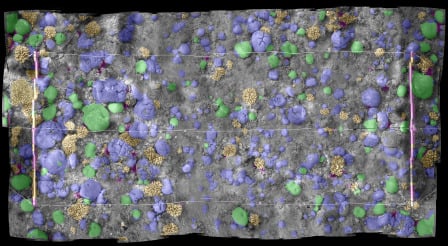
Image-Based Coral Reef Classification and Thematic Mapping
Abstract
:1. Introduction
2. Review of Existing Algorithms for Automated Benthic Classification Using Optical Imagery
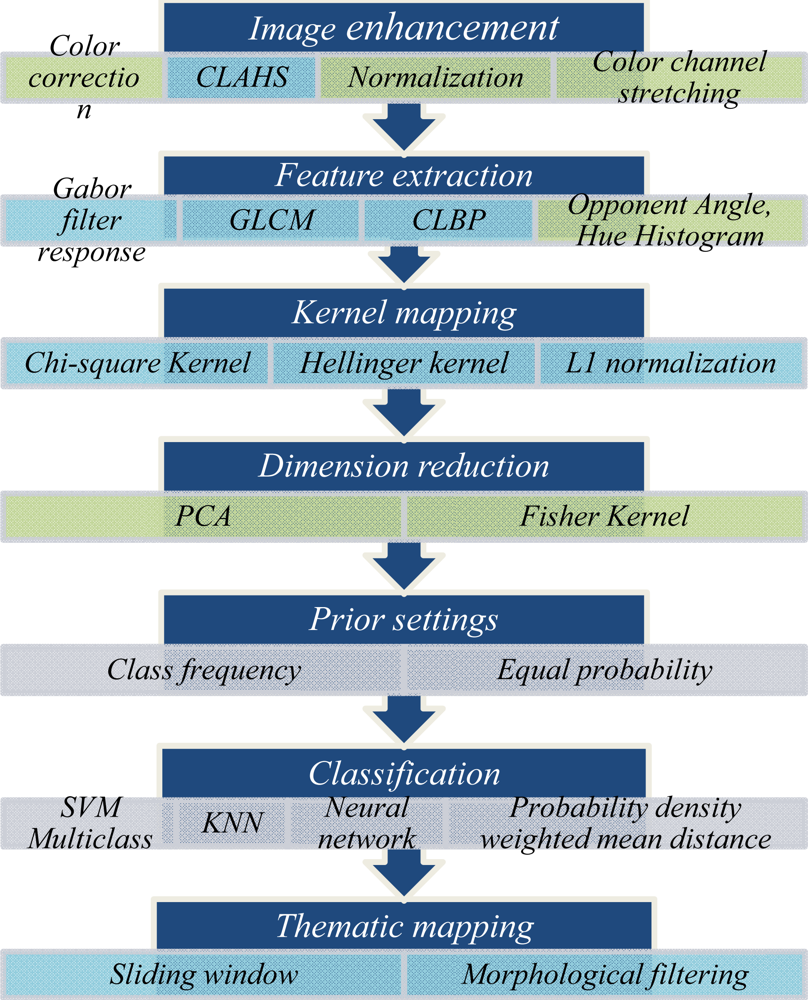
3. Proposed Algorithm
3.1. Step One: Image Enhancements
3.2. Step Two: Feature Extraction
3.3. Step Three: Kernel Mapping
3.4. Step Four: Dimension Reduction (Optional)
3.5. Step Five: Prior Settings
3.6. Step Six: Classification
3.7. Step Seven: Map Classification
3.8. Parameter Tuning
4. Methodology for Evaluating the Classifiers
4.1. Choosing Different Configurations
4.2. Datasets
4.3. Criteria for Evaluation and Comparison with Other Methods
4.4. Mosaic Image Classification
5. Results
5.1. Evaluation of Different Configurations
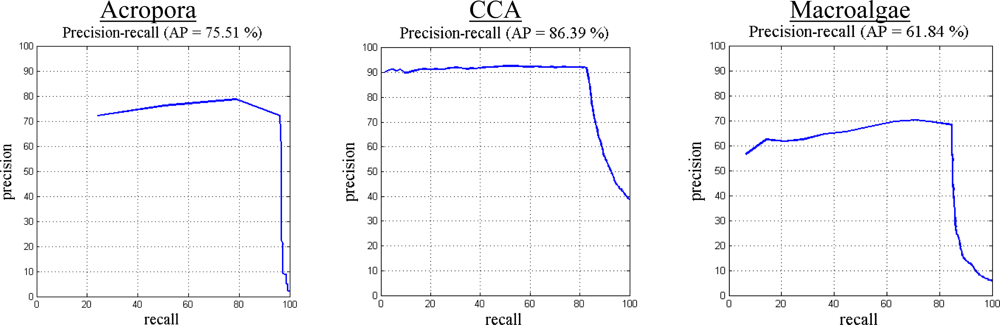
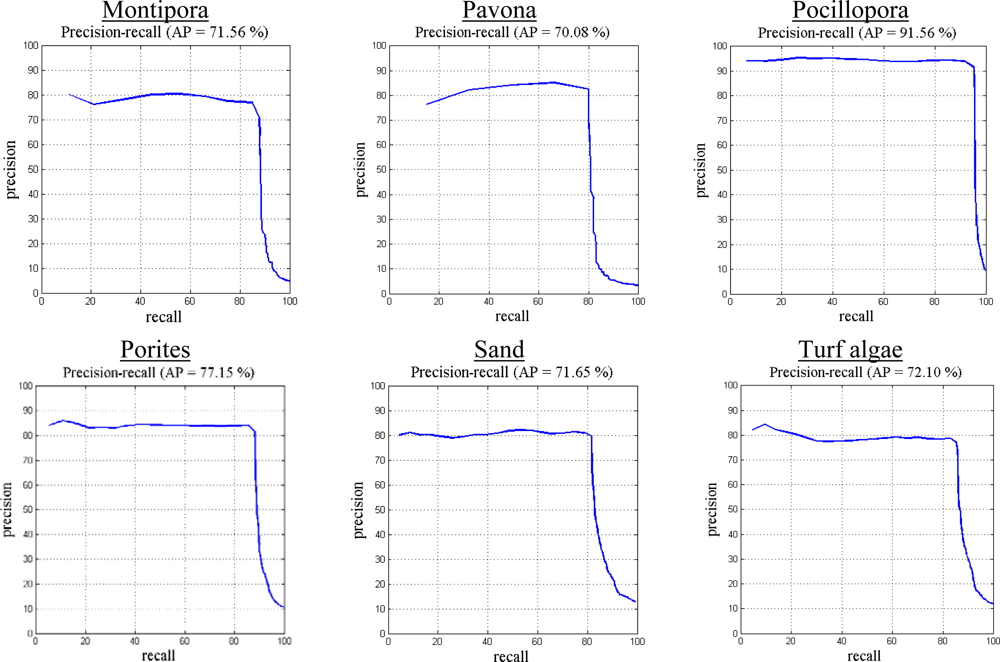
5.2. Evaluation of Proposed Classification Method
5.3. Comparison with Other Methods
5.4. Classifying Image Mosaics
5.5. Recommendations for Classifying Future Datasets
- If the dataset contains low contrast or blurred images, CLAHS works very well as an image enhancement step. If color markers are available in raw images, color correction can be performed to enhance the color constancy.
- For texture description, the concatenation of GLCM, Gabor filter response and CLBP works consistently well. If images have good reliable color information, then opponent angle and hue color channel histograms can be added, with the texture descriptor assigning equal weights to both color and texture descriptors.
- In all the cases of image patch classification, sparsely populated bins within histograms possess higher distinctive information than the high frequency bins. This statement is based on the assumption that high frequency bins often represent the background of the object and contain less distinctive information. The chi-square and Hellinger kernels can be used to modify bin counts and boost the population of low frequent bins. L1 normalization of the feature vector is necessary in all cases before applying the classifier for training and testing.
- If the dataset is small (datasets with training samples less than 5,000 are considered as small ones), then PCA and Fisher kernel mapping works very effectively to reduce the feature dimension. However, for large (Datasets with training samples more than 12,000 are considered as large ones) datasets, almost all the features become useful, and the dimension reduction is much less effective. In large datasets, almost all the dimensions become discriminative, and only a few dimensions are reduced with PCA. Therefore, for large datasets, this dimension reduction step can be avoided.
- Class frequency works well as prior in all the cases.
- For smaller datasets, the KNN classifier has the best performance. However, as the datasets get larger, the effectiveness of this method reduces, owing to the higher storage requirements, lower efficiency in classification response and lower noise tolerance. Some recent works [38] address this problem of KNN. However, SVM (linear SVM with one against the rest scheme) and neural networks (multilayer perception with back projection configuration) can be appropriate classifiers for bigger datasets. For underwater images, the classification based on probability density weighted mean distance (PDWMD) from the tail of the distribution by Stokes and Deane [10] works efficiently, both in terms of time and efficiency.
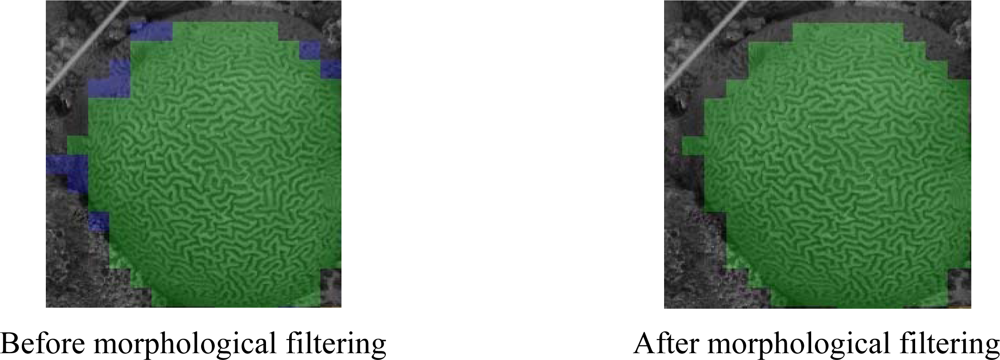
- Morphological filtering can increase the accuracy of the classification results.
- For smaller datasets, the KNN classifier has the best performance. However, as the datasets get larger, the effectiveness of this method reduces because of high storage requirements, low efficiency in classification response and low noise tolerance. Some recent works [38] address this problem of KNN. However, SVM (linear SVM with one against the rest scheme) and neural networks (multilayer perception with back projection configuration) can be appropriate classifiers for bigger datasets. For underwater images, the classification based on probability density weighted mean distance (PDWMD) from the tail of the distribution [10] works efficiently both in terms of time and efficiency.
- In our method, all the features used are partially or completely scale and rotation invariant. These features are therefore able to mitigate the effects of limited scale variation of individual classes. For larger scale variation, it is important to have enough training examples of individual classes at different scales. In the future, multi-resolution mapping might be useful for benthic habitat covering large scale variations of individual classes.
6. Conclusion
Acknowledgments
References
- Turner, W.; Spector, S.; Gardiner, N.; Fladeland, M.; Sterling, E.; Steininger, M. Remote sensing for biodiversity science and conservation. Trends Ecol. Evol 2003, 18, 306–314. [Google Scholar]
- Kohler, K.E.; Gill, S.M. Coral Point Count with Excel extensions (CPCe): A Visual Basic program for the determination of coral and substrate coverage using random point count methodology. Comput. Geosci 2006, 32, 1259–1269. [Google Scholar]
- Pican, N.; Trucco, E.; Ross, M.; Lane, D.; Petillot, Y.; Tena Ruiz, I. Texture Analysis for Seabed Classification: Co-Occurrence Matrices vs. Self-Organizing Maps. Proceedings of OCEANS ’98, Nice, France, 28 September–01 October 1998; 1, pp. 424–428.
- Beijbom, O.; Edmunds, P.J.; Kline, D.I.; Mitchell, B.G.; Kriegman, D. Automated Annotation of Coral Reef Survey Images. Proceedings of IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Providence, Rhode Island, 16–21 June 2012.
- Gracias, N.; Negahdaripour, S.; Neumann, L.; Prados, R.; Garcia, R. A motion compensated filtering approach to remove sunlight flicker in shallow water images. Oceans 2008, 2008, 1–7. [Google Scholar]
- Schechner, Y.Y.; Karpel, N. Attenuating Natural Flicker Patterns. Proceedings of IEEE/MTS OCEANS 04 Conference, Kobe, Japan, 9–12 November 2004.
- Shihavuddin, A.S.M.; Gracias, N.; Garcia, R. Online Sunflicker Removal using Dynamic Texture Prediction. Proceedings of International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications, Rome, Italy, 24–26 February, 2012; pp. 161–167.
- Pizarro, O.; Rigby, P.; Johnson-Roberson, M.; Williams, S.; Colquhoun, J. Towards Image-Based Marine Habitat Classification. Proceedings of OCEANS 08, Quebec City, QC, Canada, 15–18 September 2008.
- Marcos, M.; Shiela, A.; David, L.; Peñaflor, E.; Ticzon, V.; Soriano, M. Automated benthic counting of living and non-living components in Ngedarrak Reef, Palau via subsurface underwater video. Environ. Monit. Assess 2008, 145, 177–184. [Google Scholar]
- Stokes, M.D.; Deane, G.B. Automated processing of coral reef benthic images. Limnol. Oceanogr. Methods 2009, 7, 157–168. [Google Scholar]
- Padmavathi, G.; Muthukumar, M.; Thakur, S. Kernel Principal Component Analysis Feature Detection and Classification for Underwater Images. Proceedings of 3rd International Congress on Image and Signal Processing (CISP), Yantai, China, 16–18 October 2010; 2, pp. 983–988.
- Marcos, M.; Angeli, S.; David, L.; Peaflor, E.; Ticzon, V.; Soriano, M. Automated benthic counting of living and non-living components in Ngedarrak Reef, Palau via subsurface underwater video. Environ. Monit. Assess 2008, 145, 177–184. [Google Scholar]
- Mehta, A.; Ribeiro, E.; Gilner, J.; van Woesik, R. Coral Reef Texture Classification using Support Vector Machines. Proceedings of International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISAPP), Barcelona, Spain, 8–11 March 2007; pp. 302–310.
- Gleason, A.; Reid, R.; Voss, K. Automated Classification of Underwater Multispectral Imagery for Coral Reef Monitoring. Proceedings of OCEANS 07, Miami, FL, USA, 29 September–4 October 2007; pp. 1–8.
- Johnson-Roberson, M.; Kumar, S.; Willams, S. Segmentation and Classification of Coral for Oceanographic Surveys: A Semi-Supervised Machine Learning Approach. Proceedings of OCEANS 2006, Asia Pacific, Singapore, 16–19 May 2007.
- Johnson-Roberson, M.; Kumar, S.; Pizarro, O.; Willams, S. Stereoscopic Imaging for Coral Segmentation and Classification. Proceedings of OCEANS 06, Singapore, 18–21 September 2006; pp. 1–6.
- Clement, R.; Dunbabin, M.; Wyeth, G. Toward Robust Image Detection of Crown-of-thorns Starfish for Autonomous Population Monitoring. In Australasian Conference on Robotics & Automation; Sammut, C., Ed.; Australian Robotics and Automation Association Inc.: Sydney, Australia, 2005. [Google Scholar]
- Soriano, M.; Marcos, S.; Saloma, C.; Quibilan, M.; Alino, P. Image Classification of Coral Reef Components from Underwater Color Video. Proceedings of 2001 MTS/IEEE Conference and Exhibition OCEANS, Honolulu, HI, USA, 5–8 November 2001; 2, pp. 1008–1013.
- Caputo, B.; Hayman, E.; Fritz, M.; Eklundh, J.O. Classifying materials in the real world. Image Vis. Comput 2010, 28, 150–163. [Google Scholar]
- Zhang, J.; Lazebnik, S.; Schmid, C. Local features and kernels for classification of texture and object categories: A comprehensive study. Int. J. Comput. Vis 2007, 73, 213–238. [Google Scholar]
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization. Graphics GEMs IV; Academic Press Professional, Inc.: San Diego, CA, USA, 1994; pp. 474–485. [Google Scholar]
- Finlayson, G.D.; Schiele, B.; Crowley, J.L. Comprehensive Colour Image Normalization. Proceedings of ECCV'98 Fifth European Conference on Computer Vision, Freiburg, Germany, 2–6 June 1998; 1406, pp. 475–490.
- Gehler, P.; Nowozin, S. On Feature Combination for Multiclass Object Classification. Proceedings of IEEE 12th International Conference on Computer Vision (ICCV), Kyoto, Japan, 29 September–2 October 2009; pp. 221–228.
- Haley, G.; Manjunath, B. Rotation-invariant texture classification using a complete space-frequency model. IEEE Trans. Image Process 1999, 8, 255–269. [Google Scholar]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar]
- Soh, L.; Tsatsoulis, C. Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans. Geosci. Remote Sens 1999, 37, 780–795. [Google Scholar]
- Clausi, D.A. An analysis of co-occurrence texture statistics as a function of grey level quantization. Can. J. Remote Sens 2002, 28, 45–62. [Google Scholar]
- Guo, Z.; Zhang, L.; Zhang, D. A completed modeling of local binary pattern operator for texture classification. IEEE Trans. Pattern Anal. Mach. Intell 2010, 19, 1657–1663. [Google Scholar]
- Van de Weijer, J.; Schmid, C. Coloring Local Feature Extraction. Proceedings of the 9th European Conference on Computer Vision ECCV’06, Graz, Austria, 7–13 May 2006; pp. 334–348.
- Mika, S.; Ratsch, G.; Weston, J.; Scholkopf, B.; Mullers, K. Fisher Discriminant Analysis with Kernels. Proceedings of the 1999 IEEE Signal Processing Society Workshop, Neural Networks for Signal Processing IX, 1999, Berlin, Germany, 25 August 1999; pp. 41–48.
- Ruderman, D.L. The statistics of natural images. Netw. Comput. Nat. Syst 1994, 4, 517–548. [Google Scholar]
- Lirman, D.; Gracias, N.; Gintert, B.; Gleason, A.C.R.; Deangelo, G.; Gonzalez, M.; Martinez, E.; Reid, R.P. Damage and Recovery Assessment of Vessel Grounding Injuries on Coral Reef Habitats Using Georeferenced Landscape Video Mosaics. Limnol. Oceanogr.: Methods 2010, 8, 88–97. [Google Scholar]
- Escartin, J.; Garcia, R.; Delaunoy, O.; Ferrer, J.; Gracias, N.; Elibol, A.; Cufi, X.; Neumann, L.; Fornari, D.J.; Humphris, S.E. Globally aligned photomosaic of the Lucky Strike hydrothermal vent field (Mid-Atlantic Ridge, 37°18.5′N): Release of georeferenced data, mosaic construction, and viewing software. Geochem. Geophys. Geosyst. 2008, 9. [Google Scholar] [CrossRef]
- Lirman, D.; Gracias, N.; Gintert, B.; Gleason, A.; Reid, P.; Negahdaripour, S.; Kramer, P. Development and application of a video-mosaic survey technology to document the status of coral reef communities. Environ. Monit. Assess 2007, 125, 59–73. [Google Scholar]
- Kohavi, R. A. Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. Proceedings of the Fourteenth International Joint Conference on Artificial Intelligence, IJCAI 95, Montréal, QC, Canada, 20–25 August 1995; pp. 1137–1143.
- Congalton, R.G.; Green, K. Assessing the Accuracy of Remotely Sensed Data; Lewis Publishers: Boca Raton, FL, USA, 1999; p. 137. [Google Scholar]
- Loya, Y. The Coral Reefs of Eilat; Past, Present and Future: Three Decades of Coral Community Structure Studies. In Coral Reef Health and Disease; Springer-Verlag: Berlin/Heidelberg, Germany, 2004; pp. 1–34. [Google Scholar]
- Garcia, S.; Derrac, J.; Cano, J.; Herrera, F. Prototype Selection for Nearest Neighbor Classification: Taxonomy and Empirical Study. IEEE Trans. Pattern Anal. Mach. Intell 2012, 34, 417–435. [Google Scholar]
Appendix 1
| Co-occurrence indicator used in the features | |
|---|---|
| Statistics | Formulas | |
|---|---|---|
| 1 | Maximum Probability [33] | |
| 2 | Uniformity [12,33] | |
| 3 | Entropy [33] | |
| 4 | Dissimilarity [33] | |
| 5 | Contrast [12,33] | |
| 6 | Inverse Difference [33] | |
| 7 | Inverse Difference moment [33] | |
| 8 | Correlation 1 [12,33] | |
| 9 | Inverse Difference Normalized [27] | |
| 10 | Inverse Difference Moment Normalized [27] | |
| 11 | Sum of Squares: Variance [12] | |
| 12 | Sum Average [12] | |
| 13 | Sum Entropy [12] | |
| 14 | Sum Variance [12] | |
| 15 | Difference Variance [12] | |
| 16 | Difference Entropy [12] | |
| 17 | Info. measure of correlation 1 [12] | |
| 18 | Info. measure of correlation 2 [12] | |
| 19 | Autocorrelation [33] | |
| 20 | Correlation 2 [12,33] | |
| 21 | Cluster Shade [33] | |
| 22 | Cluster Prominence [33] |
Appendix 2
A2.1. EILAT Dataset
A2.2. RSMAS Dataset

A2.3. Moorea-Labeled Corals (MLC) dataset

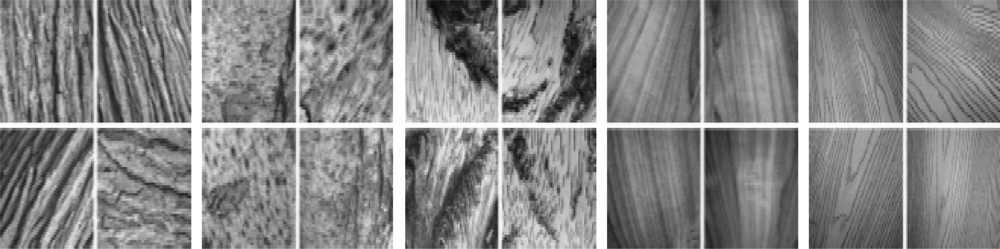
A2.4. UIUCtex Dataset
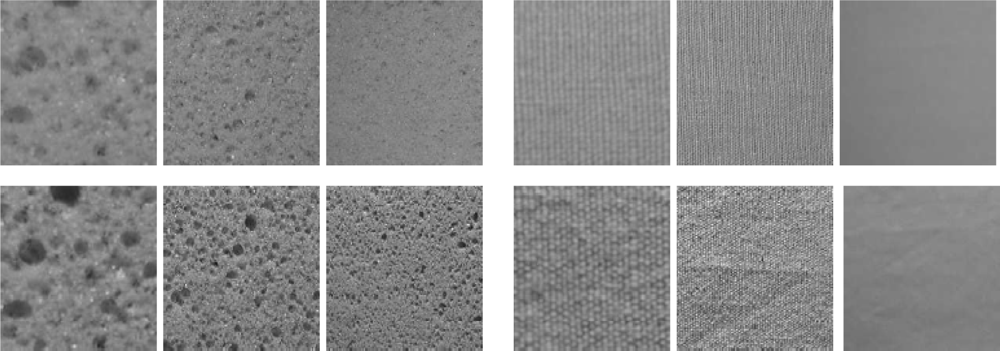
A2.5. CURET Texture Dataset

A2.6. KTH-TIPS Dataset
A2.7. EILAT 2 Dataset

References
- Lazebnik, S.; Schmid, C.; Ponce, J. A sparse texture representation using local affine regions. IEEE Trans. Pattern Anal. Mach. Intell 2005, 27, 1265–1278. [Google Scholar]
- Dana, K.J.; van Ginneken, B.; Nayar, S.K.; Koenderink, J.J. Reflectance and texture of real-world surfaces. ACM Trans. Graph 1999, 18, 1–34. [Google Scholar]
- Hayman, E.; Caputo, B.; Fritz, M.; Eklundh, J.O. On the significance of real-world conditions for material classification. Eur. Conf. Comput. Vis 2004, 3024, 253–266. [Google Scholar]






| Authors | Features | Classifiers | N |
|---|---|---|---|
| Beijbom [4] | Maximum response (MR) filter bank | Library for support vector machines (LIBSVM) | 9 |
| Padmavathi [11] | Bag of words using scale-invariant feature transform (SIFT) and kernel principal component analysis (KPCA) | Probabilistic neural network (PNN) | 3 |
| Stokes & Deane [10] | Color: (RGB histogram) Texture: discrete cosine transform (DCT) | Probability density weighted mean distance (PDWMD) | 18 |
| Marcos [12] | Texture: local binary pattern (LBP) Color: normalized chromaticity coordinate (NCC) histogram | Linear discriminant analysis (LDA) | 2 |
| Pizarro [8] | Color: normalized chromaticity coordinate (NCC) histogram Texture: bag of words using scale-invariant feature transform (SIFT), Saliency: Gabor filter response | Voting of the best matches | 8 |
| Mehta [13] | Pixel intensity | Support vector machines (SVM) | 2 |
| Gleason [14] | Multi-spectral data Texture: grey level co-occurrence matrix (GLCM) | Distance measurement | 3 |
| Johnson-Roberson [15,16] | Texture: Gabor filter response Acoustic | Support vector machines (SVM) | 4 |
| Marcos [9] | Color: normalized chromaticity coordinate (NCC) histogram Texture: local binary pattern (LBP) | 3-layer feed-forward back projection neural network | 3 |
| Clement [17] | Texture: local binary pattern (LBP) | Log-likelihood measure | 2 |
| Soriano [18] | Color: normalized chromaticity coordinate (NCC) histogram Texture: local binary pattern (LBP) | Log-likelihood measure | 5 |
| Pican [3] | Texture: grey level co-occurrence matrix (GLCM) self-organizing map | Kohonen-Map | 30 |
| Kernel Name | Formulation |
|---|---|
| Chi-square | |
| Hellinger |
| Name | Classes | N | Resolution | Color |
|---|---|---|---|---|
| EILAT | Sand, urchin, branches type I, brain coral, favid coral, branches type II, dead coral and branches type III | 1,123 | 64 × 64 | Yes |
| RSMAS | Acropora cervicornis, Acropora palmata, Colpophyllia natans, Diadema antillarum, Diploria strigosa, Gorgonians, Millepora alcicornis, Montastraea cavernosa, Meandrina meandrites, Montipora spp., Palythoas palythoa, Sponge fungus, Siderastrea siderea and tunicates | 766 | 256 × 256 | Yes |
| MLC 2008 | Crustose coralline algae, turf algae, macroalgae, sand, Acropora, Pavona, Montipora, Pocillopora, Porites | 18,872 | 312 × 312 | Yes |
| UIUCtex | bark I, bark II, bark III, wood I, wood II, wood III, water, granite, marble, stone I, stone II, gravel, wall, brick I, brick II, glass I, glass II, carpet I, carpet II, fabric I, paper, fur, fabric II, fabric III and fabric IV | 1,000 | 640 × 480 | No |
| CURET | 61 texture materials imaged over varying pose and illumination, but at constant viewing distance. | 5,612 | 200 × 200 | No |
| KTH-TIPS | sandpaper, crumpled aluminums foil, Styrofoam, sponge, corduroy, linen, cotton, brown bread orange peel, cracker B | 810 | 200 × 200 | No |
| EILAT 2 | Sand, urchin, branching coral, brain coral and favid coral | 303 | 128 × 128 | Yes |
| Red Sea mosaic | Sand, urchin, branching coral, brain coral, favid coral, background objects | 73,600 | 3,257 × 5,937 | Yes |
| EILAT | RSMAS | EILAT 2 | MLC 2008 | |
|---|---|---|---|---|
| No pre-processing | 90.7 | 70.1 | 80.1 | 64.0 |
| Color correction | NA | NA | NA | 63.8 |
| CLAHS | 92.9 | 85.8 | 87.4 | 69.3 |
| Normalization | 70.7 | 64.5 | 58.8 | 58.2 |
| Color channel stretching | 67.1 | 58.8 | 62.9 | 70.5 |
| CLAHS + Color correction | NA | NA | NA | 70.9 |
| CLAHS + Normalization | 91.0 | 85.2 | 87.3 | 68.1 |
| CLAHS + Color channel stretching | 91.4 | 82.4 | 81.7 | 72.7 |
| CLAHS + Color correction + Color channel stretching | NA | NA | NA | 73.2 |
| EILAT | RSMAS | EILAT 2 | MLC 2008 | |
|---|---|---|---|---|
| CLBP | 91.3 | 74.7 | 84.8 | 55.1 |
| Gabor | 85.7 | 61.2 | 65.3 | 39.4 |
| GLCM | 70.9 | 62.9 | 58.2 | 46.8 |
| Color histogram (hue + opponent angle) | 64.2 | 81.7 | 53.0 | 41.3 |
| CLBP + Gabor | 90.5 | 75.0 | 87.7 | 54.9 |
| CLBP + GLCM | 92.2 | 75.8 | 86.4 | 57.1 |
| Gabor + GLCM | 87.6 | 72.1 | 77.5 | 46.3 |
| CLBP + GLCM + Gabor filter response | 93.4 | 83.5 | 91.2 | 62.4 |
| CLBP + GLCM + Gabor filter response + color histogram (hue + opponent angle) | 94.7 | 89.6 | 87.3 | 65.7 |
| EILAT | RSMAS | EILAT 2 | MLC 2008 | |
|---|---|---|---|---|
| Nothing | 87.5 | 84.8 | 87.3 | 61.9 |
| L1 normalization | 91.9 | 87.6 | 87.4 | 64.0 |
| Chi-square kernel mapping | 85.7 | 87.6 | 88.1. | 63.3 |
| Hellinger kernel mapping | 85.8 | 87.9 | 88.5 | 62.2 |
| Kernel mapping (chi-square, Hellinger) | 89.2 | 88.1 | 89.3 | 65.7 |
| L1 normalization + kernel mapping (chi-square, Hellinger) | 93.4 | 89.7 | 91.1 | 66.5 |
| EILAT | RSMAS | EILAT 2 | MLC 2008 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ND | PCA | F | P+F | ND | PCA | F | P+F | ND | PCA | F | P+F | ND | PCA | F | P+F | |
| S | 94.3 | 91.9 | 86.7 | 90.2 | 92.1 | 89.0 | 81.8 | 88.2 | 93.1 | 91,5 | 88.9 | 90.7 | 75.2 | 71.7 | 61.2 | 70.1 |
| K | 91.7 | 93.4 | 85.4 | 94.9 | 91.4 | 92.8 | 87.4 | 93.5 | 88.0 | 92.2 | 85.1 | 92.7 | 69.5 | 73.9 | 64.5 | 74.9 |
| N | 89.9 | 88.1 | 83.2 | 79.7 | 88.4 | 86.4 | 86.5 | 80.2 | 92.5 | 91.1 | 86.3 | 89.2 | 77.4 | 75.5 | 67.9 | 73.0 |
| P | 91.2 | 90.5 | 84.9 | 87.1 | 87.9 | 86.5 | 85.5 | 84.3 | 89.2 | 89.4 | 80.7 | 85.1 | 79.8 | 73.3 | 66.6 | 71.3 |
| Steps | EILAT/RSMAS | MLC 2008 | CURET/KTH/UIUC |
|---|---|---|---|
| Image enhancement | CLAHS | CLAHS, color correction, color channel stretching | CLAHS |
| Feature extraction | Opponent angle histogram, hue channel histogram, CLBP, GLCM, Gabor filter response | CLBP, GLCM, Gabor filter response, Opponent angle histogram, hue channel histogram | CLBP, GLCM, Gabor filter response |
| Kernel mapping | L1 normalization, chi-square kernel, Hellinger kernel (for CLBP, color histogram) | L1 normalization, chi-square kernel, Hellinger kernel (for CLBP, color histogram) | L1 normalization, chi-square kernel, Hellinger kernel (for CLBP, color histogram) |
| Dimension reduction Prior | PCA, Fisher kernel Class frequency | None Class frequency | PCA, Fisher kernel Class frequency |
| Classifier | KNN | Probability density weighted mean distance | KNN |
| Target Class | ||||||||||
| A | C | MA | MO | PA | PP | P | S | T | ||
| Output Class | A | 146 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 1 |
| C | 9 | 2408 | 139 | 51 | 29 | 17 | 38 | 116 | 55 | |
| MA | 4 | 59 | 372 | 0 | 2 | 10 | 5 | 11 | 4 | |
| MO | 0 | 19 | 3 | 336 | 2 | 6 | 2 | 9 | 5 | |
| PA | 0 | 31 | 13 | 4 | 202 | 0 | 0 | 4 | 2 | |
| PP | 14 | 0 | 6 | 3 | 0 | 691 | 11 | 2 | 8 | |
| P | 9 | 32 | 7 | 5 | 3 | 11 | 727 | 11 | 40 | |
| S | 0 | 67 | 3 | 6 | 4 | 1 | 10 | 650 | 60 | |
| T | 6 | 18 | 16 | 11 | 6 | 6 | 24 | 45 | 817 | |
| Ground Truth | Ground truth | ||||||||||||||||||||
| A | C | MA | MO | PA | PP | P | S | T | A | C | MA | MO | PA | PP | P | S | T | ||||
| Output Class | A | 68 | 28 | 23 | 12 | 0 | 32 | 17 | 1 | 7 | Output Class | A | 112 | 17 | 12 | 1 | 0 | 21 | 12 | 0 | 13 |
| C | 1 | 2226 | 94 | 30 | 24 | 30 | 35 | 84 | 111 | C | 3 | 2366 | 62 | 22 | 25 | 19 | 28 | 67 | 43 | ||
| MA | 4 | 251 | 228 | 2 | 8 | 29 | 16 | 0 | 22 | MA | 2 | 166 | 318 | 2 | 3 | 28 | 5 | 3 | 33 | ||
| MO | 0 | 100 | 7 | 169 | 0 | 21 | 21 | 14 | 84 | MO | 4 | 115 | 7 | 247 | 2 | 1 | 10 | 10 | 20 | ||
| PA | 0 | 33 | 0 | 0 | 197 | 0 | 8 | 4 | 6 | PA | 0 | 38 | 8 | 3 | 177 | 0 | 7 | 6 | 9 | ||
| PP | 9 | 69 | 36 | 17 | 0 | 554 | 51 | 0 | 8 | PP | 16 | 62 | 21 | 0 | 1 | 625 | 8 | 1 | 10 | ||
| P | 0 | 112 | 28 | 12 | 0 | 60 | 531 | 29 | 67 | P | 9 | 100 | 8 | 8 | 2 | 4 | 629 | 8 | 71 | ||
| S | 0 | 198 | 3 | 10 | 2 | 5 | 25 | 600 | 105 | S | 0 | 178 | 4 | 5 | 2 | 3 | 18 | 686 | 52 | ||
| T | 2 | 156 | 32 | 34 | 0 | 8 | 38 | 94 | 628 | T | 2 | 94 | 12 | 9 | 1 | 4 | 53 | 46 | 771 | ||
| Marcos | Stokes & Deane | ||||||||||||||||||||
| Accuracy: 68.7% | Accuracy: 78.3% | ||||||||||||||||||||
| Ground Truth | Ground Truth | ||||||||||||||||||||
| A | C | MA | MO | PA | PP | P | S | T | A | C | MA | MO | PA | PP | P | S | T | ||||
| Output Class | A | 74 | 37 | 9 | 5 | 6 | 22 | 17 | 2 | 16 | Output Class | A | 148 | 24 | 9 | 2 | 2 | 27 | 16 | 0 | 8 |
| C | 38 | 1888 | 106 | 55 | 31 | 94 | 84 | 178 | 161 | C | 6 | 2790 | 68 | 35 | 25 | 28 | 66 | 129 | 148 | ||
| MA | 26 | 140 | 301 | 3 | 10 | 26 | 13 | 16 | 25 | MA | 2 | 194 | 421 | 15 | 10 | 21 | 10 | 1 | 28 | ||
| MO | 4 | 81 | 6 | 175 | 13 | 22 | 43 | 35 | 37 | MO | 1 | 128 | 15 | 309 | 4 | 4 | 20 | 15 | 25 | ||
| PA | 7 | 37 | 6 | 20 | 147 | 3 | 17 | 4 | 7 | PA | 1 | 50 | 4 | 1 | 216 | 9 | 21 | 2 | 7 | ||
| PP | 102 | 136 | 34 | 14 | 10 | 363 | 29 | 13 | 43 | PP | 5 | 52 | 17 | 5 | 6 | 815 | 16 | 1 | 13 | ||
| P | 13 | 119 | 22 | 35 | 27 | 27 | 457 | 41 | 98 | P | 8 | 150 | 17 | 13 | 10 | 20 | 740 | 31 | 69 | ||
| S | 6 | 248 | 17 | 28 | 9 | 23 | 52 | 479 | 86 | S | 0 | 255 | 1 | 4 | 2 | 4 | 36 | 823 | 61 | ||
| T | 10 | 185 | 28 | 21 | 7 | 38 | 87 | 92 | 524 | T | 3 | 320 | 25 | 13 | 1 | 24 | 69 | 61 | 724 | ||
| Pizarro | Beijbom | ||||||||||||||||||||
| Accuracy: 58.2% | Accuracy: 73.7% | ||||||||||||||||||||
| Marcos | Stokes & Deane | Pizarro | Beijbom | Caputo | Zhang | Our | |
|---|---|---|---|---|---|---|---|
| EILAT | 87.9 | 75.2 | 67.3 | 69.1 | NA | NA | 96.9 |
| RSMAS | 69.3 | 82.5 | 73.9 | 85.4 | NA | NA | 96.5 |
| MLC 2008 | 68.7 | 78.3 | 58.2 | 73.7 | NA | NA | 85.5 |
| CURET | 20.8 | 49.7 | 38.1 | 86.5 | 98.6 | 98.5 | 99.2 |
| KTH | 25.5 | 88.9 | 48.3 | 36.3 | 95.8 | 96.7 | 98.3 |
| UIUC | 14.6 | 56.9 | 19.9 | 32.2 | 98.2 | 99.0 | 97.3 |
| Marcos | Stokes & Deane | Pizarro | Beijbom | Caputo | Zhang | Our | |
|---|---|---|---|---|---|---|---|
| EILAT | 85.1 | 73.5 | 58.1 | 64.2 | NA | NA | 97.2 |
| RSMAS | 59.2 | 81.2 | 67.1 | 79.9 | NA | NA | 96.2 |
| MLC 2008 | 49.5 | 61.3 | 46.4 | 64.9 | NA | NA | 74.8 |
| CURET | 14.7 | 30.1 | 29.1 | 81.5 | 98.2 | 98.1 | 98.4 |
| KTH | 23.5 | 84.8 | 39.7 | 34.6 | 95.7 | 96.4 | 97.7 |
| UIUC | 14.7 | 43.5 | 18.2 | 30.4 | 97.2 | 98.5 | 96.9 |
Share and Cite
Shihavuddin, A.S.M.; Gracias, N.; Garcia, R.; Gleason, A.C.R.; Gintert, B. Image-Based Coral Reef Classification and Thematic Mapping. Remote Sens. 2013, 5, 1809-1841. https://doi.org/10.3390/rs5041809
Shihavuddin ASM, Gracias N, Garcia R, Gleason ACR, Gintert B. Image-Based Coral Reef Classification and Thematic Mapping. Remote Sensing. 2013; 5(4):1809-1841. https://doi.org/10.3390/rs5041809
Chicago/Turabian StyleShihavuddin, A.S.M., Nuno Gracias, Rafael Garcia, Arthur C. R. Gleason, and Brooke Gintert. 2013. "Image-Based Coral Reef Classification and Thematic Mapping" Remote Sensing 5, no. 4: 1809-1841. https://doi.org/10.3390/rs5041809