Health: New Tools for New Approaches. Nutrients. 2019; 11(5):1092. https://doi.org/10.3390/nu11051092
Chicago/Turabian StylePicó, Catalina, Francisca Serra, Ana María Rodríguez, Jaap Keijer, and Andreu Palou. 2019. "Biomarkers of Nutrition and Health: New Tools for New Approaches" Nutrients 11, no. 5: 1092. https://doi.org/10.3390/nu11051092





