A MALDI-TOF MS Approach for Mammalian, Human, and Formula Milks’ Profiling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Milk Sampling and Pre-Treatment
2.2. MALDI-TOF Spectra Acquisition
2.3. Statistical Analysis
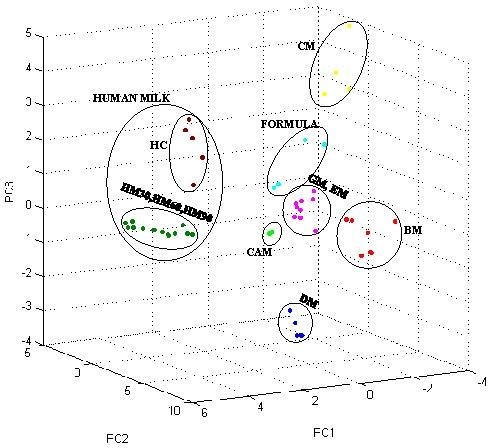
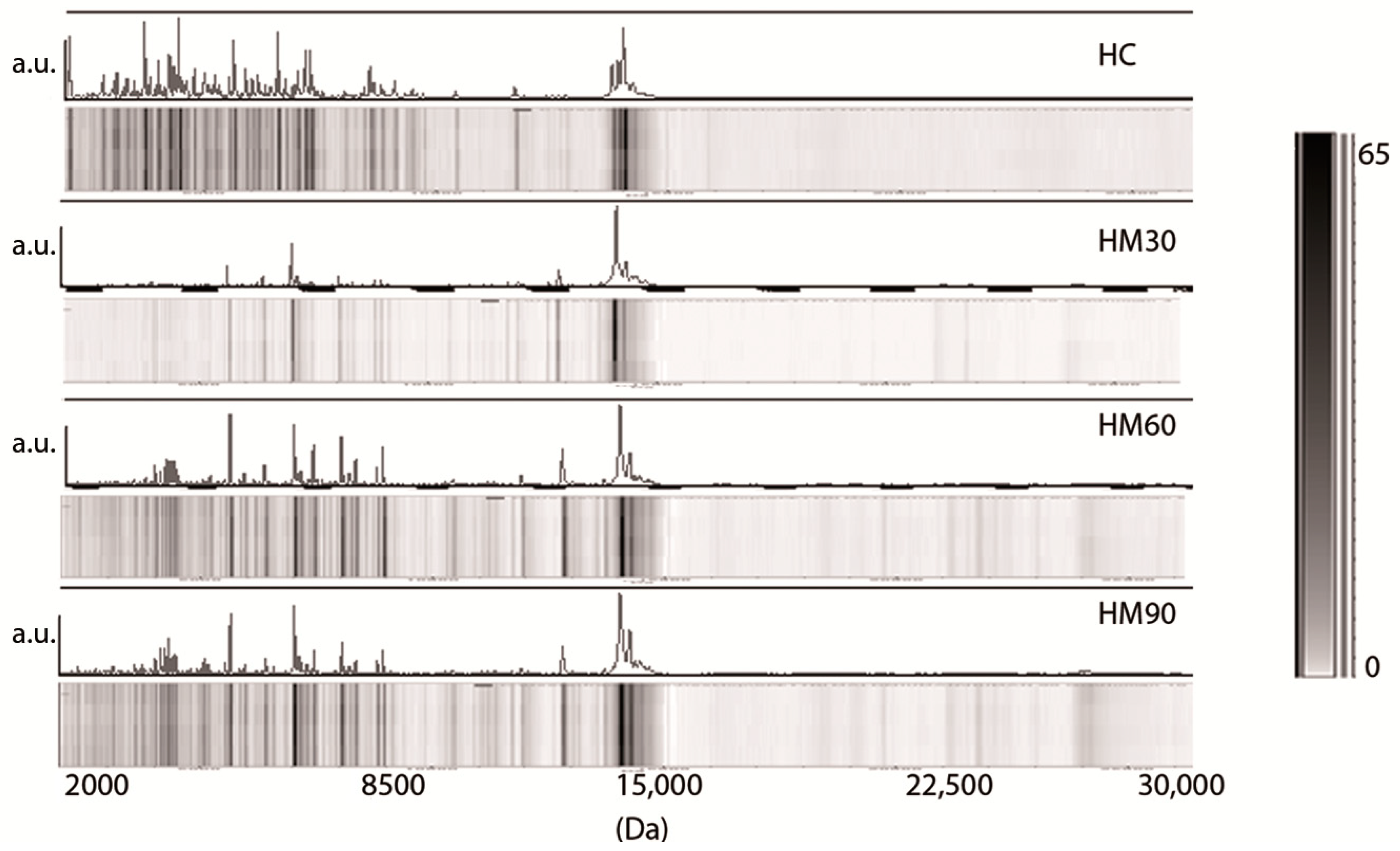
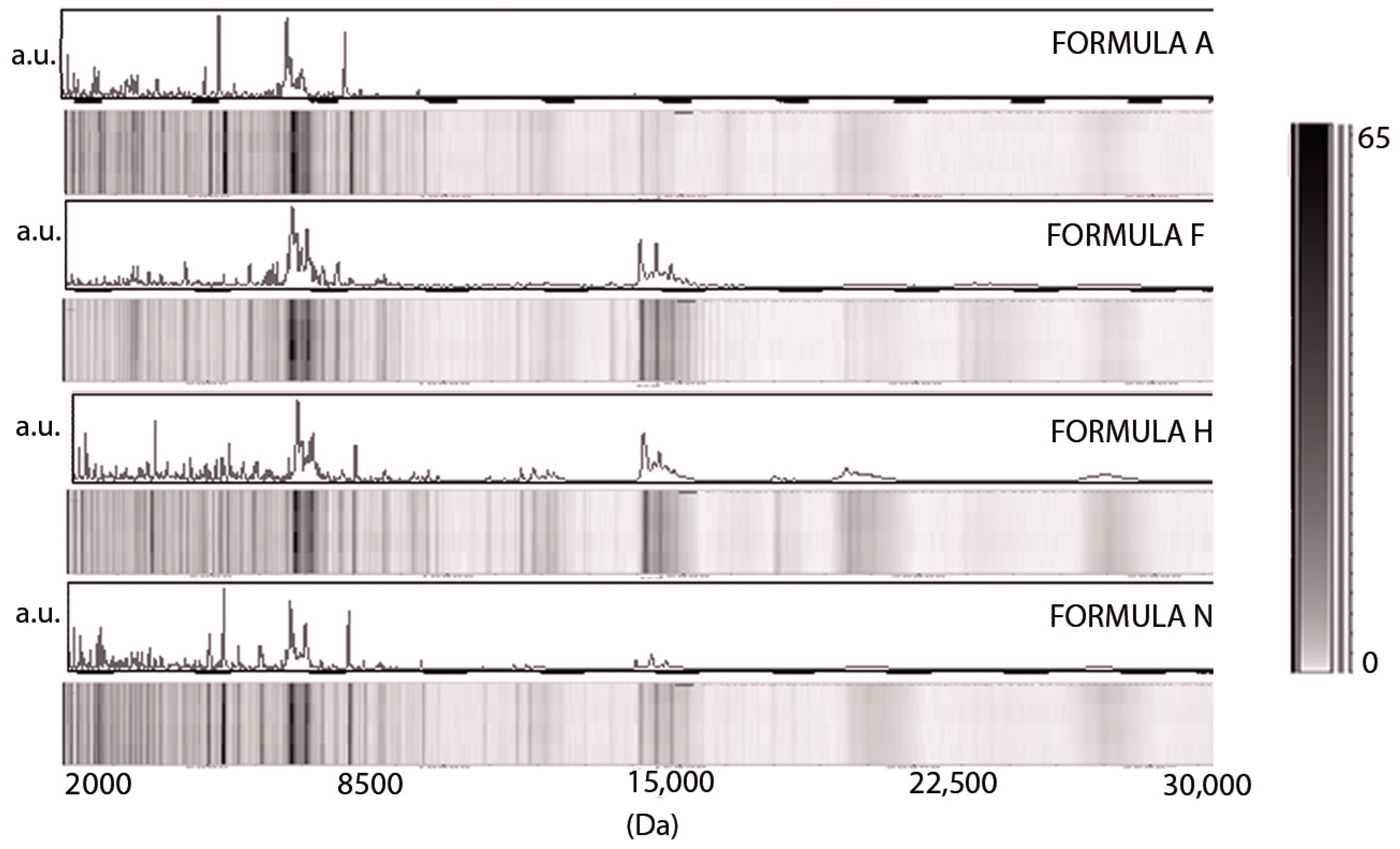
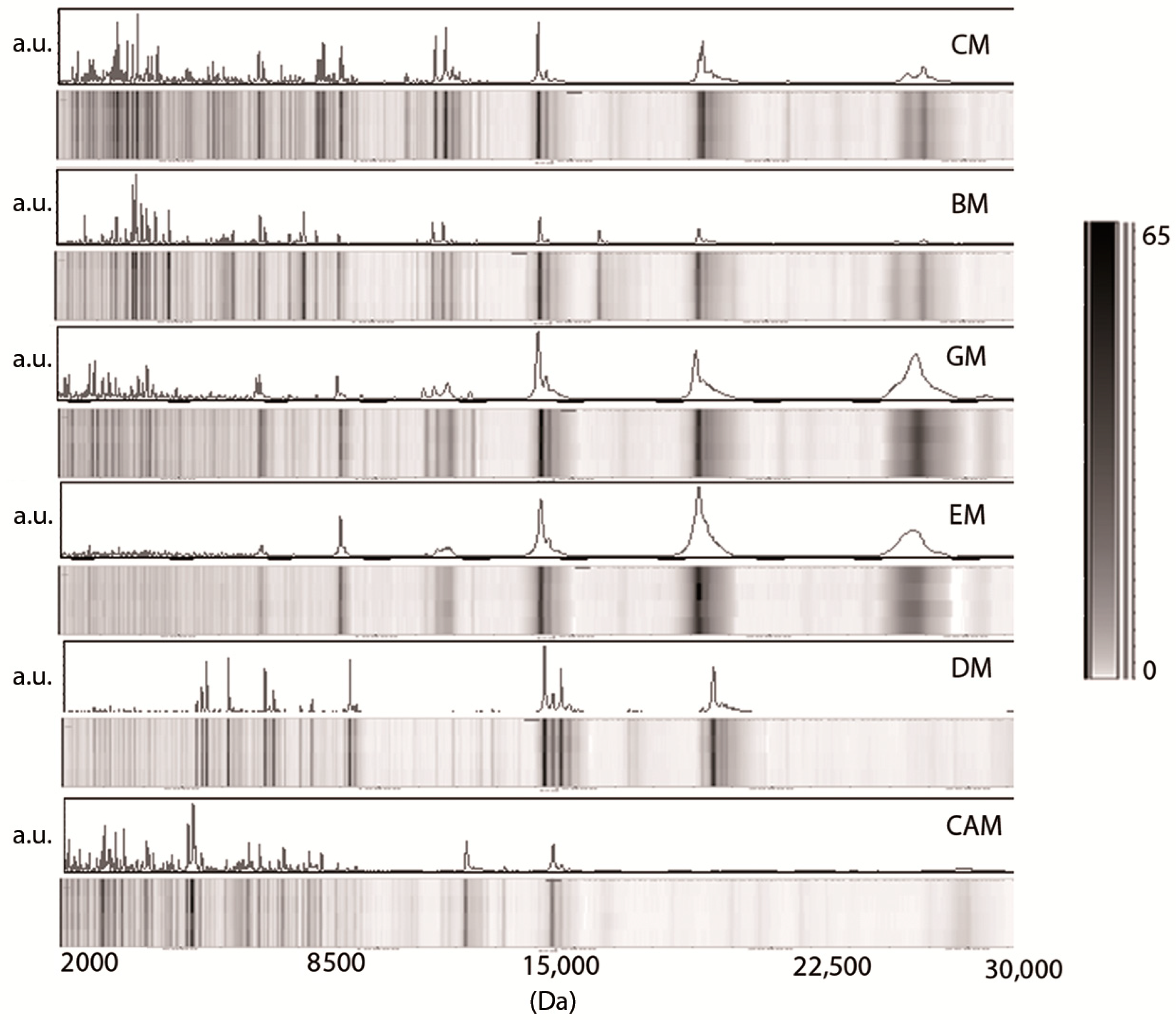
3. Results
3.1. Low-Molecular-Weight Protein Profiles from Crude Milk by MALDI-TOF MS
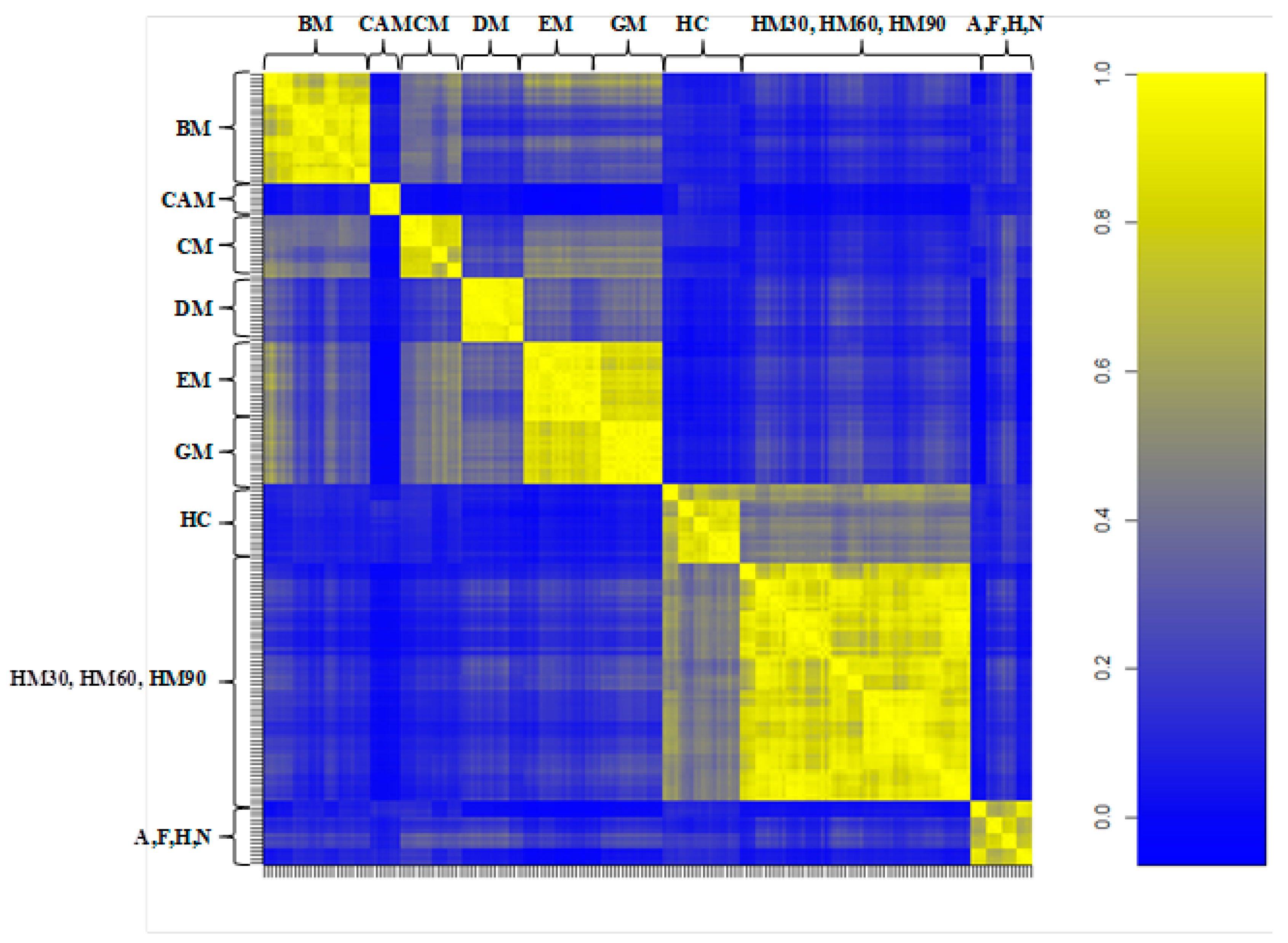
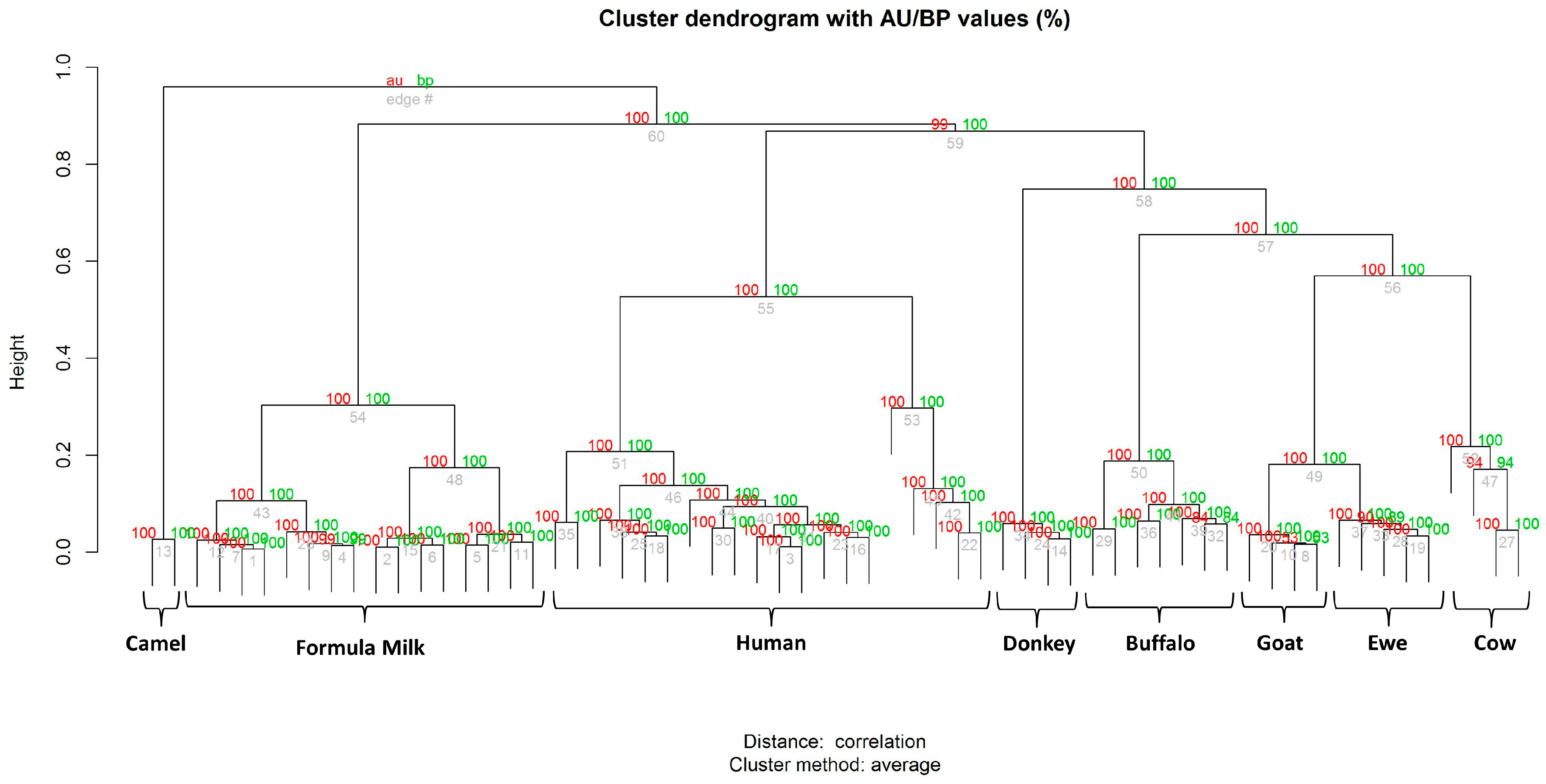
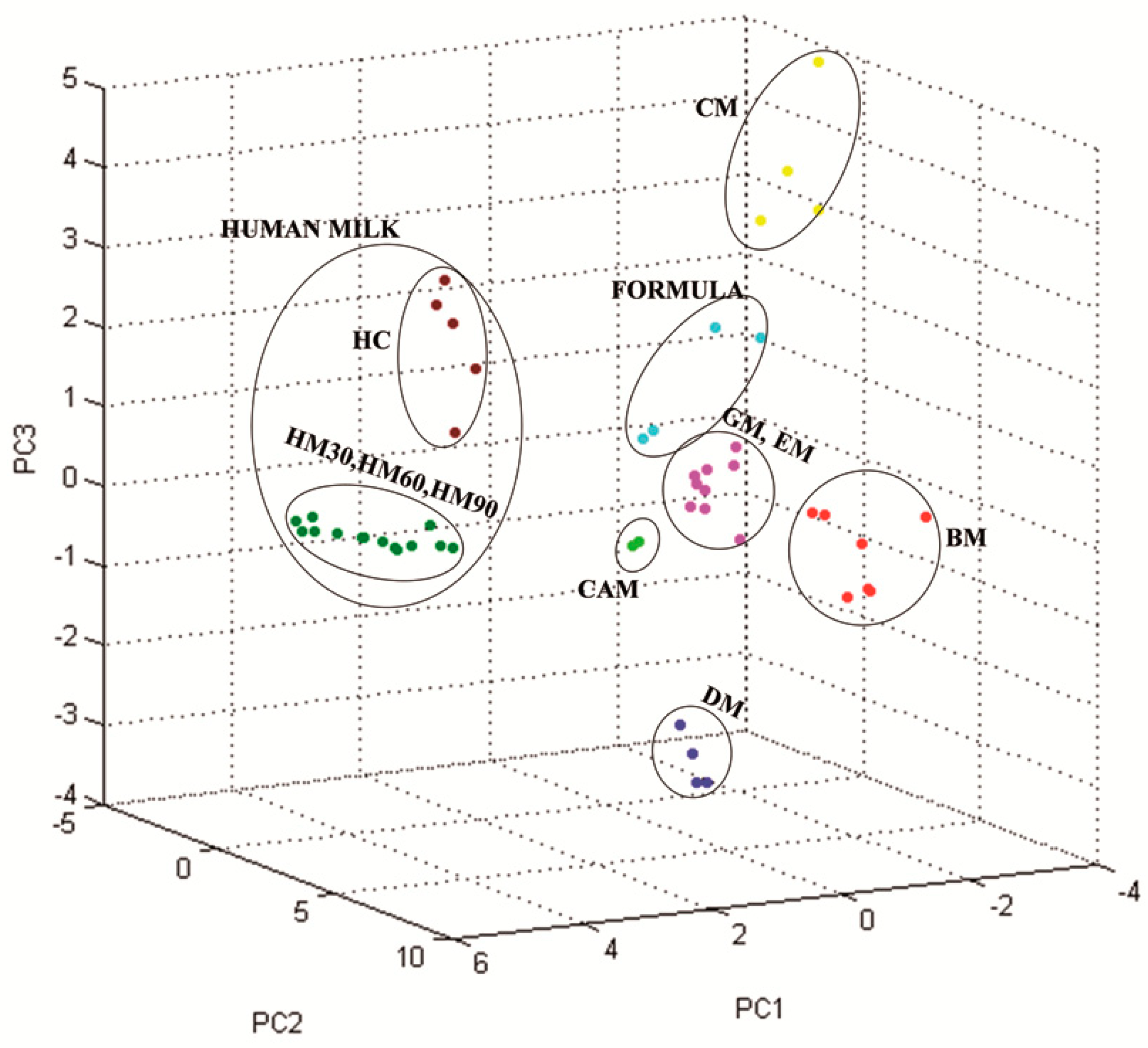
3.2. Milk Spectra Properties and Similarities
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dewey, K. Guiding Principles for Complementary Feeding of the Breastfed Child; Pan American Health Organization [PAHO], Division of Health Promotion and Protection, Food and Nutrition Program: Washington, DC, USA, 2003; p. 37. [Google Scholar]
- Lessen, R.; Kavanagh, K. Position of the academy of nutrition and dietetics: Promoting and supporting breastfeeding. J. Acad. Nutr. Diet. 2015, 115, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.R.; Hernandez, L.L. Autocrine-paracrine regulation of the mammary gland1. J. Dairy Sci. 2016, 99, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Prentice, P.; Ong, K.K.; Schoemaker, M.H.; van Tol, E.A.F.; Vervoort, J.; Hughes, I.A.; Acerini, C.L.; Dunger, D.B. Breast milk nutrient content and infancy growth. Acta Paediatr. 2016, 105, 641–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballard, O.; Morrow, A.L. Human milk composition. Pediatr. Clin. North Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [PubMed]
- Roncada, P.; Stipetic, L.H.; Bonizzi, L.; Burchmore, R.J.S.; Kennedy, M.W. Proteomics as a tool to explore human milk in health and disease. J. Proteom. 2013, 88, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Del Chierico, F.; Vernocchi, P.; Petrucca, A.; Paci, P.; Fuentes, S.; Praticò, G.; Capuani, G.; Masotti, A.; Reddel, S.; Russo, A.; et al. Phylogenetic and Metabolic Tracking of Gut Microbiota during Perinatal Development. PLoS ONE 2015, 10, e0137347. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wardlaw, T.; Brown, D.W. Global trends in exclusive breastfeeding. Int. Breastfeed. J. 2012, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Demonstration of the Quality Factor Requirements Under 21 CFR 106.96(i) for “Eligible” Infant Formulas. Available online: https://www.fda.gov/RegulatoryInformation/Guidances/ucm400036.htm (accessed on 20 May 2018).
- Food and Agriculture Organization of the United Nations World Health Organization. CODEX alimentarius: Standard for Infant Formula and Formulas for Special Medical Purposes Intended for Infants. Revised Standard for Infant Formula. Available online: http://www.fao.org/fao-who-codexalimentarius/sh-proxy/pt/?lnk=1&url=https%253A%252F%252Fworkspace.fao.org%252Fsites%252Fcodex%252FMeetings%252FCX-720-38%252FReport%252FFINAL%252FREP17_NFSDUe.pdf (accessed on 20 May 2018).
- Rossen, L.M.; Simon, A.E.; Herrick, K.A. Types of infant formulas consumed in the united states. Clin. Pediatr. (Phila.) 2016, 55, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; Baker, S.; Cleghorn, G.; Neto, U.F.; Gopalan, S.; Hernell, O.; Hock, Q.S.; Jirapinyo, P.; Lonnerdal, B.; Pencharz, P.; et al. Global standard for the composition of infant formula: Recommendations of an ESPGHAN coordinated international expert group. J. Pediatr. Gastroenterol. Nutr. 2005, 41, 584–599. [Google Scholar] [CrossRef] [PubMed]
- Stam, J.; Sauer, P.J.; Boehm, G. Can we define an infant’s need from the composition of human milk? Am. J. Clin. Nutr. 2013, 98, 521S–528S. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.S.; Hay, W.W. Challenges of infant nutrition research: A commentary. Nutr. J. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Gura, T. Nature’s first functional food. Science 2014, 345, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Grote, V.; Closa-Monasterolo, R.; Escribano, J.; Langhendries, J.P.; Dain, E.; Giovannini, M.; Verduci, E.; Gruszfeld, D.; Socha, P.; et al. Lower protein content in infant formula reduces BMI and obesity risk at school age: Follow-up of a randomized trial. Am. J. Clin. Nutr. 2014, 99, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Hidayat, K.; Du, H.-Z.; Yang, J.; Chen, G.C.; Zhang, Z.; Li, Z.N.; Qin, L.Q. Effects of milk proteins on blood pressure: A meta-analysis of randomized control trials. Hypertens. Res. 2017, 40, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; McMahon, R.J.; Woo, J.G.; Davidson, B.S.; Morrow, A.L.; Zhang, Q. Temporal changes in milk proteomes reveal developing milk functions. J. Proteome Res. 2012, 11, 3897–3907. [Google Scholar] [CrossRef] [PubMed]
- Savino, F.; Sorrenti, M.; Benetti, S.; Lupica, M.M.; Liguori, S.A.; Oggero, R. Resistin and leptin in breast milk and infants in early life. Early Hum. Dev. 2012, 88, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Restani, P.; Gaiaschi, A.; Plebani, A.; Beretta, B.; Cavagni, G.; Fiocchi, A.; Poiesi, C.; Velonà, T.; Ugazio, A.G.; Galli, C.L. Cross-reactivity between milk proteins from different animal species. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 1999, 29, 997–1004. [Google Scholar] [CrossRef]
- Fiocchi, A.; Brozek, J.; Schünemann, H.; Bahna, S.L.; von Berg, A.; Beyer, K.; Bozzola, M.; Bradsher, J.; Compalati, E.; Ebisawa, M.; et al. World Allergy Organization (WAO) Special Committee on Food Allergy World Allergy Organization (WAO) Diagnosis and Rationale for Action against Cow’s Milk Allergy (DRACMA) Guidelines. Pediatr. Allergy Immunol. 2010, 21 (Suppl 21), 1–125. [Google Scholar] [CrossRef]
- Putignani, L.; Del Chierico, F.; Onori, M.; Mancinelli, L.; Argentieri, M.; Bernaschi, P.; Coltella, L.; Lucignano, B.; Pansani, L.; Ranno, S.; et al. MALDI-TOF mass spectrometry proteomic phenotyping of clinically relevant fungi. Mol. BioSyst. 2011, 7, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Di Girolamo, F.; Masotti, A.; Salvatori, G.; Scapaticci, M.; Muraca, M.; Putignani, L. A sensitive and effective proteomic approach to identify she-donkey’s and goat’s milk adulterations by MALDI-TOF MS Fingerprinting. Int. J. Mol. Sci. 2014, 15, 13697–13719. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ketterlinus, R.; Hsieh, S.-Y.; Teng, S.-H.; Lee, H.; Pusch, W. Fishing for biomarkers: Analyzing mass spectrometry data with the new ClinProTools software. BioTechniques 2005, 38, 37–40. [Google Scholar] [CrossRef]
- Di Girolamo, F.; D’Amato, A.; Lante, I.; Signore, F.; Muraca, M.; Putignani, L. Farm animal serum proteomics and impact on human health. Int. J. Mol. Sci. 2014, 15, 15396–15411. [Google Scholar] [CrossRef] [PubMed]
- Bislev, S.L.; Deutsch, E.W.; Sun, Z.; Farrah, T.; Aebersold, R.; Moritz, R.L.; Bendixen, E.; Codrea, M.C. A Bovine Peptide Atlas of milk and mammary gland proteomes. Proteomics 2012, 12, 2895–2899. [Google Scholar] [CrossRef] [PubMed]
- Gephart, S.M.; Weller, M. Colostrum as Oral Immune Therapy to Promote Neonatal Health. Adv. Neonatal Care 2014, 14, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Bagwe, S.; Tharappel, L.J.P.; Kaur, G.; Buttar, H.S. Bovine colostrum: An emerging nutraceutical. J. Complement. Integr. Med. 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Alvarado, R.; Phinney, B.; Lönnerdal, B. Proteomic characterization of human milk whey proteins during a twelve-month lactation period. J. Proteome Res. 2011, 10, 1746–1754. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Alvarado, R.; Phinney, B.; Lönnerdal, B. Proteomic characterization of human milk fat globule membrane proteins during a 12 month lactation period. J. Proteome Res. 2011, 10, 3530–3541. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Lippolis, J.D. Developmental Changes in the milk fat globule membrane proteome during the transition from colostrum to milk. J. Dairy Sci. 2008, 91, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Sackesen, C.; Assa’ad, A.; Baena-Cagnani, C.; Ebisawa, M.; Fiocchi, A.; Heine, R.G.; Von Berg, A.; Kalayci, O. Cow’s milk allergy as a global challenge. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Shabo, Y.; Barzel, R.; Margoulis, M.; Yagil, R. Camel milk for food allergies in children. Isr. Med. Assoc. J. IMAJ 2005, 7, 796–798. [Google Scholar] [PubMed]
- Merin, U.; Bernstein, S.; Bloch-Damti, A.; Yagil, R.; van Creveld, C.; Lindner, P.; Gollop, N. A comparative study of milk serum proteins in camel (Camelus dromedarius) and bovine colostrum. Livest. Prod. Sci. 2001, 67, 297–301. [Google Scholar] [CrossRef]
- El-Agamy, E.I.; Nawar, M.; Shamsia, S.M.; Awad, S.; Haenlein, G.F.W. Are camel milk proteins convenient to the nutrition of cow milk allergic children? Small Rumin. Res. 2009, 82, 1–6. [Google Scholar] [CrossRef]
- El-Agamy, E.I. The challenge of cow milk protein allergy. Small Rumin. Res. 2007, 68, 64–72. [Google Scholar] [CrossRef]
- Järvinen, K.M.; Chatchatee, P. Mammalian milk allergy: Clinical suspicion, cross-reactivities and diagnosis. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Ehlayel, M.S.; Hazeima, K.A.; Al-Mesaifri, F.; Bener, A. Camel milk: An alternative for cow’s milk allergy in children. Allergy Asthma Proc. 2011, 32, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Piovesana, S.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Samperi, R.; Zenezini Chiozzi, R.; Laganà, A. Peptidome characterization and bioactivity analysis of donkey milk. J. Proteom. 2015, 119, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Gallina, S.; Cunsolo, V.; Saletti, R.; Muccilli, V.; Di Francesco, A.; Foti, S.; Lorenzten, A.M.; Roepstorff, P. Sequence characterization and glycosylation sites identification of donkey milk lactoferrin by multiple enzyme digestions and mass spectrometry. Amino Acids 2016, 48, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Restani, P.; Fiocchi, A.; Beretta, B.; Velonà, T.; Giovannini, M.; Galli, C.L. Meat allergy: III—Proteins involved and cross-reactivity between different animal species. J. Am. Coll. Nutr. 1997, 16, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Sabahelkhier, M.; Faten, M.; Omer, F. Comparative Determination of biochemical constituents between animals (goat, sheep, cow and camel) milk with human milk. Res. J. Recent Sci. 2012, 1, 69–71. [Google Scholar]
- Roncada, P.; Piras, C.; Soggiu, A.; Turk, R.; Urbani, A.; Bonizzi, L. Farm animal milk proteomics. J. Proteom. 2012, 75, 4259–4274. [Google Scholar] [CrossRef] [PubMed]
- D’auria, E.; Agostoni, C.; Giovannini, M.; Riva, E.; Zetterström, R.; Fortin, R.; Greppi, G.; Bonizzi, L.; Roncada, P. Proteomic evaluation of milk from different mammalian species as a substitute for breast milk. Acta Paediatr. 2005, 94, 1708–1713. [Google Scholar] [CrossRef] [PubMed]
- Lambers, T.T.; Gloerich, J.; van Hoffen, E.; Alkema, W.; Hondmann, D.H.; van Tol, E.A.F. Clustering analyses in peptidomics revealed that peptide profiles of infant formulae are descriptive. Food Sci. Nutr. 2015, 3, 81–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Milk Source | Samples (n) | Spectra Replicates (n) | Total Spectra Analyzed (n) |
|---|---|---|---|
| Human milk | |||
| Breastfeeding women at 2 days | 5 | 4 | 20 |
| Breastfeeding women at 30 days | 6 | 4 | 24 |
| Breastfeeding women at 60 days | 5 | 4 | 20 |
| Breastfeeding women at 90 days | 4 | 4 | 16 |
| Commercial brands of infant formula | 4 | 4 | 16 |
| Animal milk 1 | |||
| cows | 4 | 4 | 16 |
| buffaloes | 7 | 4 | 28 |
| goats | 4 | 4 | 16 |
| ewes | 5 | 4 | 20 |
| she-donkeys | 4 | 4 | 16 |
| camels | 2 | 4 | 8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Francesco, L.; Di Girolamo, F.; Mennini, M.; Masotti, A.; Salvatori, G.; Rigon, G.; Signore, F.; Pietrantoni, E.; Scapaticci, M.; Lante, I.; et al. A MALDI-TOF MS Approach for Mammalian, Human, and Formula Milks’ Profiling. Nutrients 2018, 10, 1238. https://doi.org/10.3390/nu10091238
Di Francesco L, Di Girolamo F, Mennini M, Masotti A, Salvatori G, Rigon G, Signore F, Pietrantoni E, Scapaticci M, Lante I, et al. A MALDI-TOF MS Approach for Mammalian, Human, and Formula Milks’ Profiling. Nutrients. 2018; 10(9):1238. https://doi.org/10.3390/nu10091238
Chicago/Turabian StyleDi Francesco, Laura, Francesco Di Girolamo, Maurizio Mennini, Andrea Masotti, Guglielmo Salvatori, Giuliano Rigon, Fabrizio Signore, Emanuela Pietrantoni, Margherita Scapaticci, Isabella Lante, and et al. 2018. "A MALDI-TOF MS Approach for Mammalian, Human, and Formula Milks’ Profiling" Nutrients 10, no. 9: 1238. https://doi.org/10.3390/nu10091238