The Effect of Epidermal Structures on Leaf Spectral Signatures of Ice Plants (Aizoaceae)
Abstract
:1. Introduction
2. Experimental Section
2.1. Sample Collection
| Epidermal Type (ET) | Species | Subfamily |
|---|---|---|
| Bladder cells | Drosanthemum curtophyllum | Ruschioideae |
| L. Bolus NM | ||
| Drosanthemum archeri | Ruschioideae | |
| L. Bolus NM | ||
| Mesembryanthemum crystallinum | Mesembryanthemoideae | |
| Linnaeus | ||
| Delosperma vogtsii | Ruschioideae | |
| L. Bolus NM | ||
| Trichodiadema rupicola | Ruschioideae | |
| L. Bolus NM | ||
| Hair cover | Gibbaeum shandii | Ruschioideae |
| N. E. Brown GC | ||
| Cheiridopsis purpurea | Ruschioideae | |
| L. Bolus NM | ||
| Braunsia apiculata | Ruschioideae | |
| (Kensit)L. Bolus | ||
| Juttadineteria albata | Ruschioideae | |
| (L. Bolus) L. Bolus | ||
| Odontophorus marlothii | Ruschioideae | |
| N. E. Brown GC | ||
| Rough | Neohenricia spiculata | Ruschioideae |
| S. A. Hammer CSJ | ||
| Hereroa puttkameriana | Ruschioideae | |
| (Dinter & A. Berger) Dinter & Schwantes | ||
| Nananthus aloides | Ruschioideae | |
| (Haworth) Schwantes GF | ||
| Rhombophyllum dolabriforme | Ruschioideae | |
| (Linnaeus) Schwantes GF | ||
| Stomatium bolusiae | Ruschioideae | |
| Schwantes MDK | ||
| Smooth | Glottiphyllum longum | Ruschioideae |
| (Haworth) N. E. Brown | ||
| Malotigena frantiskae-niederlovae | Ruschioideae | |
| Niederle | ||
| Aptenia haeckeliana | Mesembryanthemoideae | |
| (A. Berger) Bittrich ex Gerbaulet | ||
| Schlechteranthus hallii | Ruschioideae | |
| L. Bolus | ||
| Bergeranthus scapiger | Ruschioideae | |
| (Haworth) Schwantes | ||
| Wax cover | Malephora purpurea-crocea | Ruschioideae |
| (Haworth) Schwantes GF | ||
| Oscularia steenbergensis | Ruschioideae | |
| (L. Bolus) H. E. K. Hartmann Bradleya | ||
| Scopelogena verruculata | Ruschioideae | |
| (Linnaeus) L. Bolus | ||
| Amphibolia succulent | Ruschioideae | |
| (L.Bolus) H. E. K. Hartmann Bradleya | ||
| Enarganthe octonaria | Ruschioideae | |
| (L. Bolus) N. E. Brown GC |

2.2. Hyperspectral Data Collection

2.3. Data Preprocessing
2.3.1. Jump Correction
2.3.2. Standard Normal Variate Transformation
2.4. Classification
2.4.1. Partial Least Squares Discriminant Analysis
2.4.2. Validation
2.4.3. Accuracy Assessment
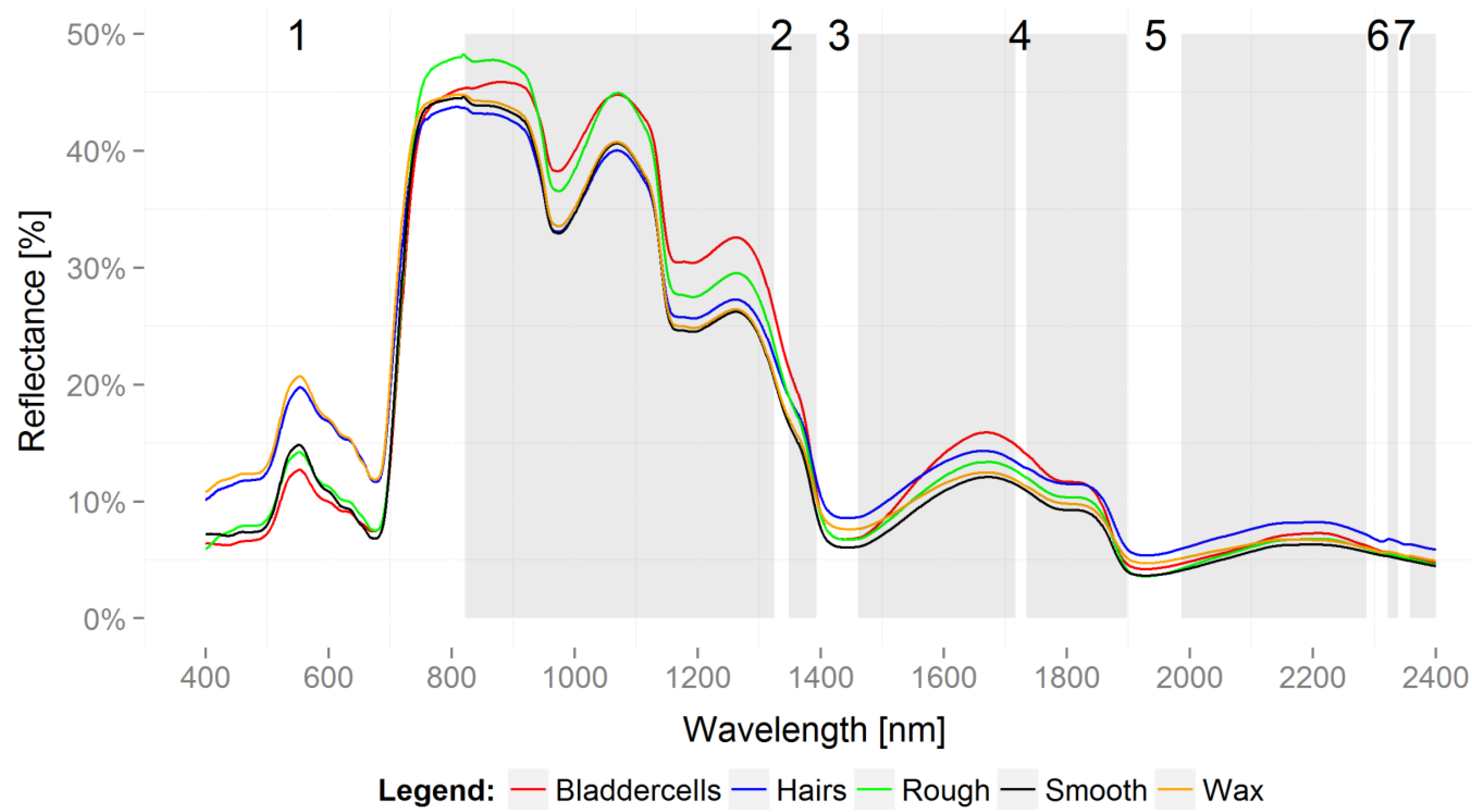
3. Results
3.1. Partial Least Squares Discriminant Analysis: Classification
| SNV/no SNV | Segment | Validation | OA | 95% CI | p-Value | Kappa | |
|---|---|---|---|---|---|---|---|
| Min | Max | ||||||
| SNV | Full | Training | 0.81 | 0.67 | 0.91 | <0.05 | 0.77 |
| SNV | Full | Cross | 0.55 | 0.48 | 0.61 | <0.05 | 0.44 |
| SNV | VIS | Training | 0.54 | 0.39 | 0.69 | <0.05 | 0.43 |
| SNV | VIS | Cross | 0.52 | 0.46 | 0.58 | <0.05 | 0.40 |
| SNV | NIR | Training | 0.52 | 0.37 | 0.67 | <0.05 | 0.40 |
| SNV | NIR | Cross | 0.50 | 0.44 | 0.57 | <0.05 | 0.38 |
| SNV | SWIR I | Training | 0.21 | 0.10 | 0.35 | 0.01 | |
| SNV | SWIR I | Cross | 0.37 | 0.31 | 0.43 | <0.05 | 0.21 |
| SNV | SWIR II | Training | 0.67 | 0.52 | 0.80 | <0.05 | 0.59 |
| SNV | SWIR II | Cross | 0.77 | 0.71 | 0.82 | <0.05 | 0.71 |
| no SNV | Full | Training | 0.60 | 0.45 | 0.74 | <0.05 | 0.51 |
| no SNV | Full | Cross | 0.92 | 0.88 | 0.95 | <0.05 | 0.90 |
| no SNV | VIS | Training | 0.46 | 0.31 | 0.61 | <0.05 | 0.32 |
| no SNV | VIS | Cross | 0.54 | 0.48 | 0.60 | <0.05 | 0.43 |
| no SNV | NIR | Training | 0.29 | 0.17 | 0.44 | 0.11 | |
| no SNV | NIR | Cross | 0.34 | 0.28 | 0.40 | <0.05 | 0.17 |
| no SNV | SWIR I | Training | 0.27 | 0.15 | 0.42 | 0.09 | |
| no SNV | SWIR I | Cross | 0.35 | 0.29 | 0.41 | 0.19 | |
| no SNV | SWIR II | Training | 0.56 | 0.41 | 0.71 | <0.05 | 0.45 |
| no SNV | SWIR II | Cross | 0.55 | 0.48 | 0.61 | <0.05 | 0.44 |
| Ground Truth (GT) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Dataset | Bladder | Hairs | Rough | Smooth | Wax | # of Classified Spectra | UA | ||
| CV/no SNV | Classification | Bladder | 47 | 0 | 1 | 2 | 0 | 50 | 94.0% |
| Hairs | 0 | 47 | 0 | 0 | 1 | 48 | 97.9% | ||
| Rough | 0 | 2 | 43 | 1 | 0 | 46 | 93.5% | ||
| Smooth | 3 | 1 | 6 | 44 | 1 | 55 | 80.0% | ||
| Wax | 0 | 0 | 0 | 2 | 47 | 49 | 95.9% | ||
| # of GT Spectra | 50 | 50 | 50 | 49 | 49 | 248 | |||
| PA | 94.0% | 94.0% | 86.0% | 89.8% | 95.9% | ||||
| Ground Truth (GT) | |||||||||
| Bladder | Hairs | Rough | Smooth | Wax | # of Classified Spectra | UA | |||
| Training/SNV | Classification | Bladder | 10 | 0 | 1 | 1 | 0 | 12 | 83.3% |
| Hairs | 0 | 7 | 1 | 0 | 0 | 8 | 87.5% | ||
| Rough | 0 | 1 | 7 | 1 | 0 | 9 | 77.8% | ||
| Smooth | 0 | 0 | 1 | 6 | 0 | 7 | 85.7% | ||
| Wax | 0 | 2 | 0 | 1 | 9 | 12 | 75.0% | ||
| # of GT Spectra | 10 | 10 | 10 | 9 | 9 | 48 | |||
| PA | 100.0% | 70.0% | 70.0% | 66.7% | 100.0% | ||||
3.2. Variable Importance for Projection
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, C.; Grosse-Stoltenberg, A.; Laustro, V.; Oldeland, J.; Werner, C. Retrieving nitrogen isotopic signatures from fresh leaf reflectance spectra: Disentangling delta δ15N from biochemical and structural leaf properties. Front. Plant Sci. 2015, 6, 307. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Carranza-Jimenez, L.; Martinez, P. Amazonian functional diversity from forest canopy chemical assembly. Proc. Natl. Acad. Sci. USA 2014, 111, 5604–5609. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.R.K.; Grosse-Stoltenberg, A.; Romer, M.; Oldeland, J. Field spectroscopy in the vnir-swir region to discriminate between mediterranean native plants and exotic-invasive shrubs based on leaf tannin content. Remote Sens. 2015, 7, 1225–1241. [Google Scholar] [CrossRef] [Green Version]
- Kalacska, M.; Bohman, S.; Sanchez-Azofeifa, G.A.; Castro-Esau, K.; Caelli, T. Hyperspectral discrimination of tropical dry forest lianas and trees: Comparative data reduction approaches at the leaf and canopy levels. Remote Sens. Environ. 2007, 109, 406–415. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Estimation of vegetation water content and photosynthetic tissue area from spectral reflectance: A comparison of indices based on liquid water and chlorophyll absorption features. Remote Sens. Environ. 2003, 84, 526–537. [Google Scholar] [CrossRef]
- Da Luz, B.R. Attenuated total reflectance spectroscopy of plant leaves: A tool for ecological and botanical studies. New Phytol. 2006, 172, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Ehleringer, J.R.; Bjorkman, O. Pubescence and leaf spectral characteristics in a desert shrub, encelia farinosa. Oecologia 1978, 36, 151–162. [Google Scholar] [CrossRef]
- Eller, B.M. Epidermis and the spectral properties of plant-surfaces. Ber. Deut. Bot. Ges. 1985, 98, 465–475. [Google Scholar]
- Eller, B.M.; Willi, P. The significance of leaf pubescence for the absorption of global radiation by tussilago farfara l. Oecologia 1977, 29, 179–187. [Google Scholar] [CrossRef]
- Ehleringer, J.R.; Mooney, H.A. Leaf hairs: Effects on physiological activity and adaptive value to a desert shrub. Oecologia 1978, 37, 183–200. [Google Scholar] [CrossRef]
- Mulroy, T.W. Spectral properties of heavily glaucous and non-glaucous leaves of a succulent rosette-plant. Oecologia 1979, 38, 349–357. [Google Scholar] [CrossRef]
- Tanner, V.; Eller, B.M. Epidermis structure and its significance for the optical-properties of leaves of the mesembryanthemaceae. J. Plant Physiol. 1986, 125, 285–294. [Google Scholar] [CrossRef]
- Huntjr, E.; Rock, B. Detection of changes in leaf water content using near- and middle-infrared reflectances. Remote Sens. Environ. 1989, 30, 43–54. [Google Scholar] [CrossRef]
- Martin, M.E.; Aber, J.D. High spectral resolution remote sensing of forest canopy lignin, nitrogen, and ecosystem processes. Ecol. Appl. 1997, 7, 431–443. [Google Scholar] [CrossRef]
- Martin, M.E.; Newman, S.D.; Aber, J.D.; Congalton, R.G. Determining forest species composition using high spectral resolution remote sensing data. Remote Sens. Environ. 1998, 65, 249–254. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf optical properties in higher plants: Linking spectral characteristics to stress and chlorophyll concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Von Willert, D.J. Life Strategies of Succulents in Deserts: With Special Reference to the Namib Desert; CUP Archive: New York, NY, USA, 1992; p. 372. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Ihlenfeldt, H.D.; Hartmann, H.E.K. Leaf surfaces in mesembryanthemaceae. In The Plant Cuticle; Cutler, D.F., Alvin, K.L., Price, C.E., Eds.; Academic Press: London, UK, 1982; pp. 397–423. [Google Scholar]
- List, T.P. The Plant List (2013). Version 1.1. Available online: http://www.theplantlist.org/ (accessed on 1 May 2014).
- Jürgens, N. Untersuchungen zur Ökologie sukkulenter Pflanzen des südlichen Afrika. Mitt. Inst. Allg. Bot. Hamburg 1986, 21, 139–365. [Google Scholar]
- Niesler, I.M. Untersuchungen zur Ontogenie und zum Bau der Blätter bei Mesembryanthemaceae Fenzl unter besonderer Berücksichtigung der Epidermis. Ph.D. Thesis, Universität Hamburg, Hamburg, Germany, 1998. [Google Scholar]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Ghiyamat, A.; Shafri, H.Z.M.; Mandiraji, G.A.; Shariff, A.R.M.; Mansor, S. Hyperspectral discrimination of tree species with different classifications using single- and multiple-endmember. Int. J. Appl. Earth Observ. Geoinf. 2013, 23, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Prospere, K.; McLaren, K.; Wilson, B. Plant species discrimination in a tropical wetland using in situ hyperspectral data. Remote Sens. 2014, 6, 8494–8523. [Google Scholar] [CrossRef]
- He, Y.; Li, X.; Deng, X. Discrimination of varieties of tea using near infrared spectroscopy by principal component analysis and bp model. J. Food Eng. 2007, 79, 1238–1242. [Google Scholar] [CrossRef]
- Edelmann, A.; Diewok, J.; Schuster, K.C.; Lendl, B. Rapid method for the discrimination of red wine cultivars based on mid-infrared spectroscopy of phenolic wine extracts. J. Agric. Food Chem. 2001, 49, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.R.; Garg, P.K.; Ghosh, S.K. Development of an agricultural crops spectral library and classification of crops at cultivar level using hyperspectral data. Precis. Agric. 2007, 8, 173–185. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gamon, J.A. Remote sensing of plant functional types. New Phytol. 2010, 186, 795–816. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, J.C.; Kent, M.; Ramsay, P.M. Plant functional types: An alternative to taxonomic plant community description in biogeography? Prog. Phys. Geogr. 2000, 24, 515–542. [Google Scholar] [CrossRef]
- Homolova, L.; Maenovsky, Z.; Clevers, J.G.P.W.; Garcia-Santos, G.; Schaeprnan, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef]
- Houborg, R.; Fisher, J.B.; Skidmore, A.K. Advances in remote sensing of vegetation function and traits. Int. J. Appl. Earth Observ. Geoinf. 2015, 43, 1–6. [Google Scholar] [CrossRef]
- Dorigo, W.; Bachmann, M.; Heldens, W. AS Toolbox & Processing of Field Spectra-user’s manual; German Aerospace Center, DLR-DFD. Imaging Spectroscopy Group: Wessling, Germany, 2006. [Google Scholar]
- Fearn, T.; Riccioli, C.; Garrido-Varo, A.; Guerrero-Ginel, J.E. On the geometry of snv and msc. Chemom. Intell. Lab. Syst. 2009, 96, 22–26. [Google Scholar] [CrossRef]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Sanchez, G.; Determan, C.; Sanchez, M.G. R Package DiscriMiner (Version 0.1–29). Available online: https://cran.r-project.org/web/packages/DiscriMiner/ (accessed on 13 May 2015).
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: Berlin, Germany, 2013. [Google Scholar]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Hyperspectral Remote Sensing of Vegetation; CRC Press: Boca Raton, FL, USA, 2011; p. 766. [Google Scholar]
- Chong, I.G.; Jun, C.H. Performance of some variable selection methods when multicollinearity is present. Chemom. Intell. Lab. Syst. 2005, 78, 103–112. [Google Scholar] [CrossRef]
- Wold, S.; Kettaneh, N.; Tjessem, K. Hierarchical multiblock pls and pc models for easier model interpretation and as an alternative to variable selection. J. Chemom. 1996, 10, 463–482. [Google Scholar] [CrossRef]
- Wold, S.; Martens, H.; Wold, H. The multivariate calibration problem in chemistry solved by the PLS method. In Matrix Pencils; Kågström, B., Ruhe, A., Eds.; Springer: Berlin, Germany, 1983; pp. 286–293. [Google Scholar]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Perez-Enciso, M.; Tenenhaus, M. Prediction of clinical outcome with microarray data: A partial least squares discriminant analysis (PLS-DA) approach. Hum. Genet 2003, 112, 581–592. [Google Scholar] [PubMed]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Leung, K.; Cheong, F.; Cheong, C.; O’Farrell, S.; Tissington, R.; Ou, C.M. A Comparison of Variable Selection Techniques for Credit Scoring. In Proceedings of the 7th International Conference on Computational Intelligence in Economics and Finance, Taiwan, 4 Augest 2008.
- Efron, B.; Tibshirani, R. Improvements on cross-validation: The 632+ bootstrap method. J. Am. Stat. Assoc. 1997, 92, 548–560. [Google Scholar]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Story, M.; Congalton, R.G. Accuracy assessment—A users perspective. Photogramm Eng. Remote Sens. 1986, 52, 397–399. [Google Scholar]
- Jones, H.G.; Vaughan, R.A. Remote Sensing of Vegetation: Principles, Techniques, and Applications; Oxford University Press: New York, NY, USA, 2010; p. 381. [Google Scholar]
- Opel, M.R. Leaf anatomy of conophytum n.E.Br. (aizoaceae). Haseltonia 2005, 11, 27–52. [Google Scholar] [CrossRef]
- Opel, M.R.; Hammer, S.A. Conophytum leaf structures. In SA Hammer, Dumpling and His Wife: New Views of the Genus Conophytum; EAE Creative Color Ltd.: Norwich, UK, 2002; pp. 300–321. [Google Scholar]
- Haberland, G. Physiologische pflanzenanatomie, Leipzig 1909, IV. Aufl. S 1909, 557, 671. [Google Scholar]
- Barthlott, W.; Wollenweber, E. Zur Feinstruktur, Chemie und taxonomischen Signifikanz epicuticularer Wachse und ähnlicher Sekrete. Trop. Subtrop. Pflanzenwelt. 1981, 32, 67. [Google Scholar]
- Roth, G.A.; Tahiliani, S.; Neu-Baker, N.M.; Brenner, S.A. Hyperspectral microscopy as an analytical tool for nanomaterials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.A.; Ustin, S.L.; Ogunjemiyo, S.; Greenberg, J.; Dobrowski, S.Z.; Chen, J.Q.; Hinckley, T.M. Spectral and structural measures of northwest forest vegetation at leaf to landscape scales. Ecosystems 2004, 7, 545–562. [Google Scholar] [CrossRef]
- Hartmann, H.E.K. Illustrated Handbook of Succulent Plants: Aizoaceae A–E; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2002; p. 456. [Google Scholar]
- Hartmann, H.E.K. Illustrated Handbook of Succulent Plants: Aizoaceae F–Z; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2002; p. 422. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heim, R.H.-J.; Jürgens, N.; Große-Stoltenberg, A.; Oldeland, J. The Effect of Epidermal Structures on Leaf Spectral Signatures of Ice Plants (Aizoaceae). Remote Sens. 2015, 7, 16901-16914. https://doi.org/10.3390/rs71215862
Heim RH-J, Jürgens N, Große-Stoltenberg A, Oldeland J. The Effect of Epidermal Structures on Leaf Spectral Signatures of Ice Plants (Aizoaceae). Remote Sensing. 2015; 7(12):16901-16914. https://doi.org/10.3390/rs71215862
Chicago/Turabian StyleHeim, René Hans-Jürgen, Norbert Jürgens, André Große-Stoltenberg, and Jens Oldeland. 2015. "The Effect of Epidermal Structures on Leaf Spectral Signatures of Ice Plants (Aizoaceae)" Remote Sensing 7, no. 12: 16901-16914. https://doi.org/10.3390/rs71215862







