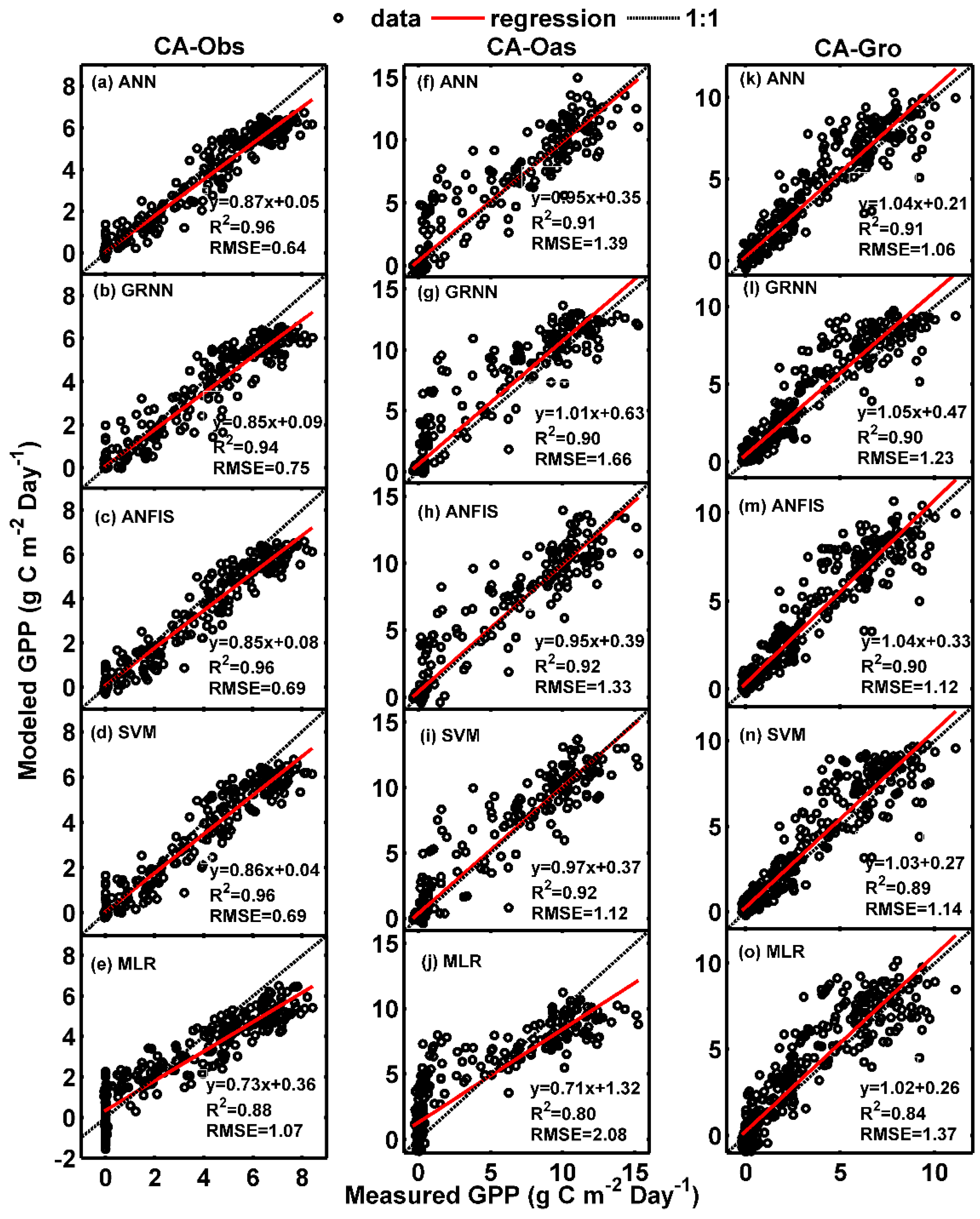
Figure 1.
Comparisons of measured and simulated daily gross primary productivity (GPP) in the testing period in the three boreal forest stands. (a–e) For CA-Obs stand; (f–j) for CA-Oas stand; and (k–o) for CA-Gro stand.
Figure 1.
Comparisons of measured and simulated daily gross primary productivity (GPP) in the testing period in the three boreal forest stands. (a–e) For CA-Obs stand; (f–j) for CA-Oas stand; and (k–o) for CA-Gro stand.
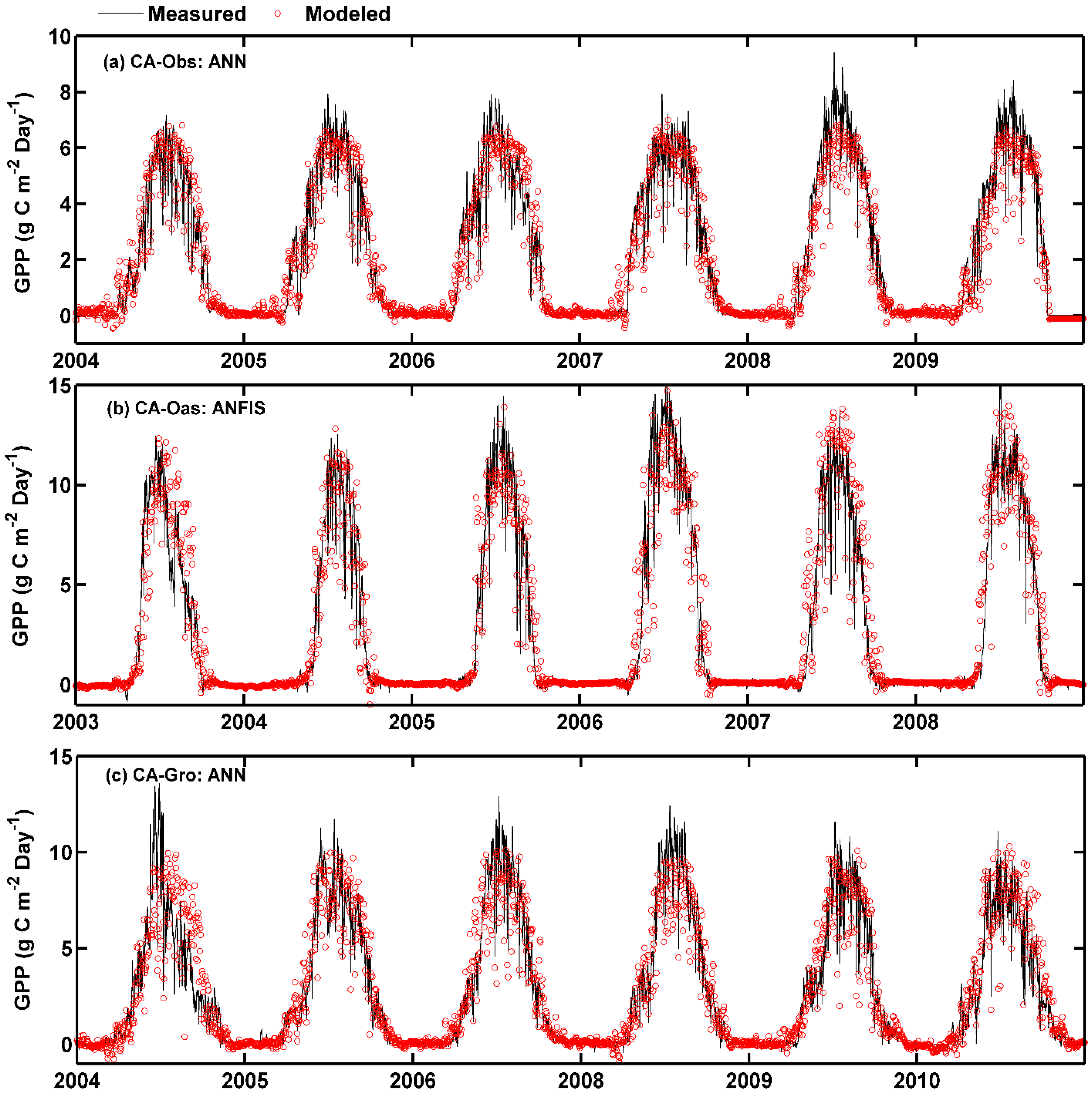
Figure 2.
Eddy covariance measured and model simulated daily gross primary productivity (GPP) in the whole period in the three boreal forest stands. (a) For CA-Obs stand using ANN model; (b) for CA-Oas stand using ANFIS model; and (c) for CA-Gro stand using ANN model.
Figure 2.
Eddy covariance measured and model simulated daily gross primary productivity (GPP) in the whole period in the three boreal forest stands. (a) For CA-Obs stand using ANN model; (b) for CA-Oas stand using ANFIS model; and (c) for CA-Gro stand using ANN model.
Figure 3.
Comparisons of measured and simulated daily ecosystem respiration (R) in the testing period in the three boreal forest stands. (a–e) For CA-Obs stand; (f–j) for CA-Oas stand; and (k–o) for CA-Gro stand.
Figure 3.
Comparisons of measured and simulated daily ecosystem respiration (R) in the testing period in the three boreal forest stands. (a–e) For CA-Obs stand; (f–j) for CA-Oas stand; and (k–o) for CA-Gro stand.
Figure 4.
Eddy covariance measured and model simulated daily ecosystem respiration (R) in the whole period in the three boreal forest stands. (a) For CA-Obs stand using ANN model; (b) for CA-Oas stand using ANFIS model; and (c) for CA-Gro stand using GRNN model.
Figure 4.
Eddy covariance measured and model simulated daily ecosystem respiration (R) in the whole period in the three boreal forest stands. (a) For CA-Obs stand using ANN model; (b) for CA-Oas stand using ANFIS model; and (c) for CA-Gro stand using GRNN model.
Figure 5.
Comparisons of measured and simulated daily net ecosystem exchange (NEE) in the testing period in the three boreal forest stands. (a–e) For CA-Obs stand; (f–j) for CA-Oas stand; and (k–o) for CA-Gro stand.
Figure 5.
Comparisons of measured and simulated daily net ecosystem exchange (NEE) in the testing period in the three boreal forest stands. (a–e) For CA-Obs stand; (f–j) for CA-Oas stand; and (k–o) for CA-Gro stand.
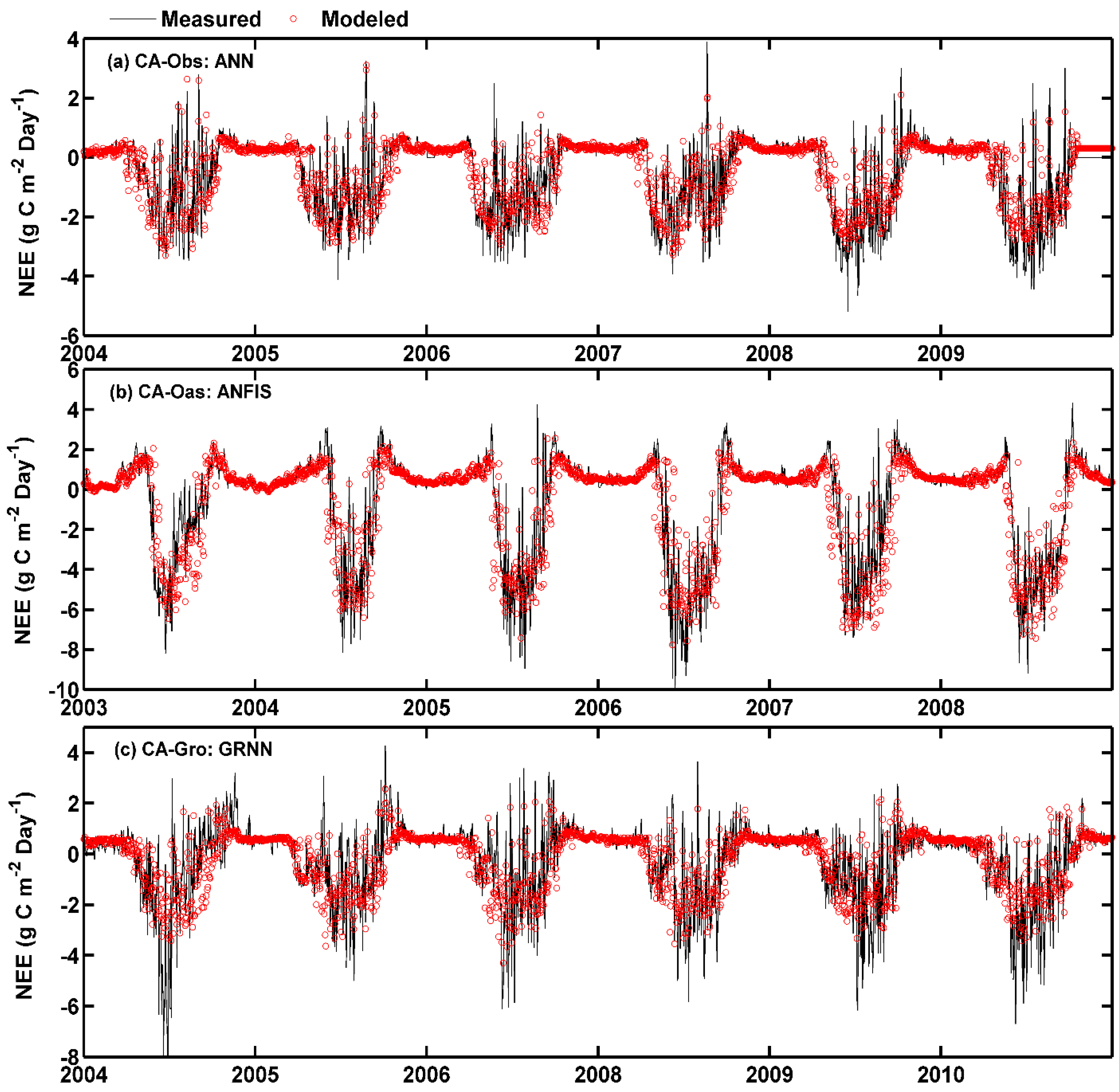
Figure 6.
Eddy covariance measured and model simulated daily net ecosystem exchange (NEE) in the whole period in the three boreal forest stands. (a) For CA-Obs stand using ANN model; (b) for CA-Oas stand using ANFIS model; and (c) for CA-Gro stand using GRNN model.
Figure 6.
Eddy covariance measured and model simulated daily net ecosystem exchange (NEE) in the whole period in the three boreal forest stands. (a) For CA-Obs stand using ANN model; (b) for CA-Oas stand using ANFIS model; and (c) for CA-Gro stand using GRNN model.
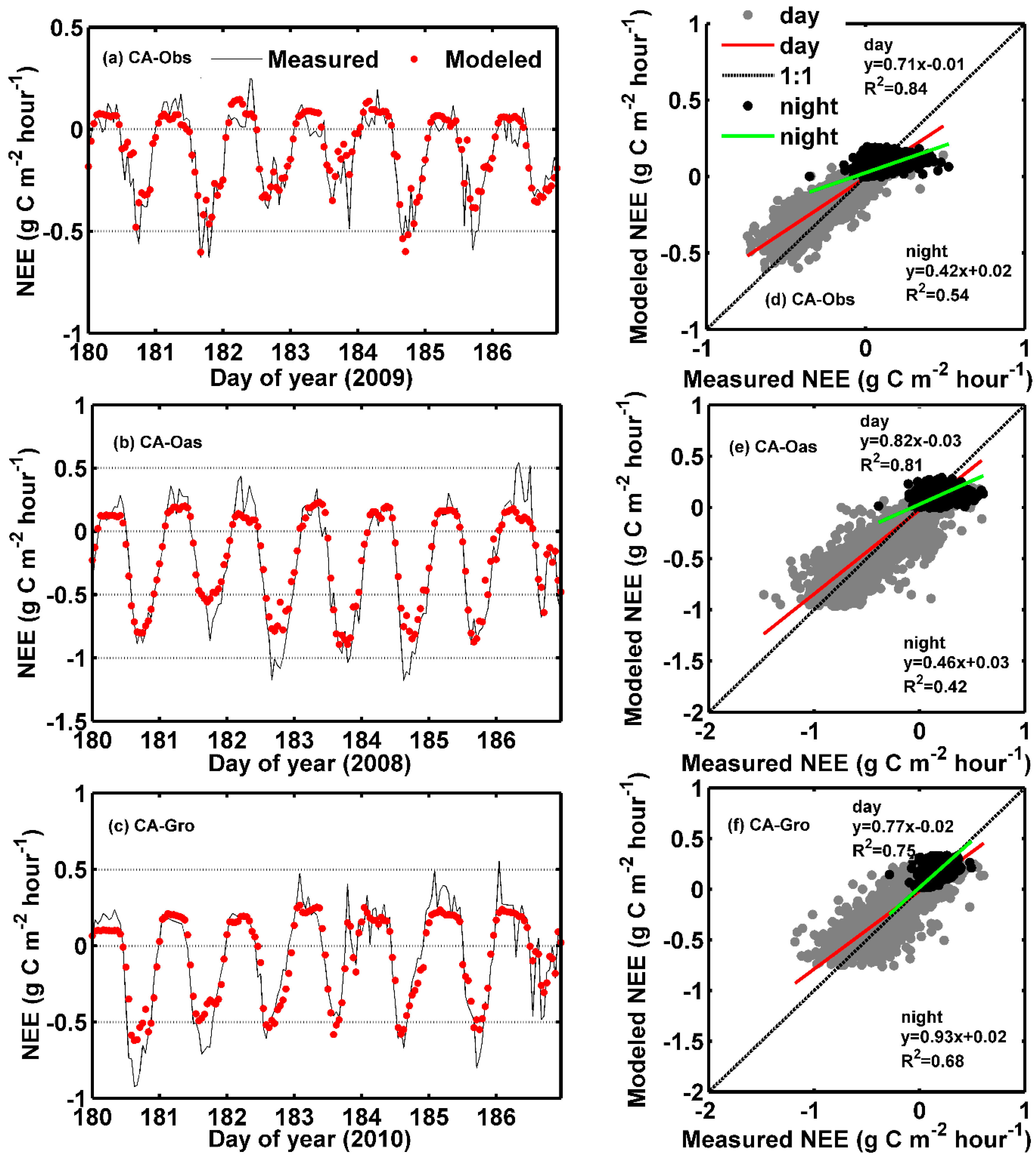
Figure 7.
Eddy covariance measured and ANFIS predicted hourly net ecosystem exchange (NEE) in the testing period. (a–c) Daily course of hourly NEE for 7 days during the mid-growing season at the three sites; and (d–f) Scatter plot of hourly NEE both in the daytime and nighttime datasets at each site.
Figure 7.
Eddy covariance measured and ANFIS predicted hourly net ecosystem exchange (NEE) in the testing period. (a–c) Daily course of hourly NEE for 7 days during the mid-growing season at the three sites; and (d–f) Scatter plot of hourly NEE both in the daytime and nighttime datasets at each site.
Table 1.
Site characteristics used in this study. The mean annual temperature (MAT, °C) and total annual precipitation (TAP, mm year−1) are for the time period (Period). Vegetation types are deciduous broadleaf forest (DBF), evergreen needle-leaf forest (ENF) and mixed forest (MF).
Table 1.
Site characteristics used in this study. The mean annual temperature (MAT, °C) and total annual precipitation (TAP, mm year−1) are for the time period (Period). Vegetation types are deciduous broadleaf forest (DBF), evergreen needle-leaf forest (ENF) and mixed forest (MF).
| Site | Latitude | Longitude | Elevation | MAT | TAP | Vegetation | Period | Reference |
|---|
| CA-Oas | 53.63 | −106.20 | 601 | 1.82 | 343 | DBF | 2003–2008 | Griffis, et al. [35] |
| CA-Obs | 53.99 | −105.12 | 629 | 1.00 | 373 | ENF | 2004–2009 | Krishnan, et al. [36] |
| CA-Gro | 48.22 | −82.16 | 341 | 3.29 | 784 | MF | 2004–2006, 2008–2010 | McCaughey, et al. [37] |
Table 2.
Daily statistical parameters of flux tower measured environmental variables including air temperature (Ta, °C), net radiation (Rn, mol m−2), relative humidity (Rh, %), soil temperature (Ts, °C), gross primary productivity (GPP, g C m−2 day−1), ecosystem respiration (R, g C m−2 day−1) and net ecosystem exchange (NEE, g C m−2 day−1) during the whole period in all three stands.
Table 2.
Daily statistical parameters of flux tower measured environmental variables including air temperature (Ta, °C), net radiation (Rn, mol m−2), relative humidity (Rh, %), soil temperature (Ts, °C), gross primary productivity (GPP, g C m−2 day−1), ecosystem respiration (R, g C m−2 day−1) and net ecosystem exchange (NEE, g C m−2 day−1) during the whole period in all three stands.
| Stand | Variable | Xmean | Xmax | Xmin | Xsd | Xku | Xsk | CC1 | CC2 | CC3 |
|---|
| CA-Oas | Ta | 1.82 | 26.46 | −35.04 | 13.02 | 2.22 | −0.40 | 0.70 | 0.82 | −0.50 |
| Rn | 5.19 | 18.94 | −4.67 | 5.66 | 2.02 | 0.45 | 0.69 | 0.68 | −0.60 |
| Rh | 69.88 | 98.78 | 21.83 | 16.15 | 2.62 | −0.52 | −0.18 | −0.21 | 0.13 |
| Ts | 4.73 | 17.31 | −5.81 | 5.81 | 1.69 | 0.30 | 0.83 | 0.93 | −0.64 |
| GPP | 2.87 | 15.99 | −0.84 | 4.29 | 2.83 | 1.17 | 1.00 | 0.91 | −0.94 |
| R | 2.25 | 8.69 | −1.17 | 2.11 | 2.34 | 0.81 | 0.91 | 1.00 | −0.70 |
| NEE | −0.62 | 4.34 | −10.41 | 2.55 | 3.69 | −1.29 | −0.94 | −0.70 | 1.00 |
| CA-Obs | Ta | 1.00 | 26.37 | −35.31 | 13.15 | 2.15 | −0.34 | 0.83 | 0.81 | −0.58 |
| Rn | 6.71 | 21.90 | −4.28 | 6.17 | 2.05 | 0.48 | 0.72 | 0.56 | −0.71 |
| Rh | 72.35 | 100.00 | 24.56 | 16.69 | 2.46 | −0.43 | −0.31 | −0.16 | 0.42 |
| Ts | 3.19 | 15.41 | −9.18 | 4.99 | 1.99 | 0.58 | 0.87 | 0.94 | −0.48 |
| GPP | 2.23 | 9.41 | −0.15 | 2.56 | 1.96 | 0.68 | 1.00 | 0.90 | −0.80 |
| R | 1.70 | 7.28 | −0.26 | 1.73 | 2.66 | 0.97 | 0.90 | 1.00 | −0.47 |
| NEE | −0.53 | 3.90 | −5.20 | 1.24 | 3.03 | −0.84 | −0.80 | −0.47 | 1.00 |
| CA-Gro | Ta | 3.29 | 27.83 | −31.66 | 12.35 | 2.26 | −0.36 | 0.78 | 0.82 | −0.46 |
| Rn | 6.60 | 21.97 | −3.42 | 5.75 | 2.13 | 0.51 | 0.69 | 0.58 | −0.64 |
| Rh | 74.98 | 98.93 | 21.54 | 15.47 | 3.27 | −0.78 | −0.25 | −0.18 | 0.27 |
| Ts | 5.56 | 17.68 | −1.21 | 5.45 | 1.65 | 0.45 | 0.87 | 0.94 | −0.48 |
| GPP | 2.88 | 13.59 | −0.45 | 3.38 | 2.70 | 0.98 | 1.00 | 0.92 | −0.80 |
| R | 2.59 | 10.62 | −0.21 | 2.31 | 2.66 | 0.90 | 0.92 | 1.00 | −0.50 |
| NEE | −0.29 | 4.27 | −8.51 | 1.56 | 5.64 | −1.42 | −0.80 | −0.50 | 1.00 |
Table 3.
The input combinations for different methods with environmental variables including air temperature (Ta, °C), net radiation (Rn, mol m−2), relative humidity (Rh, %), soil temperature (Ts, °C).
Table 3.
The input combinations for different methods with environmental variables including air temperature (Ta, °C), net radiation (Rn, mol m−2), relative humidity (Rh, %), soil temperature (Ts, °C).
| Models | Input Combinations |
|---|
| ANN | GRNN | ANFIS | SVM | MLR |
|---|
| ANN1 | GRNN1 | ANFIS1 | SVM1 | MLR1 | Ta |
| ANN2 | GRNN2 | ANFIS2 | SVM2 | MLR2 | Rn |
| ANN3 | GRNN3 | ANFIS3 | SVM3 | MLR3 | Rh |
| ANN4 | GRNN4 | ANFIS4 | SVM4 | MLR4 | Ts |
| ANN5 | GRNN5 | ANFIS5 | SVM5 | MLR5 | Ta, Rn |
| ANN6 | GRNN6 | ANFIS6 | SVM6 | MLR6 | Ta, Rn, Rh |
| ANN7 | GRNN7 | ANFIS7 | SVM7 | MLR7 | Ta, Rn, Rh, Ts |
Table 4.
Comparisons of machine learning models with different input combinations in the testing period for gross primary productivity (GPP, g C m−2 day−1) in the three boreal forest stands.
Table 4.
Comparisons of machine learning models with different input combinations in the testing period for gross primary productivity (GPP, g C m−2 day−1) in the three boreal forest stands.
| Model | CA-Obs | CA-Oas | CA-Gro |
|---|
| R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE |
|---|
| ANN1 | 0.880 | 0.854 | −0.358 | 1.046 | 0.753 | 0.742 | −0.243 | 2.279 | 0.736 | 0.682 | 0.573 | 1.705 |
| ANN2 | 0.482 | 0.472 | 0.003 | 1.989 | 0.428 | 0.420 | 0.203 | 3.417 | 0.554 | 0.529 | 0.395 | 2.076 |
| ANN3 | 0.001 | −0.088 | 0.311 | 2.854 | 0.062 | 0.062 | 0.022 | 4.347 | 0.110 | 0.072 | 0.483 | 2.914 |
| ANN4 | 0.811 | 0.788 | −0.305 | 1.259 | 0.831 | 0.830 | 0.118 | 1.848 | 0.795 | 0.778 | 0.340 | 1.425 |
| ANN5 | 0.903 | 0.883 | −0.273 | 0.937 | 0.808 | 0.806 | −0.069 | 1.975 | 0.804 | 0.763 | 0.488 | 1.473 |
| ANN6 | 0.932 | 0.917 | −0.317 | 0.789 | 0.871 | 0.870 | −0.087 | 1.618 | 0.861 | 0.825 | 0.430 | 1.267 |
| ANN7 | 0.963 | 0.944 | −0.253 | 0.645 | 0.909 | 0.905 | 0.211 | 1.386 | 0.907 | 0.877 | 0.319 | 1.060 |
| GRNN1 | 0.876 | 0.825 | −0.488 | 1.145 | 0.752 | 0.720 | −0.524 | 2.377 | 0.734 | 0.699 | 0.486 | 1.660 |
| GRNN2 | 0.485 | 0.470 | 0.056 | 1.992 | 0.434 | 0.428 | 0.261 | 3.395 | 0.564 | 0.551 | 0.325 | 2.027 |
| GRNN3 | 0.000 | −0.108 | 0.367 | 2.881 | 0.063 | 0.058 | 0.082 | 4.357 | 0.125 | 0.090 | 0.527 | 2.886 |
| GRNN4 | 0.794 | 0.770 | −0.311 | 1.311 | 0.841 | 0.822 | −0.359 | 1.894 | 0.802 | 0.787 | 0.270 | 1.396 |
| GRNN5 | 0.904 | 0.882 | −0.282 | 0.941 | 0.786 | 0.786 | 0.050 | 2.078 | 0.795 | 0.733 | 0.598 | 1.562 |
| GRNN6 | 0.926 | 0.886 | −0.402 | 0.924 | 0.870 | 0.864 | −0.105 | 1.652 | 0.838 | 0.803 | 0.465 | 1.341 |
| GRNN7 | 0.944 | 0.925 | −0.259 | 0.747 | 0.900 | 0.864 | 0.667 | 1.655 | 0.899 | 0.834 | 0.603 | 1.234 |
| ANFIS1 | 0.882 | 0.856 | −0.353 | 1.037 | 0.750 | 0.741 | −0.222 | 2.286 | 0.736 | 0.695 | 0.504 | 1.669 |
| ANFIS2 | 0.474 | 0.459 | 0.087 | 2.013 | 0.414 | 0.402 | 0.266 | 3.470 | 0.565 | 0.549 | 0.354 | 2.031 |
| ANFIS3 | 0.000 | −0.124 | 0.440 | 2.901 | 0.063 | 0.062 | −0.061 | 4.347 | 0.127 | 0.088 | 0.550 | 2.889 |
| ANFIS4 | 0.785 | 0.764 | −0.314 | 1.329 | 0.806 | 0.806 | 0.077 | 1.977 | 0.804 | 0.780 | 0.294 | 1.420 |
| ANFIS5 | 0.902 | 0.883 | −0.309 | 0.938 | 0.790 | 0.790 | −0.033 | 2.058 | 0.797 | 0.749 | 0.526 | 1.515 |
| ANFIS6 | 0.938 | 0.921 | −0.302 | 0.771 | 0.886 | 0.884 | −0.135 | 1.529 | 0.866 | 0.823 | 0.485 | 1.271 |
| ANFIS7 | 0.960 | 0.936 | −0.271 | 0.690 | 0.917 | 0.913 | 0.249 | 1.326 | 0.904 | 0.863 | 0.438 | 1.120 |
| SVM1 | 0.876 | 0.851 | −0.354 | 1.056 | 0.754 | 0.745 | −0.295 | 2.268 | 0.732 | 0.698 | 0.397 | 1.662 |
| SVM2 | 0.501 | 0.498 | 0.136 | 1.939 | 0.392 | 0.304 | 0.334 | 3.744 | 0.564 | 0.545 | 0.188 | 2.041 |
| SVM3 | 0.001 | −0.362 | 0.116 | 3.193 | 0.035 | −0.350 | −2.735 | 5.215 | 0.090 | −0.012 | −0.530 | 3.043 |
| SVM4 | 0.758 | 0.725 | −0.498 | 1.434 | 0.868 | 0.867 | −0.007 | 1.638 | 0.802 | 0.780 | 0.185 | 1.420 |
| SVM5 | 0.902 | 0.886 | −0.274 | 0.924 | 0.796 | 0.796 | 0.048 | 2.029 | 0.800 | 0.755 | 0.483 | 1.497 |
| SVM6 | 0.932 | 0.919 | −0.237 | 0.781 | 0.881 | 0.881 | −0.059 | 1.547 | 0.865 | 0.813 | 0.490 | 1.307 |
| SVM7 | 0.956 | 0.936 | −0.281 | 0.695 | 0.916 | 0.910 | 0.276 | 1.350 | 0.894 | 0.859 | 0.359 | 1.136 |
| MLR1 | 0.758 | 0.707 | −0.586 | 1.480 | 0.510 | 0.498 | −0.490 | 3.177 | 0.615 | 0.568 | 0.637 | 1.988 |
| MLR2 | 0.465 | 0.453 | 0.040 | 2.024 | 0.408 | 0.397 | 0.292 | 3.482 | 0.556 | 0.539 | 0.375 | 2.054 |
| MLR3 | 0.000 | −0.076 | 0.220 | 2.837 | 0.019 | 0.018 | −0.124 | 4.443 | 0.033 | −0.029 | 0.499 | 3.067 |
| MLR4 | 0.778 | 0.746 | −0.350 | 1.379 | 0.752 | 0.738 | 0.108 | 2.297 | 0.766 | 0.723 | 0.351 | 1.591 |
| MLR5 | 0.759 | 0.731 | −0.387 | 1.419 | 0.542 | 0.539 | −0.065 | 3.044 | 0.699 | 0.654 | 0.583 | 1.779 |
| MLR6 | 0.786 | 0.744 | −0.551 | 1.384 | 0.604 | 0.601 | −0.187 | 2.832 | 0.738 | 0.702 | 0.517 | 1.650 |
| MLR7 | 0.882 | 0.846 | −0.262 | 1.074 | 0.805 | 0.785 | 0.430 | 2.081 | 0.843 | 0.793 | 0.318 | 1.375 |
Table 5.
Comparisons of different models in the three stands for gross primary productivity (GPP, g C m−2 day−1), ecosystem respiration (R, g C m−2 day−1) and net ecosystem exchange (NEE, g C m−2 day−1) in the testing period.
Table 5.
Comparisons of different models in the three stands for gross primary productivity (GPP, g C m−2 day−1), ecosystem respiration (R, g C m−2 day−1) and net ecosystem exchange (NEE, g C m−2 day−1) in the testing period.
| Model | GPP | R | NEE |
|---|
| R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE |
|---|
| ANN | 0.926 | 0.909 | 0.092 | 1.030 | 0.944 | 0.818 | 0.236 | 0.717 | 0.789 | 0.754 | 0.160 | 0.889 |
| GRNN | 0.914 | 0.874 | 0.337 | 1.212 | 0.936 | 0.887 | 0.148 | 0.624 | 0.770 | 0.731 | −0.017 | 0.950 |
| ANFIS | 0.927 | 0.904 | 0.138 | 1.045 | 0.944 | 0.816 | 0.243 | 0.725 | 0.785 | 0.751 | 0.132 | 0.889 |
| SVM | 0.922 | 0.901 | 0.118 | 1.060 | 0.941 | 0.812 | 0.260 | 0.737 | 0.779 | 0.738 | 0.173 | 0.914 |
| MLR | 0.843 | 0.808 | 0.162 | 1.510 | 0.915 | 0.799 | 0.256 | 0.807 | 0.569 | 0.523 | 0.094 | 1.258 |
Table 6.
Comparisons of different stands using all the five models for gross primary productivity (GPP, g C m−2 day−1), ecosystem respiration (R, g C m−2 day−1) and net ecosystem exchange (NEE, g C m−2 day−1) in the testing period.
Table 6.
Comparisons of different stands using all the five models for gross primary productivity (GPP, g C m−2 day−1), ecosystem respiration (R, g C m−2 day−1) and net ecosystem exchange (NEE, g C m−2 day−1) in the testing period.
| Stand | GPP | R | NEE |
|---|
| R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE |
|---|
| CA-Obs | 0.941 | 0.918 | −0.265 | 0.770 | 0.937 | 0.925 | −0.027 | 0.496 | 0.764 | 0.714 | 0.208 | 0.701 |
| CA-Oas | 0.889 | 0.875 | 0.367 | 1.56 | 0.919 | 0.917 | 0.070 | 0.622 | 0.769 | 0.756 | −0.188 | 1.272 |
| CA-Gro | 0.889 | 0.845 | 0.407 | 1.185 | 0.952 | 0.637 | 0.643 | 1.047 | 0.682 | 0.628 | 0.305 | 0.966 |
Table 7.
Comparisons of machine learning models with different input combinations in the testing period for ecosystem respiration (R, g C m−2 day−1) in the three boreal forest stands.
Table 7.
Comparisons of machine learning models with different input combinations in the testing period for ecosystem respiration (R, g C m−2 day−1) in the three boreal forest stands.
| Model | CA-Obs | CA-Oas | CA-Gro |
|---|
| R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE |
|---|
| ANN1 | 0.829 | 0.821 | −0.032 | 0.776 | 0.837 | 0.830 | −0.116 | 0.892 | 0.847 | 0.487 | 0.802 | 1.258 |
| ANN2 | 0.258 | 0.234 | 0.275 | 1.606 | 0.378 | 0.364 | 0.122 | 1.725 | 0.421 | 0.246 | 0.677 | 1.525 |
| ANN3 | 0.011 | −0.148 | 0.428 | 1.967 | 0.055 | 0.053 | 0.011 | 2.105 | 0.070 | −0.173 | 0.799 | 1.902 |
| ANN4 | 0.904 | 0.881 | 0.076 | 0.634 | 0.904 | 0.902 | 0.073 | 0.675 | 0.938 | 0.618 | 0.721 | 1.085 |
| ANN5 | 0.842 | 0.832 | −0.083 | 0.752 | 0.830 | 0.828 | −0.074 | 0.897 | 0.853 | 0.467 | 0.849 | 1.282 |
| ANN6 | 0.913 | 0.907 | 0.023 | 0.560 | 0.893 | 0.892 | −0.048 | 0.710 | 0.908 | 0.573 | 0.740 | 1.148 |
| ANN7 | 0.954 | 0.946 | −0.033 | 0.428 | 0.927 | 0.927 | 0.027 | 0.585 | 0.950 | 0.580 | 0.714 | 1.138 |
| GRNN1 | 0.827 | 0.807 | −0.081 | 0.807 | 0.822 | 0.821 | 0.007 | 0.914 | 0.852 | 0.414 | 0.952 | 1.345 |
| GRNN2 | 0.272 | 0.252 | 0.236 | 1.588 | 0.381 | 0.368 | 0.134 | 1.719 | 0.418 | 0.284 | 0.604 | 1.487 |
| GRNN3 | 0.012 | −0.141 | 0.394 | 1.961 | 0.047 | 0.032 | 0.267 | 2.128 | 0.080 | −0.166 | 0.848 | 1.897 |
| GRNN4 | 0.905 | 0.889 | −0.059 | 0.610 | 0.904 | 0.901 | 0.082 | 0.679 | 0.950 | 0.680 | 0.672 | 0.993 |
| GRNN5 | 0.837 | 0.816 | −0.101 | 0.788 | 0.828 | 0.812 | 0.249 | 0.937 | 0.848 | 0.532 | 0.794 | 1.202 |
| GRNN6 | 0.895 | 0.886 | −0.020 | 0.620 | 0.873 | 0.872 | 0.062 | 0.775 | 0.902 | 0.595 | 0.759 | 1.118 |
| GRNN7 | 0.941 | 0.929 | −0.037 | 0.488 | 0.914 | 0.914 | 0.032 | 0.635 | 0.954 | 0.818 | 0.448 | 0.749 |
| ANFIS1 | 0.828 | 0.821 | −0.029 | 0.776 | 0.834 | 0.830 | −0.091 | 0.890 | 0.849 | 0.500 | 0.804 | 1.242 |
| ANFIS2 | 0.243 | 0.219 | 0.280 | 1.622 | 0.381 | 0.364 | 0.170 | 1.725 | 0.422 | 0.251 | 0.676 | 1.520 |
| ANFIS3 | 0.014 | −0.176 | 0.426 | 1.991 | 0.050 | 0.049 | 0.013 | 2.109 | 0.082 | −0.135 | 0.775 | 1.872 |
| ANFIS4 | 0.904 | 0.894 | −0.010 | 0.596 | 0.913 | 0.910 | 0.081 | 0.650 | 0.947 | 0.621 | 0.671 | 1.081 |
| ANFIS5 | 0.846 | 0.840 | −0.050 | 0.735 | 0.839 | 0.839 | −0.035 | 0.868 | 0.853 | 0.479 | 0.837 | 1.268 |
| ANFIS6 | 0.912 | 0.906 | −0.007 | 0.563 | 0.903 | 0.901 | −0.093 | 0.682 | 0.910 | 0.532 | 0.788 | 1.201 |
| ANFIS7 | 0.947 | 0.935 | −0.019 | 0.468 | 0.930 | 0.930 | 0.040 | 0.574 | 0.954 | 0.584 | 0.709 | 1.132 |
| SVM1 | 0.826 | 0.818 | −0.062 | 0.783 | 0.837 | 0.835 | −0.098 | 0.878 | 0.851 | 0.546 | 0.743 | 1.183 |
| SVM2 | 0.248 | 0.244 | 0.062 | 1.596 | 0.380 | 0.333 | 0.104 | 1.766 | 0.426 | 0.235 | 0.514 | 1.536 |
| SVM3 | 0.025 | −0.253 | −0.143 | 2.055 | 0.049 | −0.101 | −0.773 | 2.269 | 0.072 | −0.095 | 0.091 | 1.838 |
| SVM4 | 0.907 | 0.898 | −0.049 | 0.588 | 0.912 | 0.908 | 0.116 | 0.654 | 0.945 | 0.646 | 0.645 | 1.045 |
| SVM5 | 0.843 | 0.832 | −0.108 | 0.752 | 0.840 | 0.838 | −0.077 | 0.869 | 0.855 | 0.534 | 0.766 | 1.199 |
| SVM6 | 0.910 | 0.901 | −0.052 | 0.577 | 0.899 | 0.895 | −0.107 | 0.699 | 0.916 | 0.516 | 0.803 | 1.222 |
| SVM7 | 0.947 | 0.935 | −0.017 | 0.469 | 0.925 | 0.923 | 0.088 | 0.601 | 0.951 | 0.579 | 0.709 | 1.140 |
| MLR1 | 0.697 | 0.686 | −0.182 | 1.028 | 0.659 | 0.647 | −0.208 | 1.285 | 0.734 | 0.435 | 0.874 | 1.321 |
| MLR2 | 0.242 | 0.227 | 0.221 | 1.613 | 0.377 | 0.356 | 0.196 | 1.736 | 0.418 | 0.252 | 0.665 | 1.520 |
| MLR3 | 0.026 | −0.110 | 0.282 | 1.933 | 0.026 | 0.025 | 0.008 | 2.135 | 0.011 | −0.224 | 0.732 | 1.944 |
| MLR4 | 0.877 | 0.868 | −0.041 | 0.667 | 0.898 | 0.885 | 0.127 | 0.733 | 0.945 | 0.629 | 0.660 | 1.070 |
| MLR5 | 0.684 | 0.677 | −0.153 | 1.043 | 0.654 | 0.647 | −0.081 | 1.284 | 0.742 | 0.442 | 0.861 | 1.312 |
| MLR6 | 0.734 | 0.702 | −0.316 | 1.002 | 0.700 | 0.691 | −0.133 | 1.202 | 0.792 | 0.522 | 0.810 | 1.215 |
| MLR7 | 0.893 | 0.882 | −0.031 | 0.629 | 0.902 | 0.891 | 0.163 | 0.715 | 0.951 | 0.625 | 0.636 | 1.076 |
Table 8.
Comparisons of machine learning models with different input combinations in the testing period for net ecosystem exchange (NEE, g C m−2 day−1) in the three boreal forest stands.
Table 8.
Comparisons of machine learning models with different input combinations in the testing period for net ecosystem exchange (NEE, g C m−2 day−1) in the three boreal forest stands.
| Model | CA-Obs | CA-Oas | CA-Gro |
|---|
| R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE |
|---|
| ANN1 | 0.502 | 0.438 | 0.313 | 0.998 | 0.553 | 0.536 | 0.165 | 1.782 | 0.387 | 0.310 | 0.358 | 1.321 |
| ANN2 | 0.514 | 0.465 | 0.222 | 0.974 | 0.376 | 0.362 | −0.151 | 2.090 | 0.507 | 0.475 | 0.250 | 1.152 |
| ANN3 | 0.054 | 0.026 | −0.073 | 1.314 | 0.067 | 0.064 | 0.065 | 2.532 | 0.107 | 0.098 | 0.126 | 1.510 |
| ANN4 | 0.423 | 0.332 | 0.382 | 1.088 | 0.614 | 0.614 | 0.026 | 1.625 | 0.412 | 0.333 | 0.301 | 1.299 |
| ANN5 | 0.736 | 0.701 | 0.207 | 0.728 | 0.638 | 0.636 | 0.039 | 1.580 | 0.605 | 0.559 | 0.315 | 1.056 |
| ANN6 | 0.775 | 0.719 | 0.270 | 0.705 | 0.744 | 0.744 | 0.004 | 1.325 | 0.674 | 0.631 | 0.301 | 0.966 |
| ANN7 | 0.830 | 0.785 | 0.232 | 0.618 | 0.817 | 0.815 | −0.073 | 1.125 | 0.719 | 0.663 | 0.322 | 0.923 |
| GRNN1 | 0.515 | 0.433 | 0.330 | 1.003 | 0.558 | 0.538 | 0.134 | 1.779 | 0.402 | 0.346 | 0.276 | 1.286 |
| GRNN2 | 0.528 | 0.464 | 0.271 | 0.975 | 0.376 | 0.352 | −0.387 | 2.106 | 0.507 | 0.481 | 0.249 | 1.146 |
| GRNN3 | 0.050 | 0.035 | 0.124 | 1.309 | 0.073 | 0.066 | 0.034 | 2.529 | 0.107 | 0.067 | −0.311 | 1.537 |
| GRNN4 | 0.368 | 0.297 | 0.313 | 1.117 | 0.685 | 0.627 | −0.557 | 1.597 | 0.420 | 0.337 | 0.344 | 1.295 |
| GRNN5 | 0.739 | 0.685 | 0.235 | 0.748 | 0.637 | 0.606 | −0.344 | 1.642 | 0.614 | 0.274 | 0.927 | 1.355 |
| GRNN6 | 0.779 | 0.725 | 0.079 | 0.699 | 0.714 | 0.694 | −0.339 | 1.447 | 0.661 | 0.554 | 0.436 | 1.062 |
| GRNN7 | 0.805 | 0.766 | 0.055 | 0.644 | 0.787 | 0.755 | −0.353 | 1.294 | 0.718 | 0.672 | 0.246 | 0.911 |
| ANFIS1 | 0.510 | 0.438 | 0.327 | 0.998 | 0.535 | 0.525 | 0.147 | 1.803 | 0.396 | 0.335 | 0.304 | 1.296 |
| ANFIS2 | 0.531 | 0.482 | 0.193 | 0.959 | 0.354 | 0.345 | −0.106 | 2.117 | 0.510 | 0.468 | 0.304 | 1.160 |
| ANFIS3 | 0.052 | 0.038 | −0.008 | 1.307 | 0.071 | 0.066 | 0.067 | 2.529 | 0.129 | 0.109 | 0.223 | 1.501 |
| ANFIS4 | 0.338 | 0.281 | 0.297 | 1.130 | 0.694 | 0.692 | 0.026 | 1.453 | 0.433 | 0.336 | 0.377 | 1.296 |
| ANFIS5 | 0.732 | 0.678 | 0.240 | 0.756 | 0.640 | 0.639 | −0.028 | 1.572 | 0.609 | 0.565 | 0.304 | 1.049 |
| ANFIS6 | 0.804 | 0.739 | 0.281 | 0.681 | 0.761 | 0.759 | 0.032 | 1.285 | 0.683 | 0.628 | 0.322 | 0.970 |
| ANFIS7 | 0.808 | 0.758 | 0.264 | 0.655 | 0.830 | 0.824 | −0.158 | 1.097 | 0.716 | 0.670 | 0.289 | 0.914 |
| SVM1 | 0.490 | 0.449 | 0.262 | 0.988 | 0.552 | 0.539 | 0.184 | 1.776 | 0.424 | 0.348 | 0.336 | 1.284 |
| SVM2 | 0.539 | 0.482 | 0.253 | 0.958 | 0.315 | 0.241 | −0.289 | 2.279 | 0.493 | 0.413 | 0.388 | 1.218 |
| SVM3 | 0.031 | −0.063 | 0.137 | 1.373 | 0.012 | −0.192 | 1.178 | 2.857 | 0.078 | −0.058 | 0.583 | 1.636 |
| SVM4 | 0.244 | 0.133 | 0.432 | 1.240 | 0.708 | 0.701 | 0.052 | 1.430 | 0.446 | 0.325 | 0.468 | 1.307 |
| SVM5 | 0.737 | 0.697 | 0.202 | 0.733 | 0.642 | 0.640 | −0.051 | 1.569 | 0.630 | 0.564 | 0.318 | 1.050 |
| SVM6 | 0.795 | 0.755 | 0.212 | 0.659 | 0.737 | 0.737 | 0.023 | 1.343 | 0.666 | 0.597 | 0.355 | 1.010 |
| SVM7 | 0.818 | 0.768 | 0.258 | 0.641 | 0.811 | 0.809 | −0.089 | 1.142 | 0.707 | 0.636 | 0.351 | 0.959 |
| MLR1 | 0.408 | 0.305 | 0.404 | 1.109 | 0.307 | 0.294 | 0.282 | 2.196 | 0.298 | 0.254 | 0.237 | 1.372 |
| MLR2 | 0.524 | 0.467 | 0.182 | 0.972 | 0.345 | 0.342 | −0.096 | 2.120 | 0.496 | 0.455 | 0.289 | 1.173 |
| MLR3 | 0.057 | 0.049 | 0.062 | 1.298 | 0.011 | 0.008 | 0.132 | 2.603 | 0.052 | 0.025 | 0.233 | 1.569 |
| MLR4 | 0.273 | 0.201 | 0.310 | 1.190 | 0.495 | 0.481 | 0.018 | 1.883 | 0.349 | 0.292 | 0.309 | 1.337 |
| MLR5 | 0.557 | 0.491 | 0.234 | 0.949 | 0.379 | 0.378 | −0.016 | 2.060 | 0.503 | 0.464 | 0.278 | 1.163 |
| MLR6 | 0.557 | 0.492 | 0.235 | 0.949 | 0.443 | 0.443 | 0.054 | 1.951 | 0.518 | 0.475 | 0.293 | 1.151 |
| MLR7 | 0.560 | 0.493 | 0.231 | 0.948 | 0.598 | 0.576 | −0.267 | 1.703 | 0.548 | 0.499 | 0.319 | 1.124 |
Table 9.
Comparisons of different models with four input variables for predicting hourly gross primary productivity (GPP, g C m−2 hour−1), ecosystem respiration (R, g C m−2 hour−1) and net ecosystem exchange (NEE, g C m−2 hour−1) in the testing period in the three boreal forest stands.
Table 9.
Comparisons of different models with four input variables for predicting hourly gross primary productivity (GPP, g C m−2 hour−1), ecosystem respiration (R, g C m−2 hour−1) and net ecosystem exchange (NEE, g C m−2 hour−1) in the testing period in the three boreal forest stands.
| Site | Model | GPP | R | NEE |
|---|
| R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE |
|---|
| CA-Obs | ANN | 0.932 | 0.910 | −0.015 | 0.056 | 0.843 | 0.832 | 0.001 | 0.036 | 0.850 | 0.821 | 0.013 | 0.060 |
| GRNN | 0.924 | 0.905 | −0.009 | 0.057 | 0.827 | 0.577 | −0.040 | 0.057 | 0.819 | 0.789 | 0.015 | 0.065 |
| ANFIS | 0.914 | 0.890 | −0.016 | 0.061 | 0.838 | 0.823 | −0.002 | 0.037 | 0.844 | 0.811 | 0.013 | 0.061 |
| SVM | 0.925 | 0.904 | −0.015 | 0.057 | 0.833 | 0.826 | 0.001 | 0.037 | 0.842 | 0.814 | 0.011 | 0.061 |
| MLR | 0.690 | 0.671 | −0.008 | 0.106 | 0.723 | 0.714 | 0.001 | 0.047 | 0.544 | 0.526 | 0.008 | 0.097 |
| CA-Oas | ANN | 0.893 | 0.893 | 0.006 | 0.094 | 0.767 | 0.766 | 0.001 | 0.049 | 0.834 | 0.832 | −0.008 | 0.096 |
| GRNN | 0.885 | 0.877 | −0.024 | 0.101 | 0.755 | 0.729 | −0.016 | 0.053 | 0.828 | 0.828 | 0.001 | 0.097 |
| ANFIS | 0.886 | 0.886 | 0.005 | 0.097 | 0.769 | 0.768 | 0.001 | 0.049 | 0.819 | 0.819 | −0.004 | 0.100 |
| SVM | 0.893 | 0.892 | 0.006 | 0.095 | 0.766 | 0.765 | 0.002 | 0.049 | 0.834 | 0.832 | −0.008 | 0.096 |
| MLR | 0.606 | 0.601 | 0.017 | 0.182 | 0.737 | 0.730 | 0.005 | 0.053 | 0.460 | 0.457 | −0.012 | 0.173 |
| CA-Gro | ANN | 0.859 | 0.844 | 0.016 | 0.088 | 0.887 | 0.563 | 0.030 | 0.051 | 0.810 | 0.803 | 0.013 | 0.087 |
| GRNN | 0.850 | 0.728 | 0.077 | 0.116 | 0.884 | 0.399 | -0.085 | 0.091 | 0.800 | 0.715 | 0.057 | 0.104 |
| ANFIS | 0.846 | 0.829 | 0.019 | 0.092 | 0.889 | 0.587 | 0.029 | 0.049 | 0.794 | 0.789 | 0.010 | 0.090 |
| SVM | 0.853 | 0.838 | 0.015 | 0.090 | 0.883 | 0.572 | 0.028 | 0.050 | 0.803 | 0.796 | 0.013 | 0.088 |
| MLR | 0.634 | 0.621 | 0.014 | 0.137 | 0.884 | 0.591 | 0.028 | 0.049 | 0.531 | 0.526 | 0.014 | 0.134 |
Table 10.
Comparisons of different models with four input variables for predicting hourly net ecosystem exchange (NEE, g C m−2 hour−1) in the testing period for daytime and nighttime datasets in the three boreal forest stands.
Table 10.
Comparisons of different models with four input variables for predicting hourly net ecosystem exchange (NEE, g C m−2 hour−1) in the testing period for daytime and nighttime datasets in the three boreal forest stands.
| Model | Dataset | CA-Obs | CA-Oas | CA-Gro |
|---|
| R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE | R2 | NSE | Bias | RMSE |
|---|
| ANN | Daytime | 0.847 | 0.809 | 0.023 | 0.076 | 0.822 | 0.820 | −0.010 | 0.121 | 0.765 | 0.761 | 0.012 | 0.111 |
| Nighttime | 0.566 | 0.533 | 0.004 | 0.041 | 0.590 | 0.580 | −0.005 | 0.052 | 0.799 | 0.674 | 0.015 | 0.041 |
| GRNN | Daytime | 0.824 | 0.784 | 0.024 | 0.081 | 0.814 | 0.814 | 0.003 | 0.123 | 0.750 | 0.677 | 0.061 | 0.129 |
| Nighttime | 0.400 | 0.385 | 0.006 | 0.047 | 0.601 | 0.589 | −0.002 | 0.051 | 0.803 | 0.251 | 0.052 | 0.062 |
| ANFIS | Daytime | 0.841 | 0.799 | 0.025 | 0.078 | 0.814 | 0.814 | −0.005 | 0.123 | 0.753 | 0.751 | 0.008 | 0.113 |
| Nighttime | 0.544 | 0.511 | 0.003 | 0.042 | 0.419 | 0.414 | −0.003 | 0.061 | 0.684 | 0.568 | 0.012 | 0.047 |
| SVM | Daytime | 0.844 | 0.807 | 0.022 | 0.077 | 0.822 | 0.820 | −0.007 | 0.121 | 0.757 | 0.753 | 0.012 | 0.113 |
| Nighttime | 0.496 | 0.476 | 0.002 | 0.043 | 0.582 | 0.569 | −0.008 | 0.053 | 0.784 | 0.645 | 0.015 | 0.043 |
| MLR | Daytime | 0.483 | 0.456 | 0.021 | 0.129 | 0.453 | 0.443 | −0.014 | 0.213 | 0.457 | 0.451 | 0.017 | 0.169 |
| Nighttime | 0.073 | 0.061 | −0.003 | 0.058 | 0.121 | 0.176 | −0.010 | 0.107 | 0.012 | 0.105 | 0.010 | 0.075 |