1. Introduction
The Normalized Difference Vegetation Index (NDVI) is one of the most commonly used vegetation indices to characterize the absorptive and reflective features of vegetation, which represent vegetation greenness and vigor [
1,
2,
3]. Therefore, NDVI time-series data derived from multi-temporal satellite images are an appropriate data source for studying the spatial and temporal dynamics of ecosystem responses to climate change [
4]. However, due to technological and budget limitations, there has been a trade-off within remote sensing instruments from high spatial resolution to more frequent coverage. Consequently, the available NDVI time-series products are generated from sensors with frequent observations (e.g., daily) but coarse spatial resolutions, ranging from 250 m to 8000 m, such as Moderate Resolution Imaging Spectroradiometer (MODIS) and NOAA Advanced Very High Resolution Radiometer (AVHRR). As a result, these products are incapable of capturing spatial details necessary for monitoring land cover and ecosystem changes in heterogeneous areas [
5]. On the other hand, NDVI data derived from Landsat sensors (e.g., TM/ETM+/OLI with 30 meters resolution at visible and near infrared bands) can successfully capture the spatial details and alleviate the spectral mixture issue to some extent [
6,
7]. However, longer observation cycles of these satellites and frequent cloud contamination in some regions limit their application for detecting rapid changes in the ecosystem [
8,
9]. Combining NDVI data from multi-sensors is a possible solution for producing high spatial and temporal resolution NDVI data, which is a critical requirement for monitoring vegetation dynamics.
Since Landsat satellites and MODIS have similar orbital parameters and less than 30 minutes difference in crossing time, methods have recently been developed and validated for blending Landsat TM/ETM+ and MODIS data [
10,
11,
12,
13]. Generally, there are two strategies to generate a fine-spatial resolution NDVI data from MODIS time series with the help of Landsat data. The first one is the indirect strategy, in which fine spatial-resolution NDVI is calculated from the reflectance data downscaled from MODIS data [
14,
15]. Up to now, there are many algorithms developed for downscaling MODIS reflectance with the help of fine-resolution sensor imagery. These algorithms include the Spatial and Temporal Adaptive Reflectance Fusion Model (STARFM) [
10], the Spatial Temporal Adaptive Algorithm for mapping Reflectance Change (STAARCH) [
11], the Semi-physical Fusion Approach [
16], the Enhanced STARFM (ESTARFM) [
13], the multi-temporal fusion method [
17], and the Spatial Temporal Data Fusion Approach (STDFA) [
18]. These methods can be categorized into two groups. STARFM, STAARCH, the Semi-physical Fusion Approach, and ESTARFM belong to the first group, which obtains the relationship of reflectance between MODIS and Landsat data from one or more pairs of images. Then this relationship is used to predict Landsat-like reflectance from MODIS data at prediction dates. The drawbacks for these methods are: (1) they often require known image pairs as close as possible to the prediction dates because the relationship between MODIS and Landsat learned from known pairs may change with time; and (2) they often have more strict assumptions when deducing the relationship between MODIS and Landsat, and these assumptions may not valid in reality. For example, STARFM assumes Landsat pixels within one MODIS pixel have the same change, while ESTARFM assumes that reflectance of each land cover has a linear change between two known dates. The second group is the unmixing-based methods, including the multi-temporal fusion method and STDFA. These algorithms predict fine-resolution reflectance from MODIS using the linear unmixing techniques, in which the fractional cover of each endmember is provided by Landsat or other ancillary data [
19]. The multi-temporal fusion method employs a soft clustering method to address the intra-class variability issue. However, considering that soft clustering is not a mature technology, the errors from soft clustering introduce extra uncertainties for predicting fine-resolution reflectance. For STDFA, it only used two Landsat data (the beginning and end date of the prediction period) to estimate fractional cover. This would lose important temporal change information and lead to an inappropriate cluster result. Besides, it generated synthetic Landsat data by adding temporal changes to original data, while temporal changes were obtained from the reflectance unmixed from the MODIS data at two dates. During this procedure, errors from two spectral unmixing steps would aggregate to the final predictions. Additionally, most of the current unmixing based methods only use fine-resolution data to provide fraction information, while other important information including spatial details are missing in these methods. More important, indirect approaches, regardless of what reflectance blending algorithms used, all suffer from heavy computation loads and errors accumulation, because both reflectance of red and NIR bands must be predicted before calculating NDVI. A recent study using three sites with contrasting spatial and temporal heterogeneity has demonstrated this limitation for nine indices including NDVI [
15]. It compares the prediction results of using the indirect approaches and the ones of directly downscaling NDVI using the blending algorithms (e.g., STARFM, ESTARFM), and concludes that directly downscaling NDVI would get better results than indirect strategy [
15].
The second strategy is to directly downscale the NDVI product, which could directly apply blending algorithms from the first strategy or use methods specifically developed for blending NDVI as well. Busetto
et al. proposed a novel approach for directly downscaling MODIS NDVI based on a Weighted Linear Mixing model (WLM) [
20,
21], which assumes that the NDVI value of a coarse pixel can be calculated as the area fraction weighted average of NDVI values of several land cover classes at a fine spatial scale within the coarse pixel [
22]. However, its basic assumption that the same land cover type has the same NDVI value is somewhat unreasonable in the real situations, because spectral variability is very common in the same land cover type, especially within larger areas [
23]. Such unreasonable assumption is likely to cause large estimation errors and significant loss of spatial details. Recently, a Spatial and Temporal Adaptive Vegetation index Fusion Model (STAVFM) was also developed based on STARFM to directly downscale MODIS NDVI with the help of HJ-1 images, which include four spectral bands with similar characteristics (
i.e., spatial resolution, band width, spectral response functions) with band 1–4 of Landsat TM sensor [
24]. To modify the original STARFM model, STAVFM defines a time window based on expert knowledge about crop phenology, which was then used to choose more appropriate data pairs for the blending process. However, the specification of crop phenology limits its applications to other regions. Therefore, the main challenge remains of directly downscaling NDVI products with higher accuracy and fewer requirements of ancillary data or prior knowledge.
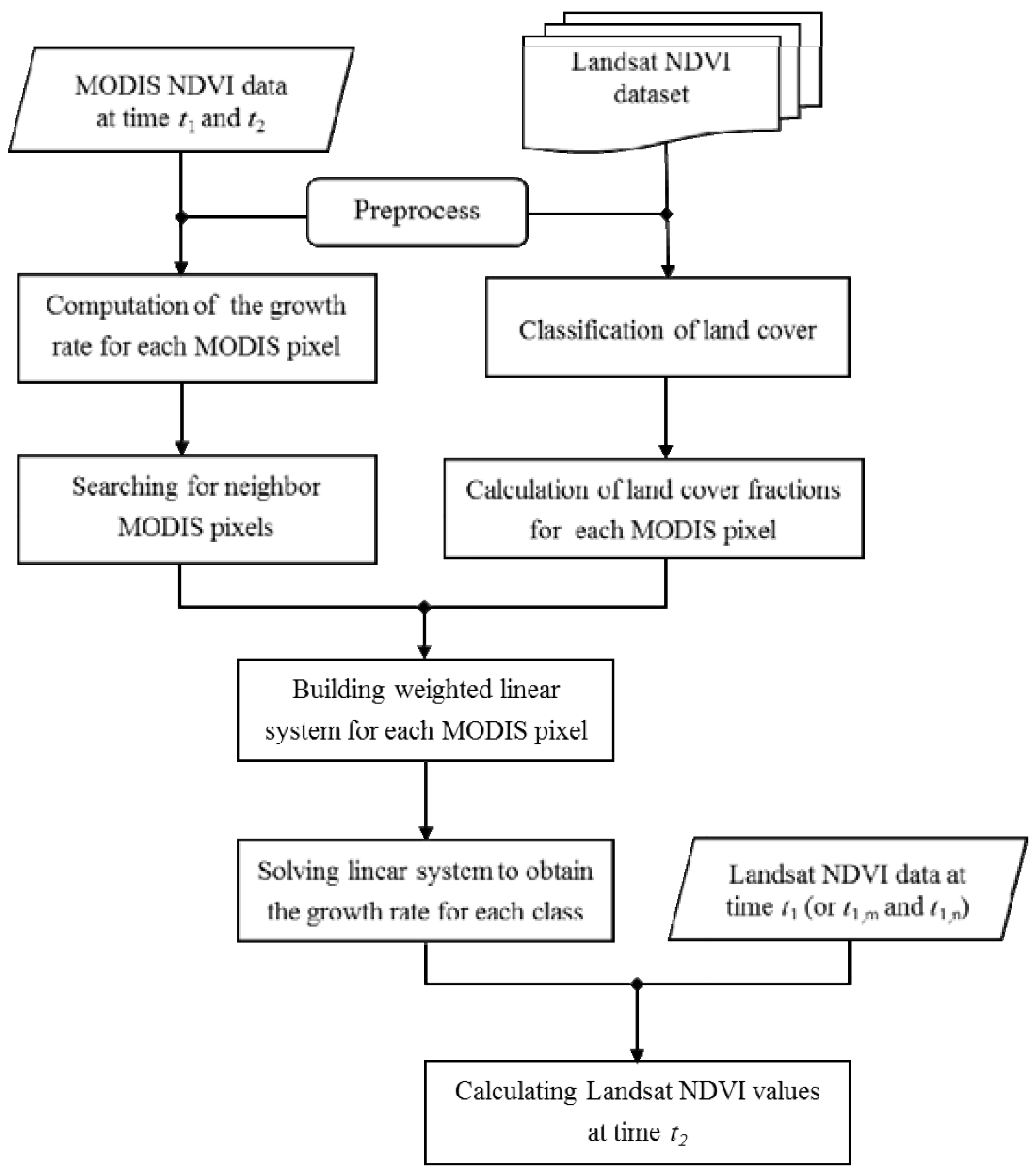
In order to address limitations of current methods for directly downscaling NDVI from MODIS, we developed the NDVI Linear Mixing Growth model (referred to hereafter as NDVI-LMGM) to directly obtain NDVI time-series data with a high spatial resolution. The main goal of the proposed approach is to achieve more accurate results but with less computational costs and prior knowledge through using the spatial details from acquired Landsat images. To test the effectiveness of the proposed approach, we compared it to several other typical and popular methods, including STARFM [
10], ESTARFM [
13], and WLM [
22].
4. Discussions
4.1. The Advantages of NDVI-LMGM
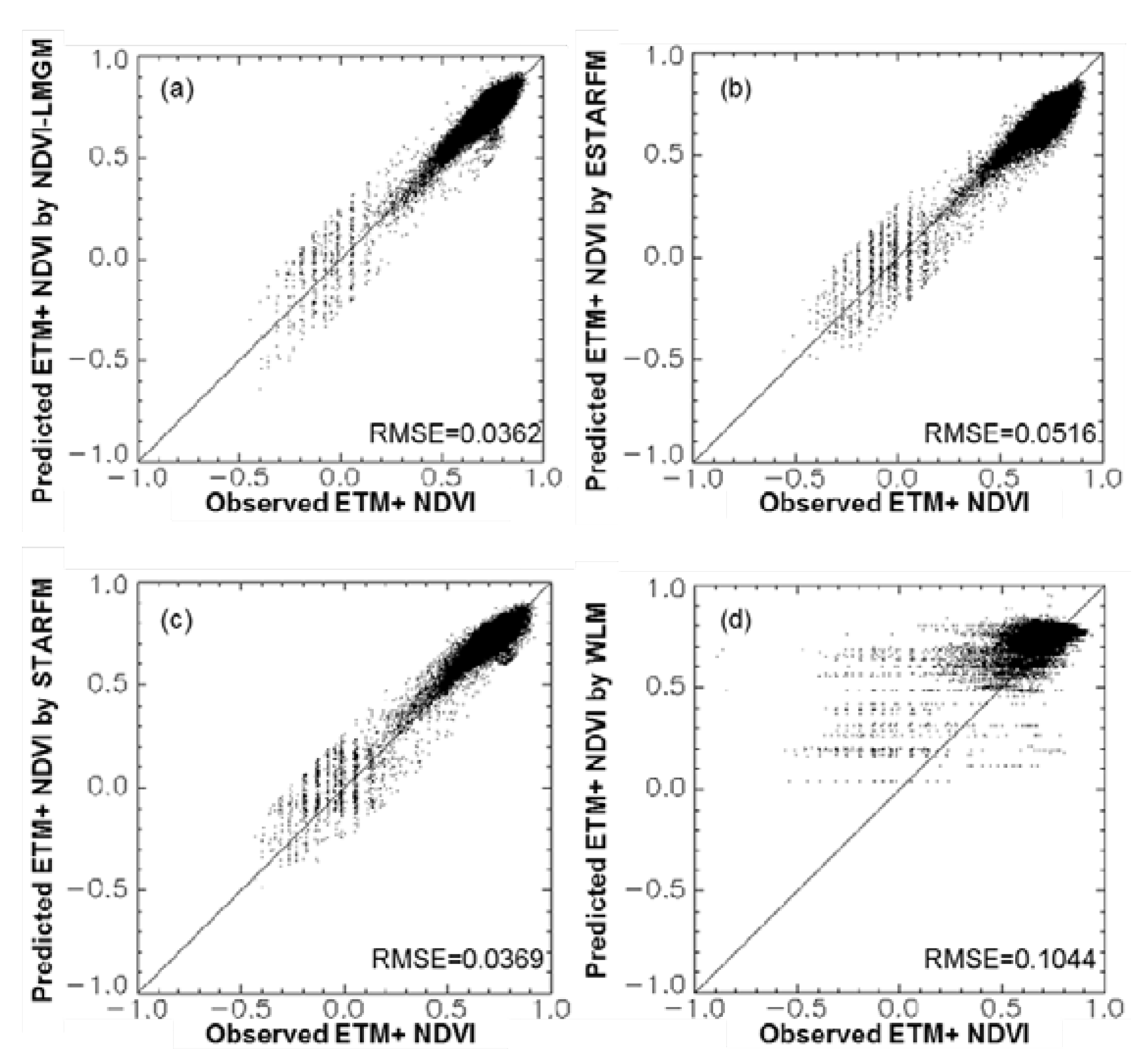
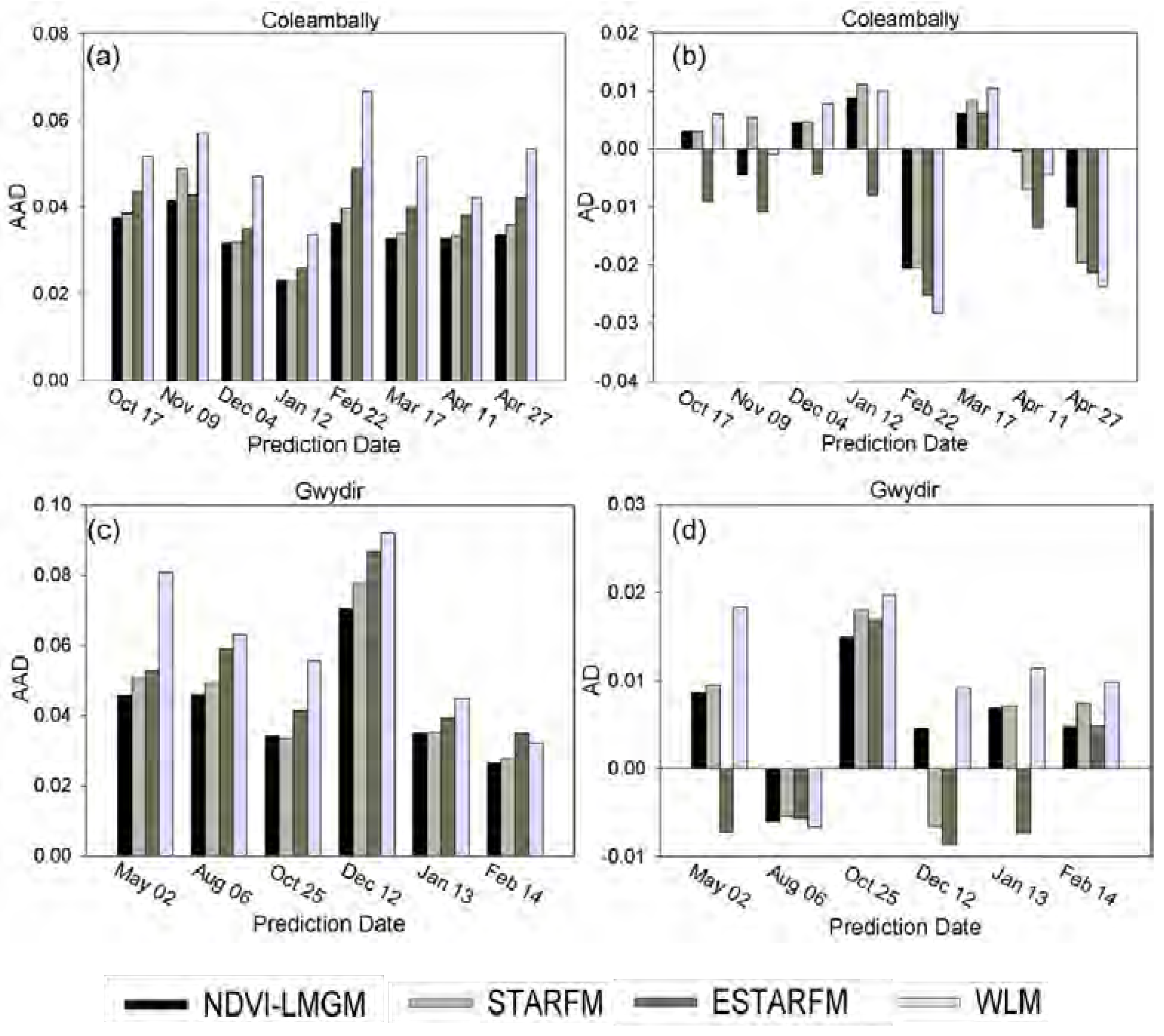
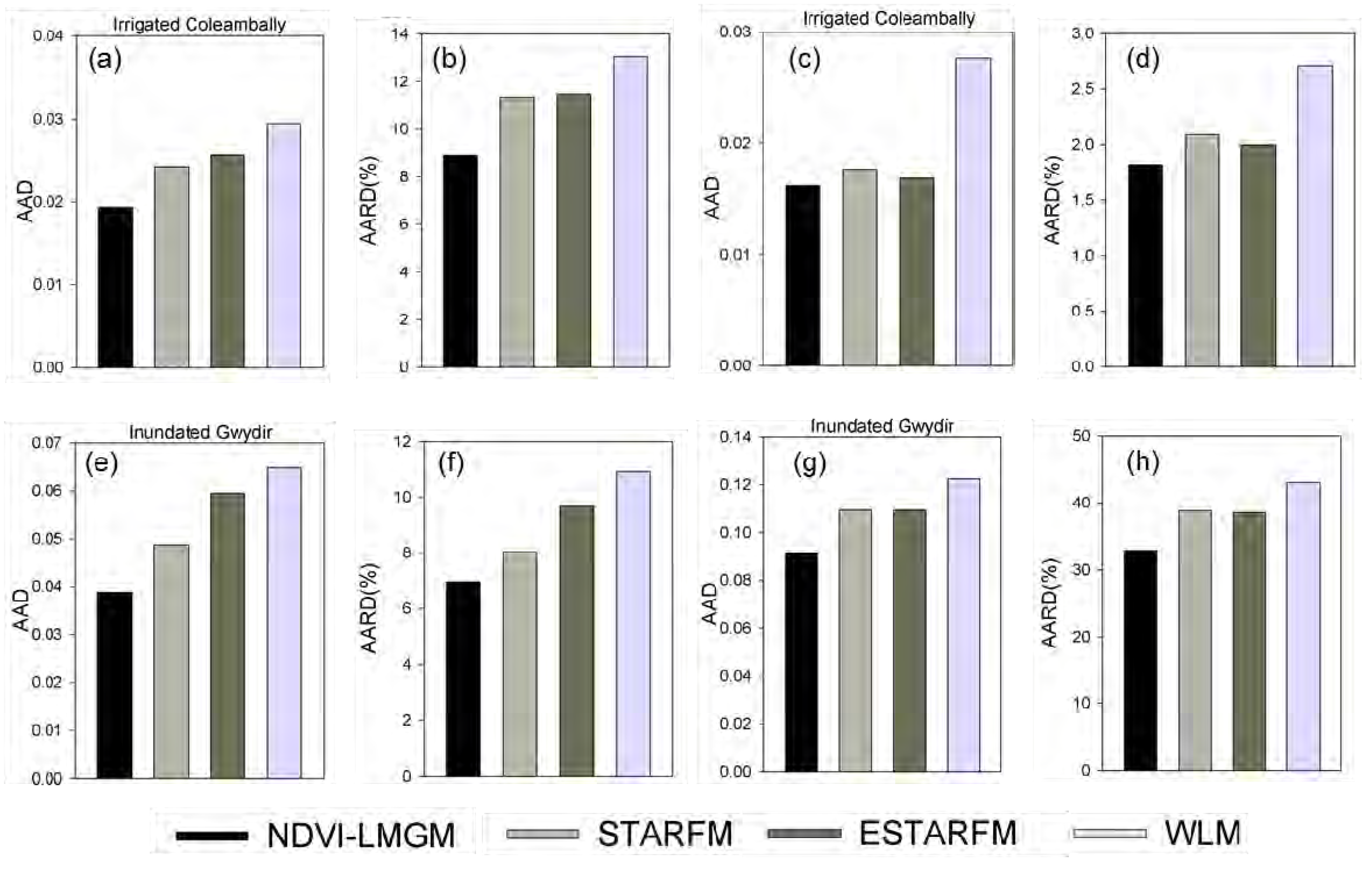
Experiments at three different sites have demonstrated that NDVI-LMGM performs better than the other three methods, STARFM, ESTARFM and WLM. Its superior performance may be attributed to the following advantages: (1) more reasonable assumptions; (2) better use of the information from base TM/ETM+ images; and (3) better selection of pixels with the same land cover type. These advantages will be explained in detail in the following sections.
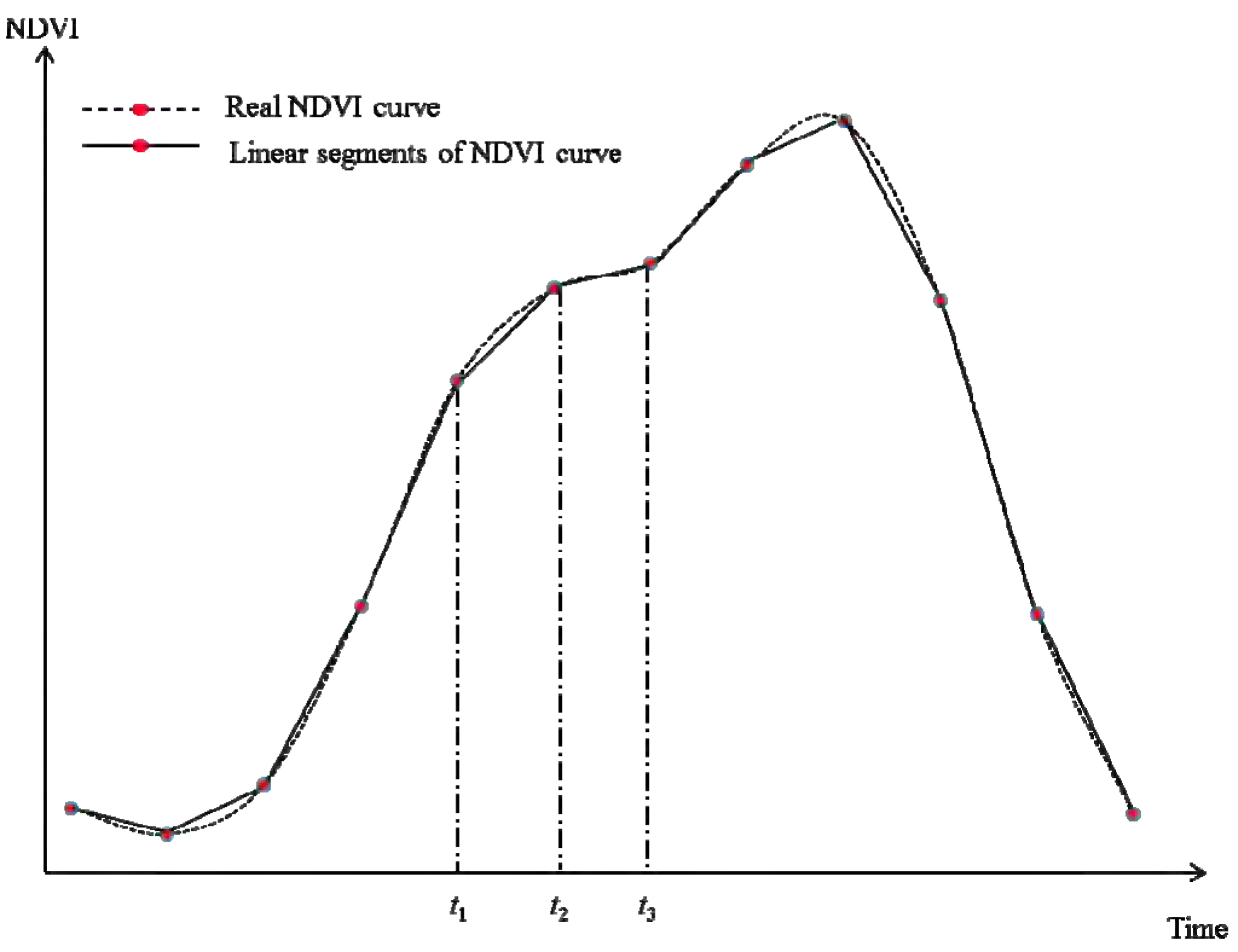
First, the most important assumption of NDVI-LMGM is that neighboring pixels with the same land cover exhibit the same linear change (
i.e., growth rate) during a short period. NDVI-LMGM has more practical and reasonable assumptions compared to the other methods. Generally, the growth of vegetation can be assumed to be under the influence of two groups of controlling factors. The first are biotic factors (e.g., vegetation type), which are not highly dependent on their geo-location. The second are the environmental factors, such as soil conditions, temperature, precipitation,
etc., which have a strong dependence on geo-location. NDVI is representative of vegetation greenness and vigor, so it is reasonable to consider NDVI as a function of these two groups of controlling factors. In the same way, the change pattern of NDVI (growth rate) is also determined by both biotic factors and environmental factors; however, environmental factors play a more significant role than biotic factors, because the variations in environmental factors over time are more considerable than that of the biotic factors alone. Consequently, NDVI change within the same land cover over a small area can be assumed to be similar, because (1) the same land cover type in a small area has similar biotic factors; and (2) environmental factors have less variation in a small area according to the Tobler’s First Law of Geography [
38]. STARFM assumes that a MODIS pixel and all Landsat pixels within that MODIS pixel share similar change patterns, which ignores the variation in change pattern among land cover types [
10]. This unreasonable assumption decreases STARFM’s performance in heterogeneous landscapes [
13]. For ESTARFM, it estimates the linear change rate between two Landsat observation dates and applies it to estimate the reflectance changes between the observation dates and any prediction dates during two observation dates [
13]. However, the assumption of ESTARFM is invalid in some real situations, especially for areas with frequent cloud contamination or temporally heterogeneous landscapes, in which rapid changes can occur in a short time. For WLM, the basic assumption is that each land cover class has the same NDVI value regardless of the differences in biotic and environmental conditions [
22]. This assumption cannot hold true in reality due to the diversity of environmental conditions and biotic characteristics, which can result in significant variation of NDVI values within the same land cover class in a larger area, also referred to as spectral variability when used in spectral mixture analysis [
23].
Second, NDVI-LMGM makes better use of the spatial information from Landsat observations. In NDVI-LMGM, the original Landsat-scale NDVI value is used (
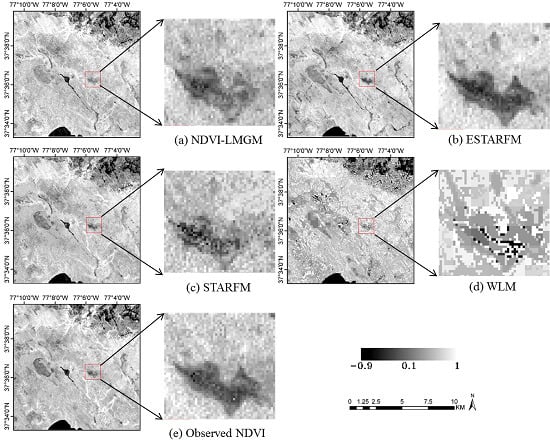
i.e., the Landsat observation) as the basic value to which the predicted Landsat-scale NDVI changes are added. This method retains spatial details very well since the base Landsat image preserves more spatial details. On the other hand, WLM only uses the Landsat images to obtain constant weights for the linear system, which ignores the within-class variations and results in a strong patchy effect (
Figure 9). In addition, STARFM and ESTARFM both employ a weighted average procedure for predictions from all neighboring similar pixels. Although it would improve the prediction results to some extent, it also might produce smoother prediction results while some spatial details could be lost.
Third, an important step for all the methods is to determine the pixels that share the same change patterns, also referred to as similar pixels or pixels with same land cover. NDVI-LMGM employs ISODATA to classify all available Landsat images to obtain the land cover map. As we know, multi-temporal images can improve the land cover classification because temporal information is an important supplement to spectral information, which helps differentiate various land covers. Therefore, it is expected that NDVI-LMGM can accurately select pixels of similar land cover. Unlike NDVI-LMGM, WLM uses existing fine-resolution land cover maps to select pixels of similar land cover, which has two limitations (1) high-quality land cover maps are often unavailable; and (2) temporal mismatching between the land cover map and prediction dates might introduce errors in the downscaling process because land cover often changes over time. Both STARFM and ESTARFM use one or two base images to select similar pixels based on a threshold of spectral distance. This procedure may introduce significant errors in similar pixel selection when the base images are further away from the prediction date. In addition, the determination of the threshold is very empirical, which might affect the robustness of STARFM and ESTARFM.
Along with the above advantages, which lead to a more accurate prediction, NDVI-LMGM is more efficient in computational time than the three other methods, especially when compared to indirect methods, such as ESTARFM. As mentioned above, all current methods (i.e., STARFM, ESTARFM and WLM) require a procedure for identifying similar pixels within each target area, which is very time-consuming. Instead, NDVI-LMGM directly employs neighboring MODIS pixels and the land cover result to build a linear model system. The high efficiency of NDVI-LMGM makes it appropriate for large-scale applications.
4.2. Influence of Parameters in NDVI-LMGM
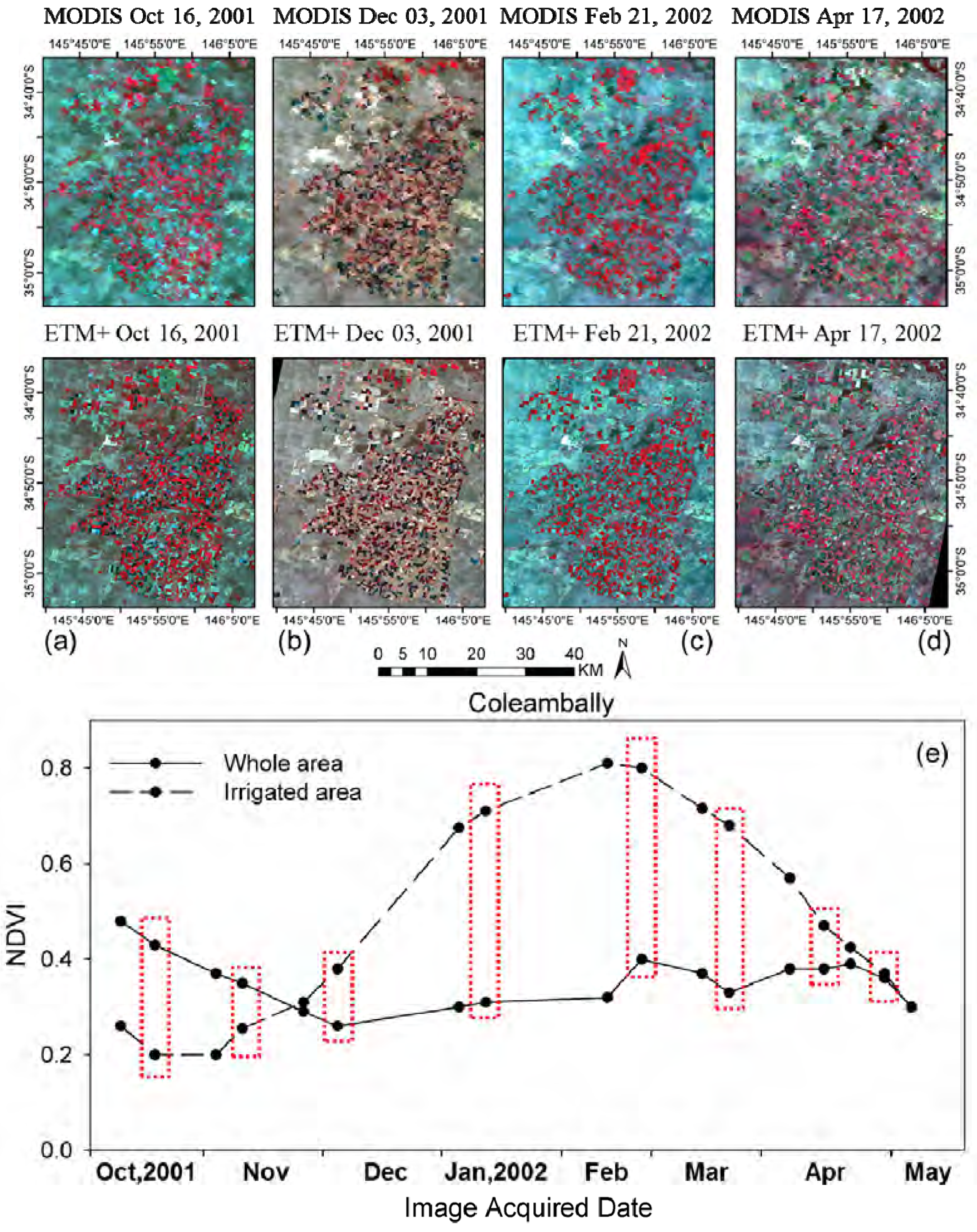
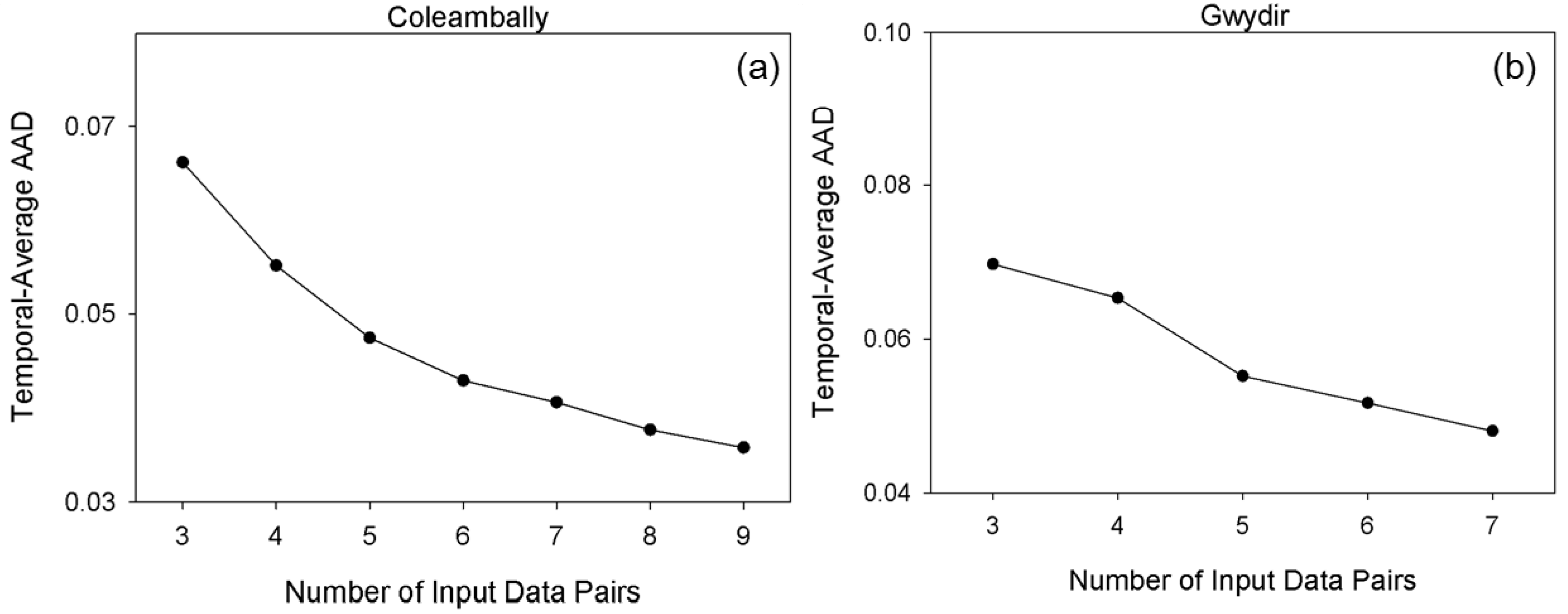
As described, the class number in unsupervised classification is empirically determined by users. Therefore, it is necessary to investigate the influence of the class numbers for prediction accuracy. Accordingly, experiments were conducted in the forest site and two Australian sites using various class numbers in unsupervised classification (i.e., two-nine classes in this study). For the forest site, 11 July 2001 was used as the prediction date and the other two pairs were used as input. For Coleambally, 12 February 2002 was selected as the prediction date and image pairs on 11 January and 21 February were used as input. Lastly, for Gwydir, 2 May 2004 was the prediction date and image pairs on 16 April and 5 July were set as inputs.
Figure 14.
The prediction error (AAD) vs. class number for three study sites, (a) the forest site (prediction date: 11 July 2001); (b) Coleambally site (prediction date: 12 February 2002); and (c) Gwydir site (prediction date: 2 May 2004).
Figure 14.
The prediction error (AAD) vs. class number for three study sites, (a) the forest site (prediction date: 11 July 2001); (b) Coleambally site (prediction date: 12 February 2002); and (c) Gwydir site (prediction date: 2 May 2004).
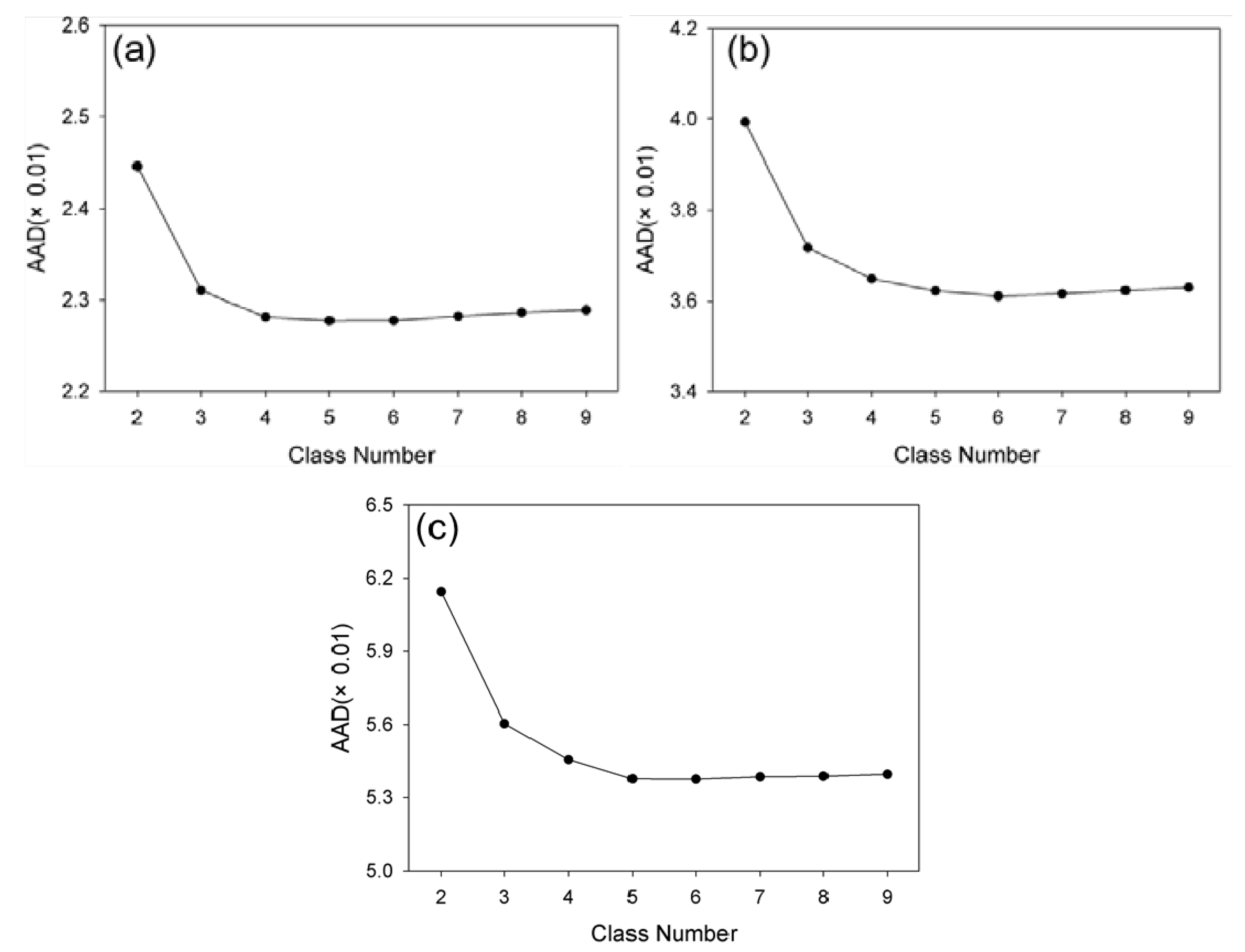
Figure 15 illustrates the prediction error (AAD) against the numbers of classes in the three test sites. Although AAD values vary according to the number of classes, these variation ranges are relatively small at 0.0017, 0.0038 and 0.0076, respectively, indicating that NDVI-LMGM is not highly sensitive to the number of classes. Moreover, there was a general trend among the three sites in which the AAD decreased at first, and then became stable followed by a slight increase as the class number steadily increased (
Figure 14). Although the trends are similar, the change points from a decreasing to an increasing curve, representing the optimal class number, are different. Specifically, the class number corresponding to the change point for the forest site was four, while it was six and five for the Coleambally and Gwydir, respectively. It is obvious that the optimal class number in a relatively homogenous site, such as the forest site, was lower than those in spatially heterogeneous sites, such as the two Australian sites. This finding substantiates the guideline of ISODATA classification in NDVI-LMGM that the more complex the landscape in the study area, the more class numbers should be established. It also indicates that the class number of four, five or six could be recommended for applications according to the complexity of landscapes, which may ensure a satisfactory prediction result of NDVI-LMGM.
The growth rate from
t1 to
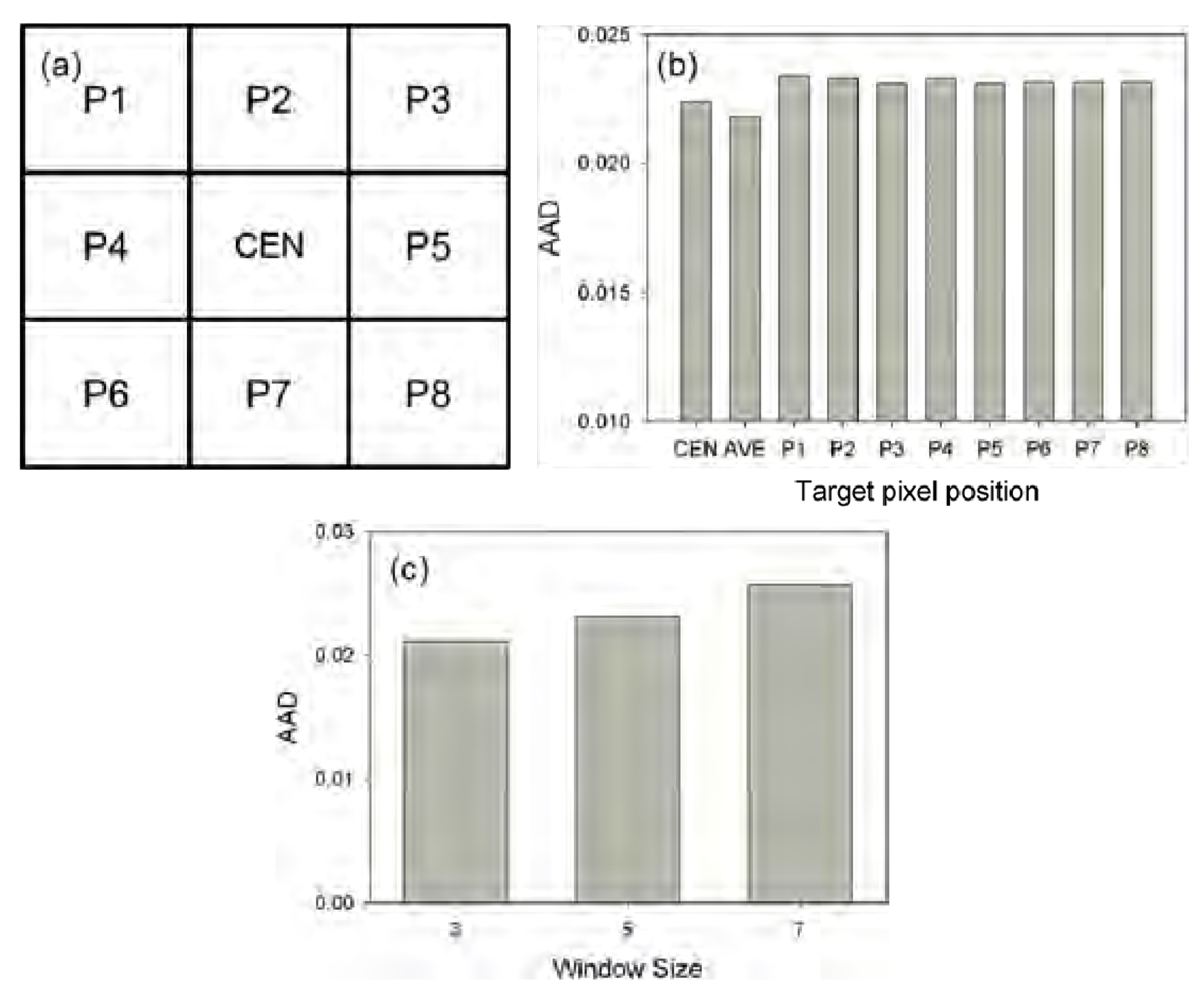
t2 at TM scale is the most important parameter required to be estimated in NDVI-LMGM. Specifically, spatially neighboring MODIS pixels have been employed to solve a linear model system (Equation (4)), resulting in a situation in which each MODIS pixel is used several times and the same number of growth rates for each MODIS pixel can be obtained when it is located in varying positions of a moving window (e.g., nine times for each MODIS pixel when the moving window size is set as 3 × 3). However, in NDVI-LMGM, only the growth rate can be estimated when the MODIS pixel located in the center of the window (
Figure 15a) is used for prediction (Equation (1)). How does the performance change if the growth rate is estimated when a given MODIS pixel is at other positions within a moving window or the average of all estimated growth rates is used? To answer this question, an experiment was conducted in the forest site using the same data and parameter settings as those shown in
Figure 8. As
Figure 15b shows, the AAD values for using the growth rate estimated when the MODIS pixel was located in the center of the window or when the average of all estimated growth rates was used, both of them were lower than those using the growth rate estimated when a given MODIS pixel was at any other position within the moving window. Accordingly, considering the simplification of NDVI-LMGM, the growth rate estimated from the center of the window was finally adopted. In addition, the moving window size in NDVI-LMGM was also investigated in the forest study site.
Figure 15c exhibits the AAD variation when three different window sizes were used. It was evident that the larger window size, after satisfying Equation (6), could introduce more significant errors into the prediction results, mainly because the larger window size may challenge the assumption underlying our method that the same land cover type has a similar growth rate in a small area Therefore, using the moving window with a size of 3 × 3 is recommended if the number of land cover classes is set to less than nine.
Figure 15.
(a) The MODIS pixels within a moving window used to solve a linear model system, CEN indicates the center of the moving window. (b) The performance comparison by using different growth rates estimated from different positions of a moving window. Here, AVE stands for averaged growth rate of all growth rates. (c) The AAD comparison using different window sizes.
Figure 15.
(a) The MODIS pixels within a moving window used to solve a linear model system, CEN indicates the center of the moving window. (b) The performance comparison by using different growth rates estimated from different positions of a moving window. Here, AVE stands for averaged growth rate of all growth rates. (c) The AAD comparison using different window sizes.
4.3. The Limitations of NDVI-LMGM
In addition to the strengths mentioned, NDVI-LMGM also has some limitations that should be noted. First, as discussed in
Section 4.2, the number of classes will affect its prediction performance. Also, the parameters of class number should be determined according to the complexity of the landscape in the study area. The presence of multi-temporal Landsat images would help to improve the classification and prediction results. However, we acknowledge here that using smaller numbers of classifications can create errors, which would also affect the NDVI prediction accuracy of our method even if the temporal TM/ETM+ images are used in classification. As an operational method, we must accept such imperfect results because obtaining a perfect classification by using any up-to-date classifier is currently unpractical. However, it is expected that any attempts to improve classification accuracy will benefit our method after it is integrated into our system.
The prediction results would be influenced by the composite process of the MODIS data. Walker
et al. has analyzed how this process would affect the results of STARFM [
14]. The study revealed that the composite product (including 8-day MOD09A1 and 16-day NBAR (MCD43A3)) could be problematic, especially for the regions that experience rapid vegetation changes. However, the use of composite datasets during the peak of the growing season has the advantage that vegetation states is not likely to change appreciably during an 8- or 16-day period. Thus, for the region with rapid vegetation changes, the daily MODIS product (
i.e., MOD09GA) is a better option for use as input. Moreover, the acquisition date of the input data also has an inevitable influence on the prediction performance (
Section 3.3), which is similar to other existing methods [
10,
13,
14].
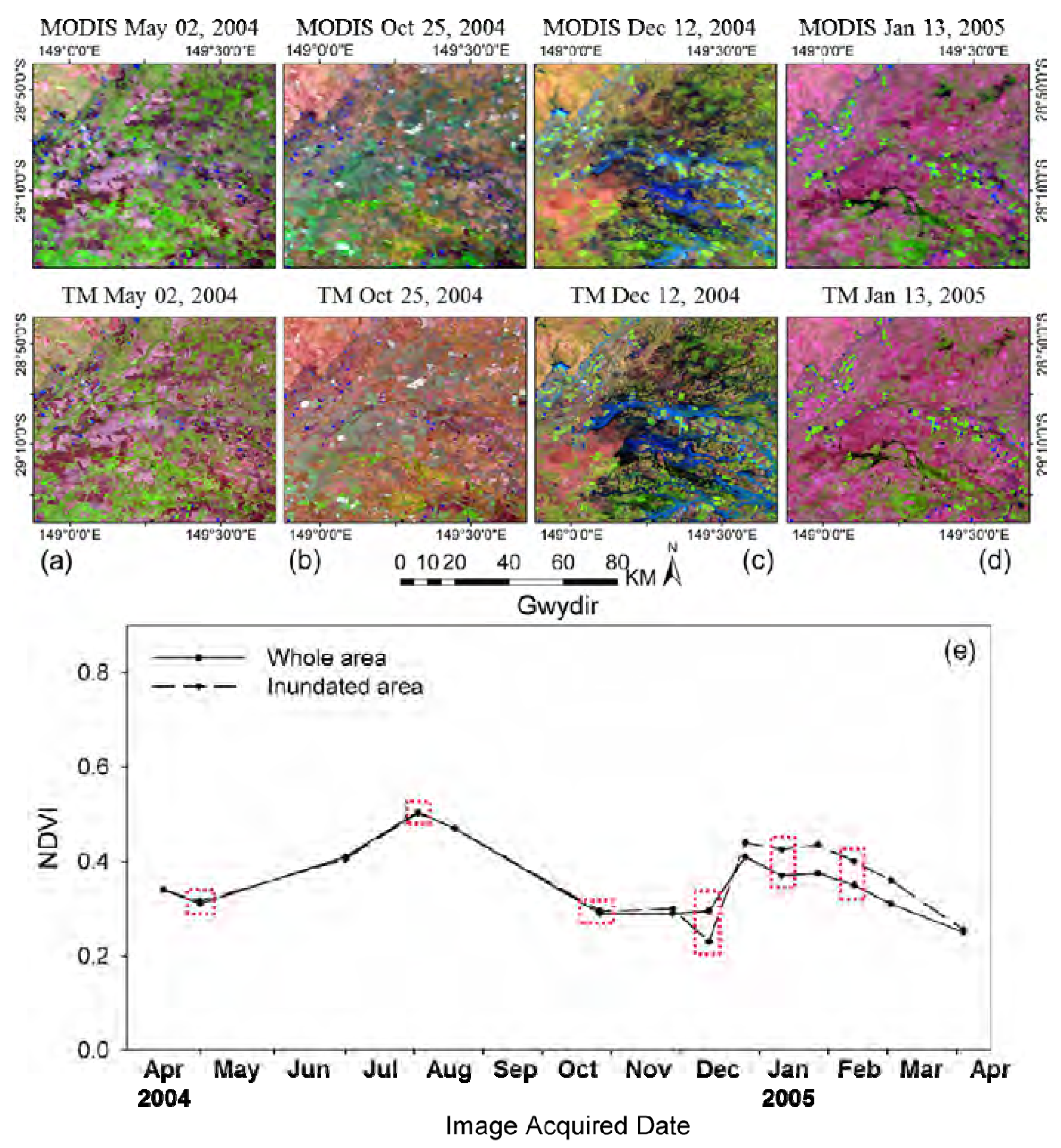
Additionally, it should be noted that small-scale land cover changes occurred between the observation date and prediction date might impact on the prediction, especially when only single observed image are used for clustering. These small-scale changes may not been detected in the classification map, thus leading to large errors for these regions. This drawback could be found in tests over Gwydir when the flood suddenly occurred to that region (
Section 3.3). Fortunately, if there are multi-temporal high spatial resolution image that can be used for clustering, these land cover changes could be captured based on their different temporal change information from other land cover types.
5. Conclusion
This study proposed a new method, NDVI-LMGM, to produce high spatial resolution NDVI time-series data by using MODIS NDVI time-series data and Landsat images. As demonstrated by experiments at three different sites with contrasting spatial and temporal heterogeneity, we found that NDVI-LMGM could produce high spatial resolution NDVI time-series dataset with smaller errors (with the average absolute difference less than 0.08 and average absolute relative difference less than 9% for three experiments) and less bias compared to STARFM, ESTARFM and WLM.
NDVI-LMGM combines the NDVI Linear Mixing model with the NDVI Linear Growth model, with which the NDVI prediction problem can be transformed into solving a linear system. Since there are several growth rates for different land cover classes at the Landsat scale, which require estimation in the linear system, the extra information from the spatially neighboring MODIS pixels was employed based on the assumption that spatially neighboring pixels with the same land cover type in a small area have similar growth rates. Compared with the assumptions needed for STARFM, ESTARFM and WLM, the NDVI-LMGM’s assumption is easier to satisfy and is more reasonable. In order to obtain more accurate land cover classification results, NDVI-LMGM employs all available multi-temporal Landsat images for ISODATA classification. Although the performance of NDVI-LMGM would be affected by various classification results and input Landsat images, it is expected that the new method can provide an alternative for directly downscaling NDVI products with higher accuracy and less computational cost. In addition, such downscaled NDVI products will benefit studies of ecosystem dynamics as one crucial aspect of understanding global climate change. In future studies, more efforts should be spent to reduce the influence of land cover classification and small-scale land cover type changes to the prediction results.