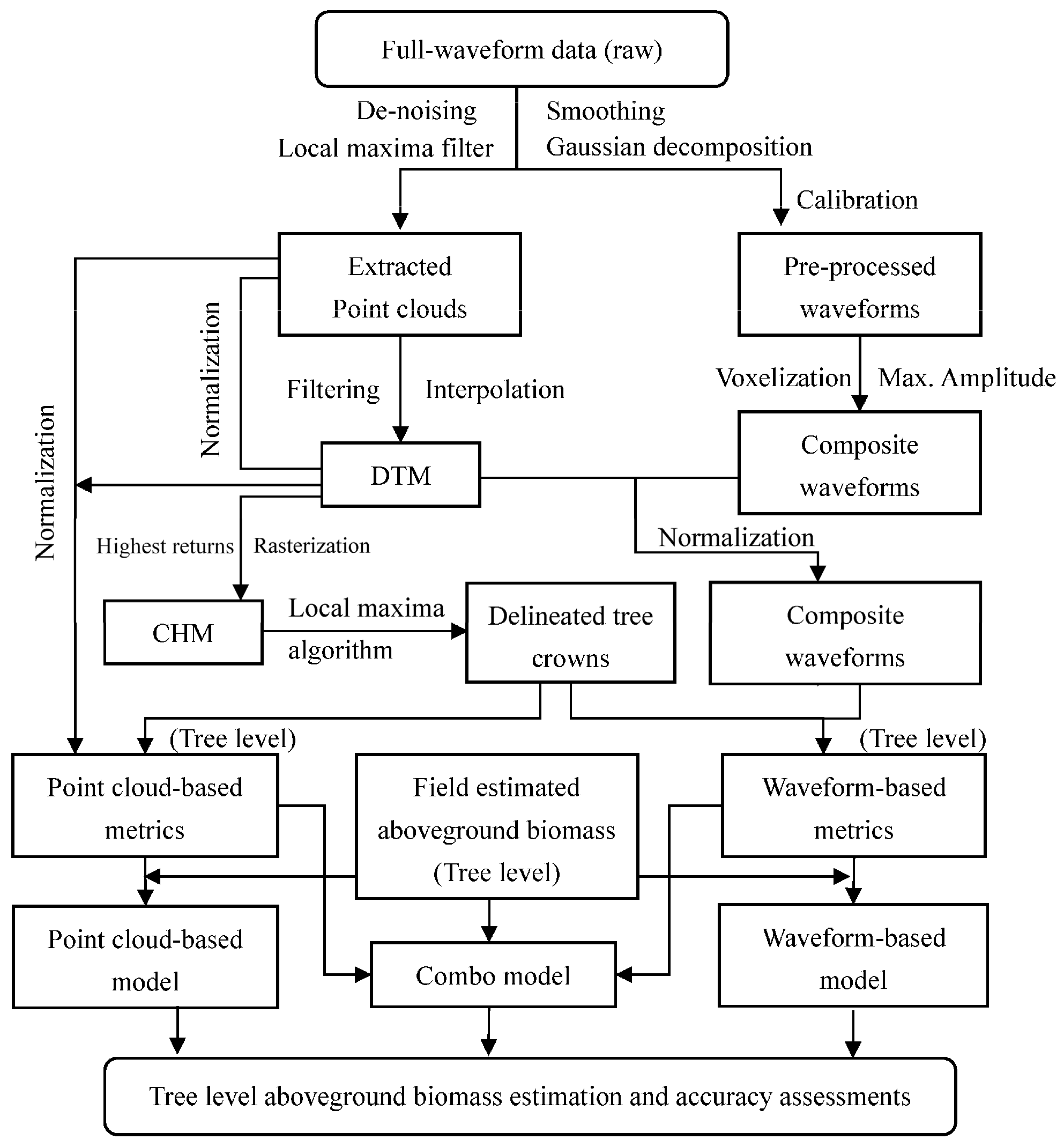
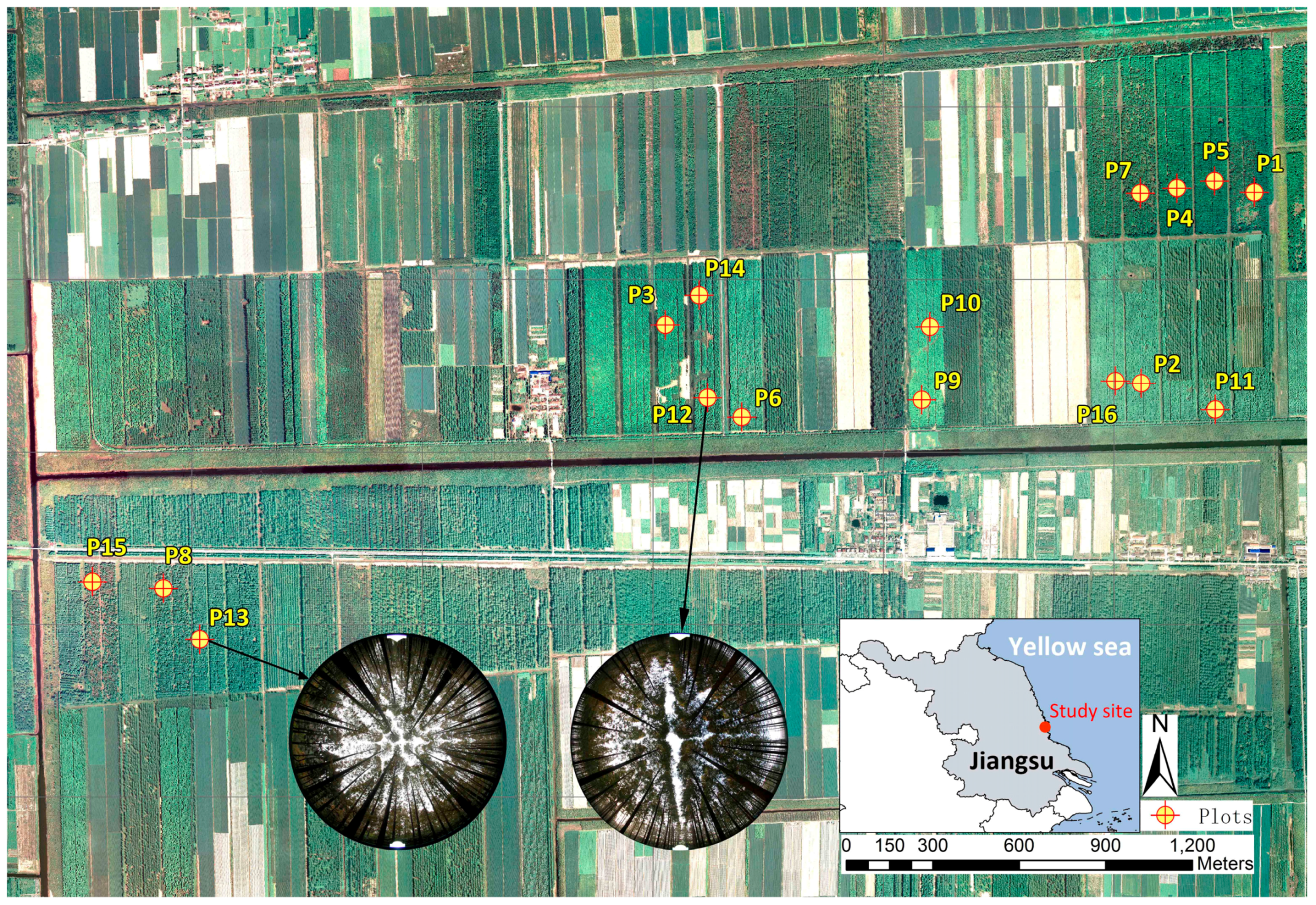
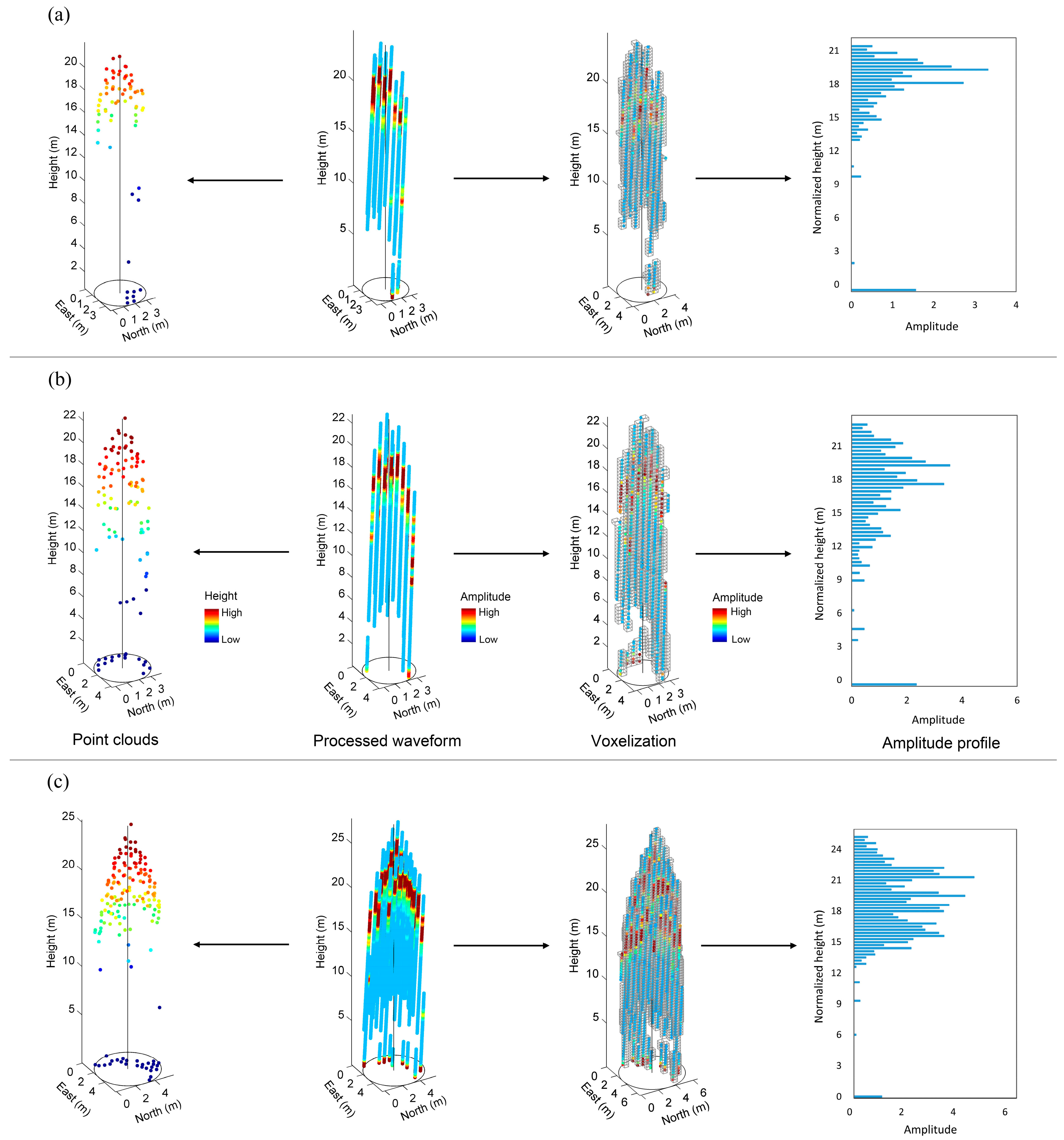
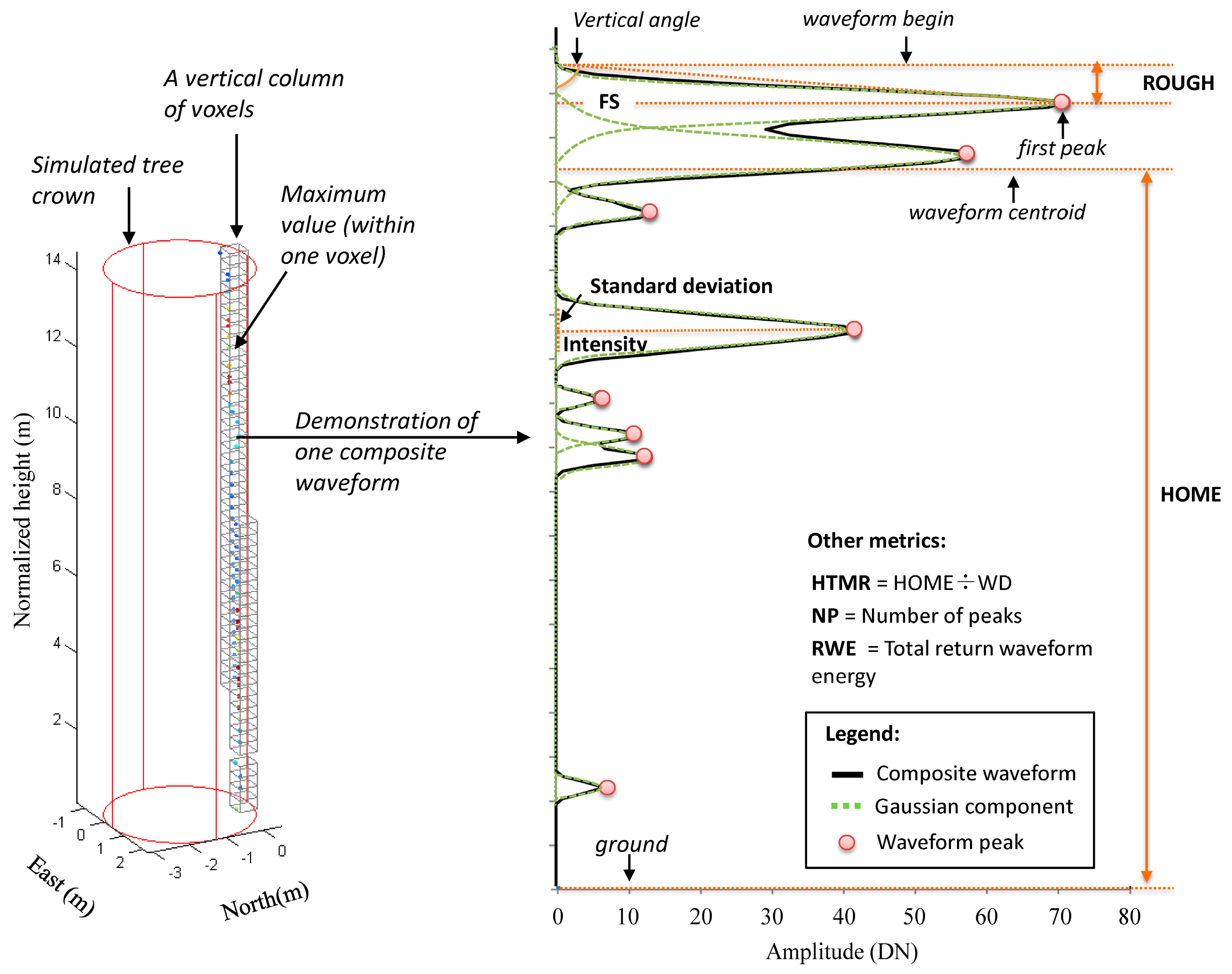
1. Introduction
Forests play a key role in global carbon (C) cycle as they contain 85%–90% of total living terrestrial biomass, and annually exchange up to 90% of total terrestrial ecosystem C with the atmosphere through photosynthesis and respiration [
1,
2]. China has the largest area of planted forests in the world (71 million ha, approximately 27% of world’s planted forests) [
3]. The increase of planted forest biomass in China contributed to approximately 30% of carbon sinks in China’s terrestrial ecosystems [
4]. Previous studies indicated that these forests play an important role in C sequestration to mitigate anthropogenic C emission [
4,
5,
6]. Thus, accurately assessing forest biomass (i.e., C sinks) of the planted forests in China is important to maintain regional and global carbon balance and mitigate climate change.
Airborne Laser Scanning (ALS) is an active remote sensing technology, which provides an advantage over most other remote sensing technologies in its ability of providing detailed three-dimensional information of forest canopy structure [
7,
8]. The laser pulses can penetrate through the forest canopy and characterize the vertical distribution of the canopy. Previous studies have found ecological and biomechanical links between biomass and vertical structure, and as a result, a strong correlation between ALS metrics and biomass can be expected [
9,
10,
11]. Generally, methodological approaches for estimating forest biomass can be classified into two categories, i.e., area-based (ABA) and individual tree crown (ITC) approaches [
12]. Most of these studies used the ABA approach to predict biomass at plot, stand or regional level, e.g., Cao et al. (2014) [
13], Li et al. (2008) [
14] and Næsset et al. (2008) [
15], etc., whereas some studies implemented the ITC approach to estimate biomass at individual tree level, e.g., Popescu 2007 [
16], Gleason and Im 2012 [
17], Hauglin et al. (2013) [
18], and Kankare et al. (2013) [
19], etc. The advantages of the ITC approach for biomass estimation (over the ABA approach) include the following: First, the ITC approach estimates biomass in minimal scale (in practical forest management), thus biomass mapping at other scales (above tree-level) can be immediately available by summing tree-level estimations up to desired scales [
16,
20]. Second, the ITC approach can take the advantage of existed species-specific allometric models from the literature. In other words, in ABA analysis, the relationship between plot-level biomass (per unit area) and ALS metrics has no practical meaning [
20]. However, it should be noted that, in ITC approach, the errors of tree detection will be transferred into the AGB summaries at plot or stand level, and can result in the estimation of AGB with considerable systematic errors.
For the past decade, most ALS systems produced point cloud (or discrete-return) data [
21]. These systems record multiple returns (
n ≤ 5) per transmitted pulse with each return representing the 3D position and intensity of the reflected light. These point cloud-based approaches have been successfully used to estimate a wide range of forest characteristics [
22]. For instance, point cloud data estimated tree heights were found to be of similar or better accuracy than corresponding field-based measurements [
23]. Additional attributes, such as leaf area index [
24], volume [
25] and biomass [
13], are also well characterized with point cloud ALS metrics at plot scale. However, the discrete return ALS systems can only distinguish surfaces that are sufficiently spaced apart, due to limitations in the electronics [
26]. Furthermore, since the returned signal is interpreted internally, the recorded information from these systems is limited for each pulse [
27]. In contrast, small-footprint full-waveform ALS systems have become available commercially since 2004 [
28]; these systems provide new opportunities for forestry studies. The full-waveform systems digitize and record the entire backscattered signal of each emitted pulse, and allow the recording of geometric and physical properties of intercepted objects [
29]. The FWF systems have advantages with regard to not limiting the number of returns for each laser pulse; providing additional investigation possibilities, e.g., point cloud density can be enhanced by processing the FWF return signal; and additional metrics can be extracted by modeling the received waveforms [
30]. The analysis of full-waveform data has improved the urban area classification [
31], and the estimations of forest structural variables, e.g., DBH [
32], canopy height [
33], number of stems [
34], stem volume [
21] and biomass [
35].
To date, most of the studies related to ALS data-based biomass estimation used the ABA approach. Published studies focused on tree-level biomass estimation by exploring ALS data (especially full-waveform data) are few. Allouis et al. (2013) [
36] assessed the capability of full-waveform data to improve the estimation of individual tree-based biomass in a black pine forest in southern France. They found that the accuracy of biomass estimation (adjusted
R2 = 0.90–0.91) was improved compared with models developed using Canopy Height Model (CHM)-only (adjusted
R2 = 0.87) or CHM + Point cloud (adjusted
R2 = 0.87–0.88) metrics. They also found that when including the waveform metric (i.e., the integral of the cumulative signal, which related to the vertical structure of a tree as a whole), the accuracy of biomass estimation was enhanced [
36]. Wu et al. (2009) [
37] extracted structural metrics (i.e., crown length and canopy height) from waveforms for estimating woody and foliar biomass in a wooded savanna in South Africa. The results showed that waveform metrics have strong potential for estimating woody (
R2 = 0.71) and foliar biomass (
R2 = 0.73) at the tree-level in a savanna environment [
37].
In this study, the capability of point cloud (extracted from the full-waveform data) and waveform (calculated based on a voxel-based composite waveform approach) metrics for estimating tree level aboveground biomass (AGB), was assessed individually and in combination, over a planted forest in the coastal region of east China. The specific objectives of this study are: (1) to investigate the importance of point cloud and waveform metrics for estimating tree-level AGB; and (2) to assess the capability of the point cloud and waveform metrics based models and combo model (including the combination of both point cloud and waveform metrics) for tree-level AGB estimation and evaluate the accuracies of these models.
3. Results
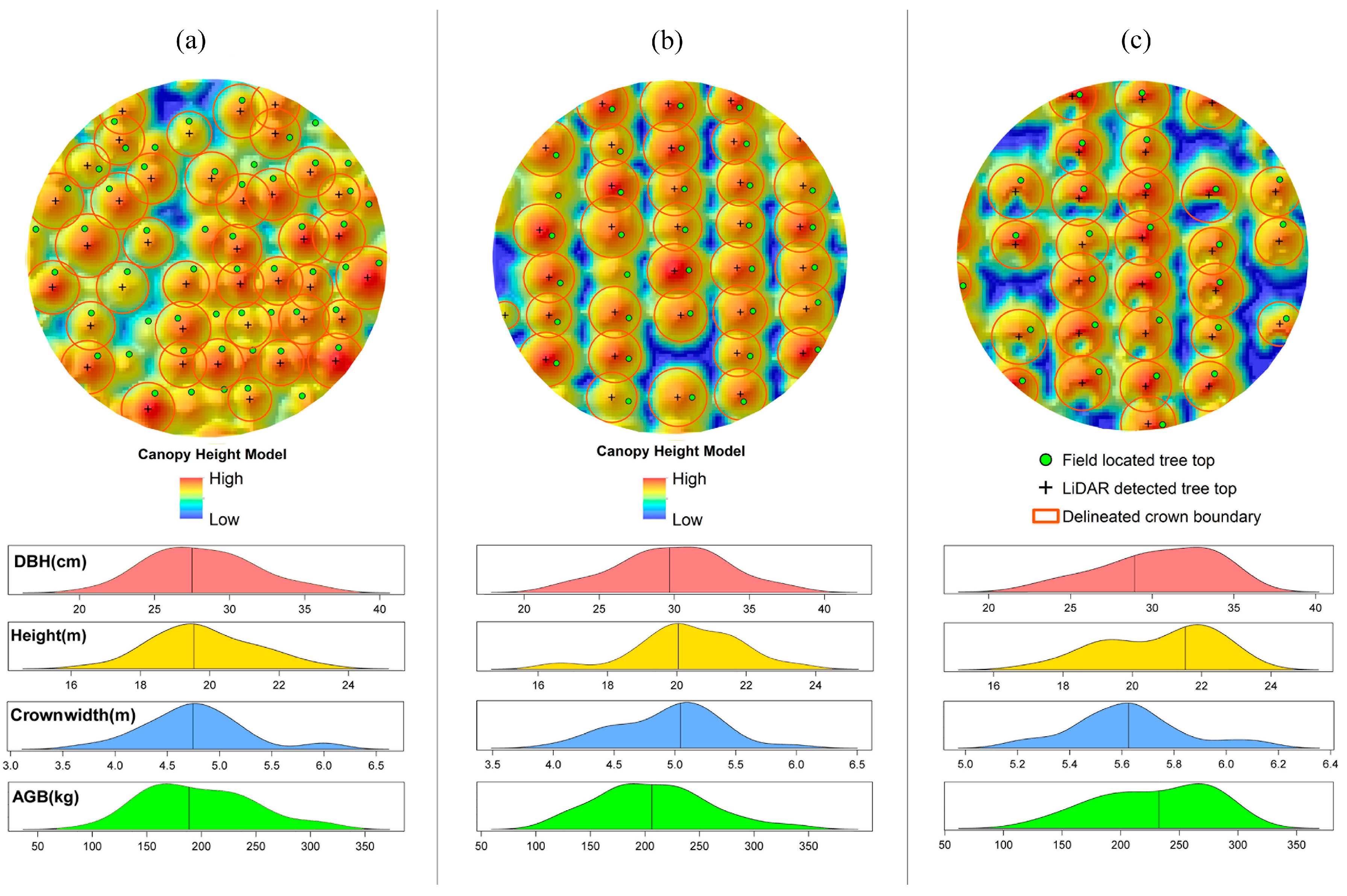
In total, 563 (=85.6% of field measured trees) were correctly detected in the plots, with an error of omission (i.e., the number of the trees which were not detected) of 95 (=14.4% of field measured trees) and an error of commission (the number of detected trees which do not exist in the field) of 53 (=8.1% of field measured trees) (
Table 4). The location of ALS-detected trees, crown boundaries and their related ground trees, and the canopy height model (CHM) of three sample plots (with different stem densities) can be seen in
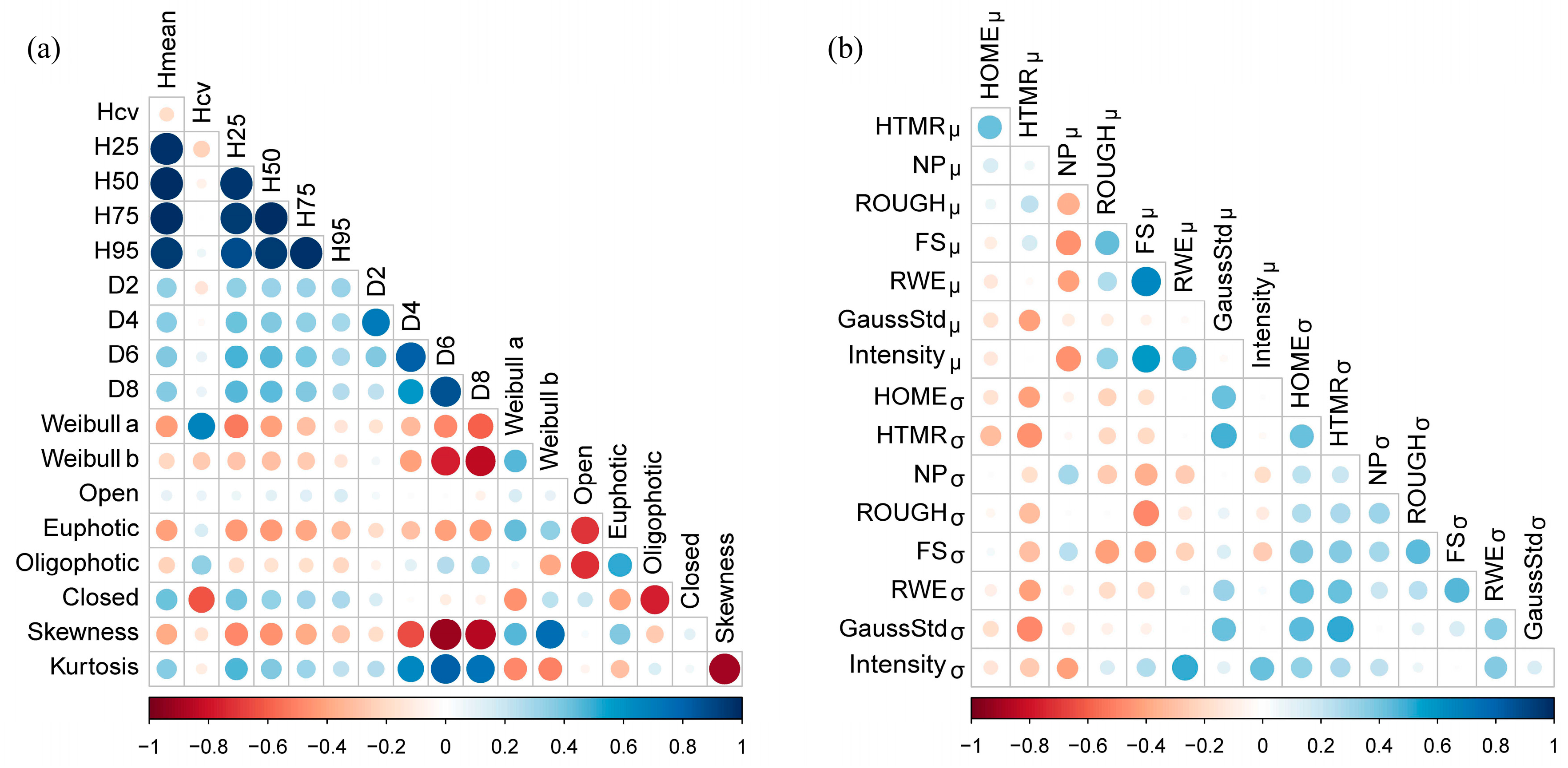
Figure 5. The correlation of the metrics is shown in
Figure 6. Compared with point cloud metrics, waveform metrics have relatively lower correlation coefficients with other metrics. Both point cloud and waveform metrics with low correlation coefficients (<0.60) (with any other metrics) were then used in regression analysis.
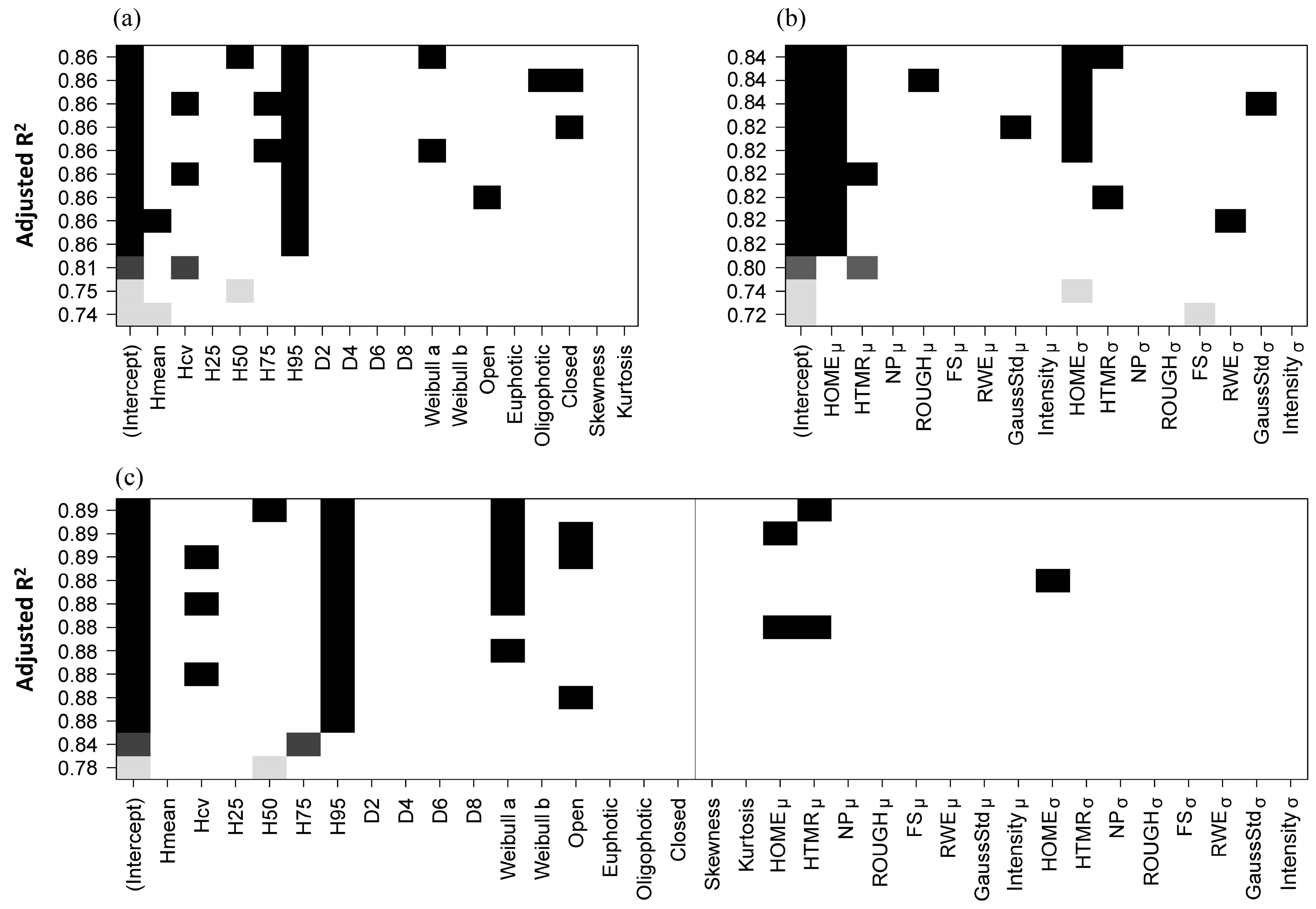
All possible independent variable combinations (size = 3, i.e., from one to three independent variables; best four models are provided for each size based on Adjusted
R2) of the AGB estimation models, fitted by point cloud and waveform metrics individually and in combination, are shown in
Figure 7. Overall, the individual tree level AGB is generally well predicted (Adjusted
R2 = 0.84–0.89). The combo models (Adjusted
R2 = 0.78–0.89), including both point cloud and waveform metrics, have a relatively higher performance than the models fitted by point cloud based metrics (Adjusted
R2 = 0.74–0.86) and waveform based metrics (Adjusted
R2 = 0.72–0.84) individually. For AGB predictive models based on point cloud metrics (
Figure 7a), the mostly selected metric is the 95th percentile height (
H95). For AGB predictive models based on waveform metrics (
Figure 7b), the mostly selected metric is the mean of height of median energy (
HOMEμ). For AGB predictive models based on both point cloud and waveform metrics (
Figure 7c), the most frequently selected metrics are the 95th percentile height (
H95), mean of height of median energy (
HOMEμ) and mean of the height/median ratio (
HTMRμ).
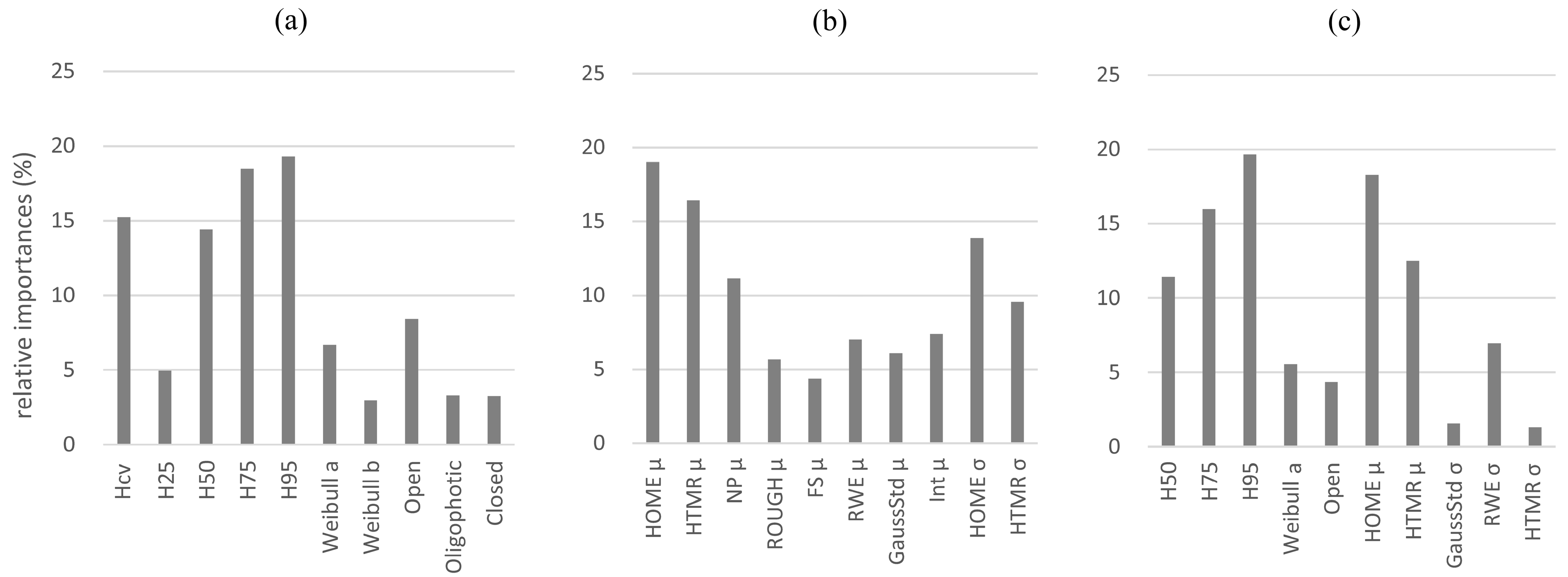
The total amount of variance of the AGB estimation models, fitted by point cloud and waveform metrics individually and in combination, have been divided among the independent variables as relative weights in
Figure 8. Based on the relative weights (i.e., the percentage of contribution for
R2) of the mostly selected metrics for all subsets, the metric of 95th percentile height (
H95) has the highest relative importance for AGB estimation (19.23%), followed by 75th percentile height (
H75) (18.02%) and coefficient of variation of heights (
Hcv) (15.18%) in the point cloud metrics based models. For the waveform metrics based models, the metric of mean of height of median energy (
HOMEμ) has the highest relative importance for AGB estimation (17.86%), followed by mean of the height/median ratio (
HTMRμ) (16.23%) and standard deviation of height of median energy (
HOMEσ) (14.78%). Similarly, for the combo model (including both point cloud and waveform based metrics), the metrics with highest importance are
H75 (15.12%),
H95 (19.42%),
HOMEμ (17.83%) and
HTMRμ (12.47%).
The best models for AGB estimation at individual tree level, based on point cloud metrics, waveform metrics and the combination of point cloud and waveform metrics are shown in
Table 5. In general, the combo model (Adjusted
R2 = 0.89, rRMSE = 5.66%), including both point cloud and waveform based metrics, has a relatively higher accuracy than the model fitted by point cloud metrics (Adjusted
R2 = 0.86, rRMSE = 6.01%) and waveform metrics (Adjusted
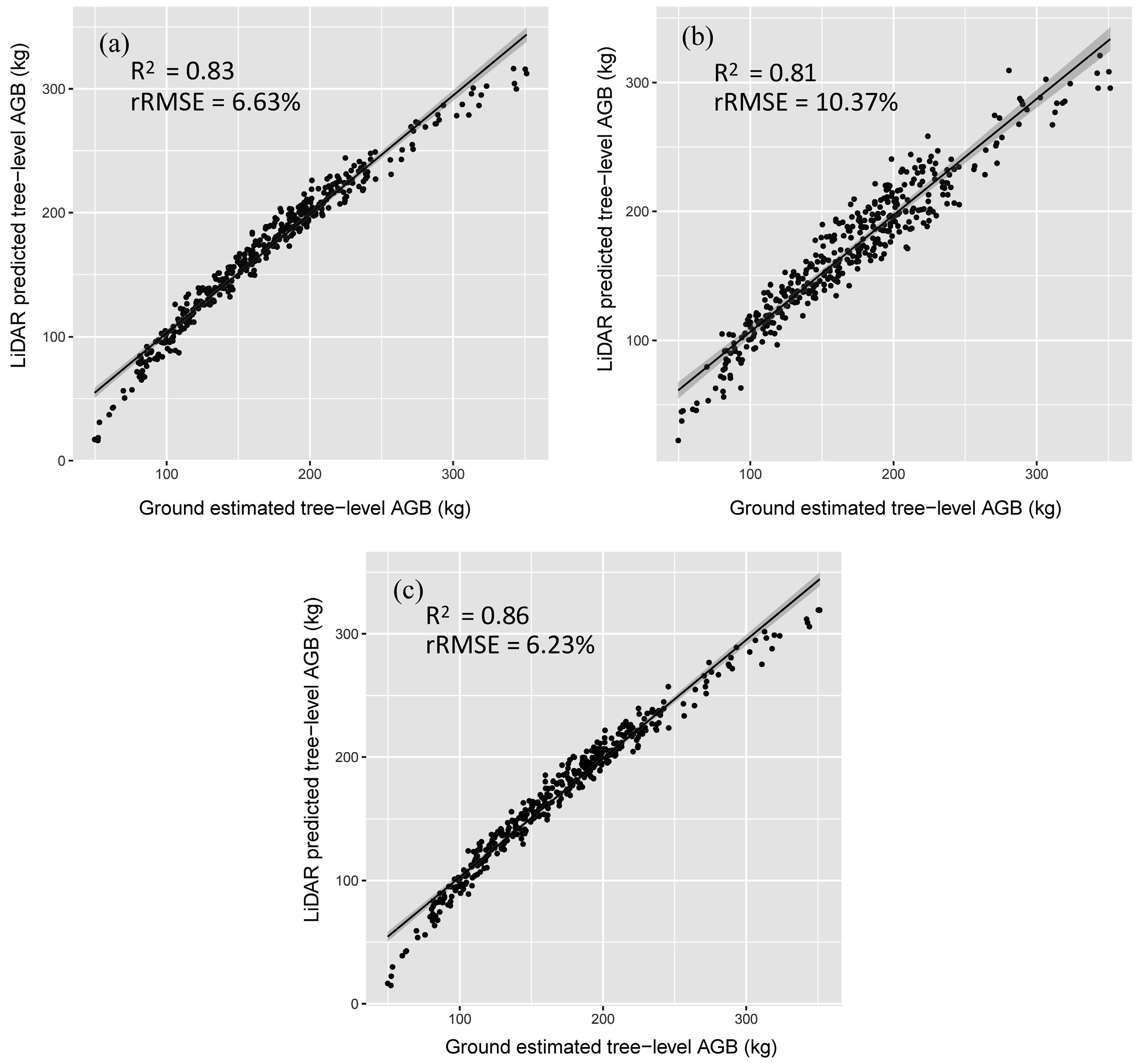
R2 = 0.84, rRMSE = 9.75%). For the AGB estimations of the correctly detected trees using the selected models, their relationships are close to the 1:1 line, according to the scatterplots of AGB between the field-estimated and model-predicted results for cross-validation (
Figure 9). The widths of the confidence intervals in the combo model are relatively narrow, indicating better quality of model fitting. In general, all the models did not have significant mean differences between field-estimated and model-predicted AGB (
Table 6). AGB predicted by the combo model (cross-validation
R2 = 0.86, rRMSE = 6.23%) has higher accuracy than point cloud metrics based model (cross-validation
R2 = 0.83, rRMSE = 6.63%) and waveform metrics based model (cross-validation
R2 = 0.81, rRMSE = 10.37%).
4. Discussion
To support sustainable forest management, high spatial resolution forest structure information is required to accurately assess the structure, composition and distribution of tree species with forest stands that, in turn, can be used as base information for management decisions across a range of spatial and temporal scales [
22]. ALS has emerged as one of the most promising remote sensing technologies, providing detailed, spatially explicit, three-dimensional information on forest structure, for operational applications in a wide range of disciplines related to the management of forest ecosystems [
70]. With increasing availability of ALS data, forest managers have seen opportunities for using ALS to meet a wider range of forest information needs [
71]. For instance, this study has shown promising results that FWF ALS data can be effectively used to estimate tree level AGB with relatively high accuracy in the study site. The ITC approach provides minimal scaling effects for investigating AGB, and the tree level AGB information (predicted by ALS data) can provide better understanding of the sources of uncertainties and errors when scaling-up (i.e., estimating AGB at larger scales using remote sensing) [
16]. This information can also be helpful for forest management, because the development and use of species-specific forest growth models or to inform a range of forest treatment schedules that are highly dependent on tree-level information. As there is no significant difference in the acquisition costs between discrete and FWF data, more contractors are promoting FWF data because of its perceived additional benefits [
12]. In previous studies, these additional information from FWF data has shown significant potential to forest management in the estimation of forest structure parameters at plot or stand scale [
34]. This study has also shown that the point cloud and waveform metrics extracted from full-waveform ALS data have strong capabilities for tree level AGB estimation in the coastal planted forests.
As reported in previous literature, the performances of ALS data based tree detection vary under different forest canopy conditions. In this research, a method similar to that of Popescu et al. (2002) and Popescu and Wynne (2004) was used to detect individual trees by the CHM, and to delineate tree crowns by fitting radius profiles and analyzing the critical points [
53,
54]. The detection rate of trees in this study was 85.6%, which is higher than that of 78.5% in a subtropical secondary forest [
72]. This is likely due to: (1) the crown shape of the coniferous trees in the study site tends to be conical and relatively isolated, which are likely to be detected by the local maxima algorithm used in this study, whereas trees in other studies may have more rounded crowns (e.g., broadleaved tree species) that are likely to overlap, or suppressed under the dominant canopies that may not be successfully detected; (2) the local maxima algorithm was applied on a CHM created from a relatively high density point clouds (approximate 9.2 pts∙m
−2 in the study site) extracted from full-waveform data by decomposition algorithms; and (3) the size of the search window was determined by a model fitted by the locally collected field measurements, which may enhance the performances of tree top identification. In this study, as most of the dominant trees were correctly detected by the tree detection algorithm, the ITC approach can be effective because dominant trees are usually the main contributors to AGB in forest stands [
73]. However, it should be noted that the errors in tree identification will be transferred into the AGB summaries at plot or stand level, and can result in the estimation of AGB with considerable systematic errors. The semi-ITC approach (or tree crown approach (TCA)) [
74], which indicates that an ALS detected tree crown can be a cluster that may contain no, one, or several field-measured trees, has shown potential to reduce the systematic errors, and can be used an alternative approach in the future to help reducing the systematic errors caused by errors from tree detection in the study site.
In this study, AGB is generally well predicted at tree level by the best regression models using point cloud and waveform metrics derived from the full-waveform ALS data. In previous studies, the discrete return data based metrics of multiple height quantiles have been commonly used for estimating forest biomass. However, these metrics could be highly correlated, especially if the data have a symmetric distribution [
75]. We found the tree level waveform-based metrics have low correlations among independent variables (
Figure 4), which agreed with the finding of Muss et al. (2011) [
76] that waveform metrics are less correlated and easier to be explained on the physical structure of the forests [
76].
Compared to the individual point cloud and waveform based models, the combo models performed the best, explaining a large amount of variability for AGB (
Table 5), and indicating significant synergies in point cloud and waveform metrics for estimating tree level AGB in the planted forests. Point cloud metrics allow the estimation of the 3-D structural characteristics of the forest canopy, which provides a good foundation for strong relationships between the metrics and forest biomass. Since most ALS point clouds are from upper tree crowns, the ALS upper percentile heights (e.g.,
H95) (
Figure 8) likely represent the tree top heights. The inclusion of the variable that describes variations (e.g.,
Hcv) accounts for the height variations of tree crowns, especially in the over- and mid-story, therefore potentially improves the characterization of vertical structure (
Table 5). The most important waveform metrics selected by the relative weight are: mean of height of median energy (
HOMEμ), mean of the height/median ratio (
HTMRμ) and standard deviation of height of median energy (
HOMEσ) (see
Figure 8 and
Table 5). The selection results of these metrics agreed with Drake et al. (2002) [
10] that the height-related metrics are the most sensitive, and highly correlated with forest age, basal area, and biomass [
10]. The metric of
HOMEμ appears to have the strongest capability for biomass estimation, and metric of
HOMEσ (can be interpreted as the variations of the vertical woody materials and foliage density within each tree crown) also appears to be sensitive to biomass as the third rank metric (
Figure 8). Drake et al. (2002) reported that
HOME is sensitive to canopy openness and the vertical arrangement of canopy elements footprint (0.05 ha) and plot (0.25–0.5 ha) level of the Laser Vegetation Imaging Sensor (LVIS) full-waveform data [
10]. In this study, we found similar principles at the tree level by using small-footprint (0.3 m) full-waveform ALS data. Tree crowns with densely packed woody materials and foliage reduce the number of laser pulses that can penetrate and reach the ground, thereby increasing
HOME. Conversely, for more transparent crowns, more laser pulses can pass through, thus, reducing
HOME.
HOME is sensitive to these differences and as a result makes it a potentially good indicator for biomass estimation in tree level as well. The second ranked metric is
HTMRμ, describes the change of
HOME relative to the canopy height. Drake et al. (2002) [
10] demonstrated that
HTMR (i.e.,
HTRT in their study) is sensitive to changes in forest structure, especially to plot-level basal area (with
R2 = 0.72) as the best single-term predictor [
10]. Neuenschwander et al. (2012) [
59] reported that the metric of vertical distribution ratio (
VDR, indicates height differences through the canopy which are likely related to energy distributed in the canopy, can be directly calculated from
HTMR as: 1-
HTMR) is sensitive to variations of forest canopy structures (i.e., what height the energy is distributed in the canopy), and is evenly distributed throughout the entire height profile of the forest [
59]. Their findings agreed with results of metrics selection in this study.
Tree-level AGB estimation has previously been studied by Popescu 2007 [
16] and Hauglin et al. (2013) [
18] using discrete return ALS data, and assessed by Allouis et al. (2013) [
36] and Wu et al. (2009) [
37] using full-waveform ALS data [
16,
17,
18,
19,
36,
37]. Popescu (2007) used ALS data to estimate the AGB and biomass component for loblolly pine (
Pinus taeda L.) in southeastern United States. ALS-derived single tree-level height and crown diameter were used to estimate tree DBH, AGB and component biomass using regression models. The results showed that 93% of the variability of individual tree biomass, 90% of DBH, and 79%–80% of components biomass could be explained by the predictive models. Compared to their results, the accuracy of AGB (adjusted
R2 = 0.84–0.89) estimations in the present study was relatively lower. Potential sources of error could include field estimates of biomass components (including errors in filed measurements of DBH and height, and errors in allometric equations), identification of individual trees, statistical error associated with estimating coefficients, and data processing errors [
16]. Allouis et al. (2013) [
36] assessed the capability of full-waveform data to improve the estimation of individual tree-based biomass in a black pine forest in southern France. They found that including the full-waveform metrics improved the accuracy of biomass estimation [
36]. The results showed that CHM (generated from point clouds) + FWF metrics models (adjusted
R2 = 0.90–0.91) has higher accuracy than CHM-only model (adjusted
R2 = 0.87), which has a general agreement of our results that the combo model (Adjusted
R2 = 0.89, rRMSE = 5.66%), including both point cloud (similar to the CHM that it also characterizes the 3-D structure of the tree crown) and waveform based metrics, has a relatively higher accuracy than the point cloud-only model (Adjusted
R2 = 0.86).
In this study, composite waveforms were constructed by directly georeferecing each bin of the raw waveform within the voxels. As a result, incomplete composite waveforms could be created if they were located around the edge of tree boundaries. Although these incomplete composite waveforms were not used for calculating final metrics, future work could focus on how to improve the waveform synthesis approach in order to construct improved composite waveforms. Nevertheless, the exploration of spectral, textural or shape metrics from hyperspectral and high-resolution data might further extend the range of preferred metrics and enhance the capability of tree level AGB estimation in planted forests. Building links between the inherent architectural differences of tree crown and how they are manifested in waveform metrics remains a critical endeavor for remote sensing scientists and ecologists. In small footprint ALS studies, one footprint (e.g., diameter = 30 cm) only describes a portion of the tree structure. As a result, gathering field measurements (potentially, terrestrial laser scanning (TLS) data that can be used as a precise and efficient tool to directly measure detailed 3D models of vegetation elements) and developing relationships between crown structural characteristics and waveform metrics (aggregated or calculated variability within tree crown) is important to improve the direct interpretability of the waveform metrics [
59].
5. Conclusions
This study contributes to the research community for assessing the capability of the metrics of point clouds, extracted from the full-waveform Airborne Laser Scanning (ALS) data, and of composite waveforms, calculated based on a voxel-based approach, for estimating tree level Aboveground biomass (AGB) individually and in combination, over a planted forest in the coastal region of east China. The importance of point cloud and waveform metrics for estimating tree-level AGB were investigated by all subsets models and relative weight indices. The capability of the point cloud and waveform metrics based models and combo model (including the combination of both point cloud and waveform metrics) for tree-level AGB estimation was assessed and the accuracies of these models were evaluated.
The results demonstrated that most of the waveform metrics have relatively low correlation coefficients (<0.60) with other metrics. The combo models (Adjusted R2 = 0.78–0.89), including both point cloud and waveform metrics, have a relatively higher performance than the models fitted by point cloud metrics-only (Adjusted R2 = 0.74–0.86) and waveform metrics-only (Adjusted R2 = 0.72–0.84), with the mostly selected metrics of the 95th percentile height (H95), mean of height of median energy (HOMEμ) and mean of the height/median ratio (HTMRμ). Based on the relative weights (i.e., the percentage of contribution for R2) of the mostly selected metrics for all subsets, the metric of 95th percentile height (H95) has the highest relative importance for AGB estimation (19.23%), followed by 75th percentile height (H75) (18.02%) and coefficient of variation of heights (Hcv) (15.18%) in the point cloud metrics based models. For the waveform metrics based models, the metric of mean of height of median energy (HOMEμ) has the highest relative importance for AGB estimation (17.86%), followed by mean of the height/median ratio (HTMRμ) (16.23%) and standard deviation of height of median energy (HOMEσ) (14.78%).
However, although this study showed significant synergies in point cloud and waveform metrics for estimating tree level AGB in planted forests, future works should focus on building links between the inherent architectural differences of tree crown and how they are manifested in waveform metrics, to improve the direct interpretability of these metrics. China has the largest area of planted forest in the world, and these forests contribute to a large amount of carbon sequestration in terrestrial ecosystems. Thus, this study demonstrated benefits of using full-waveform ALS data for estimating biomass at tree level, for sustainable forest management and mitigating climate change by planted forest.