LMP1 and Dynamic Progressive Telomere Dysfunction: A Major Culprit in EBV-Associated Hodgkin’s Lymphoma
Abstract
:1. Introduction
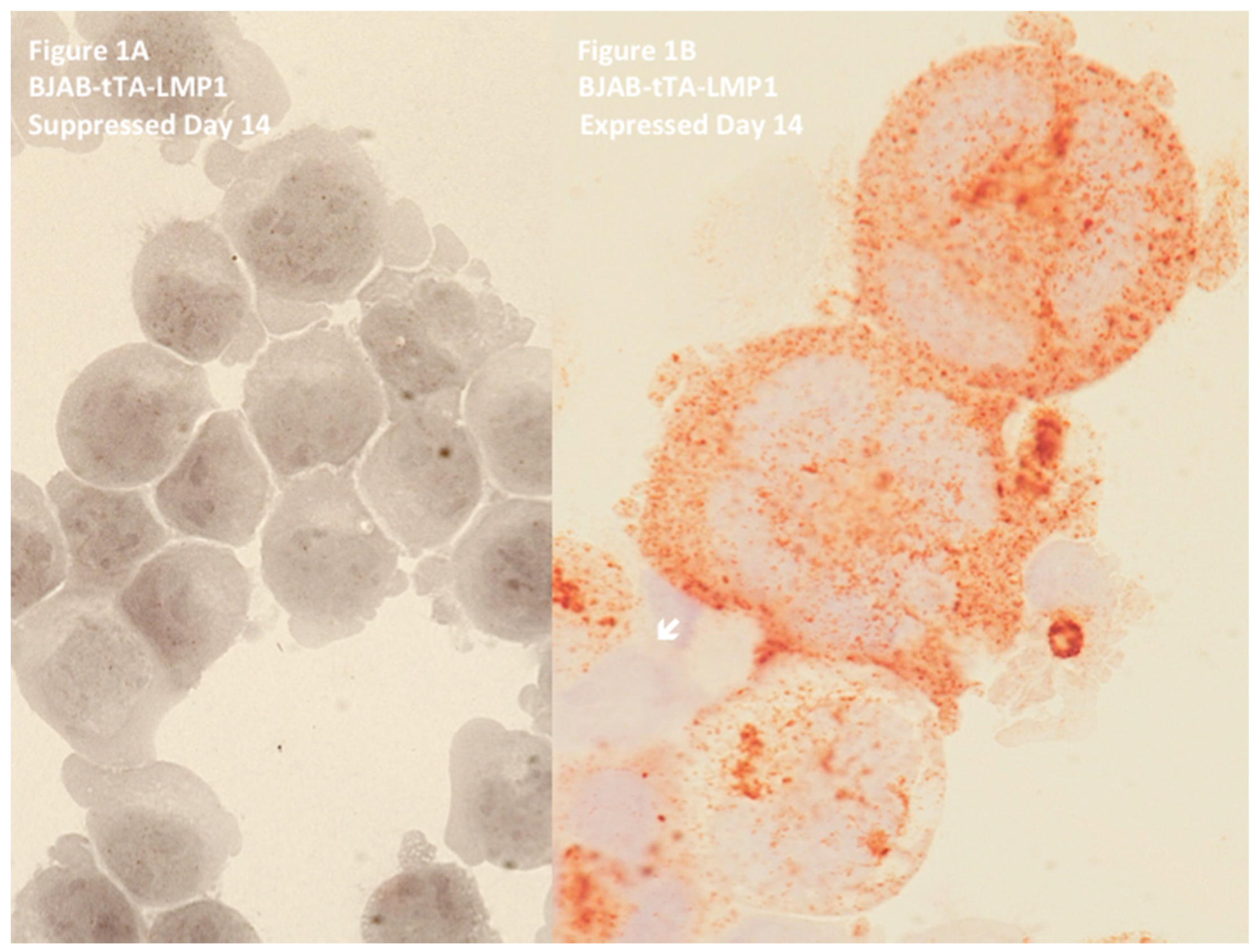
2. Latent Membrane Protein 1 (LMP1) Induces Multinuclearity
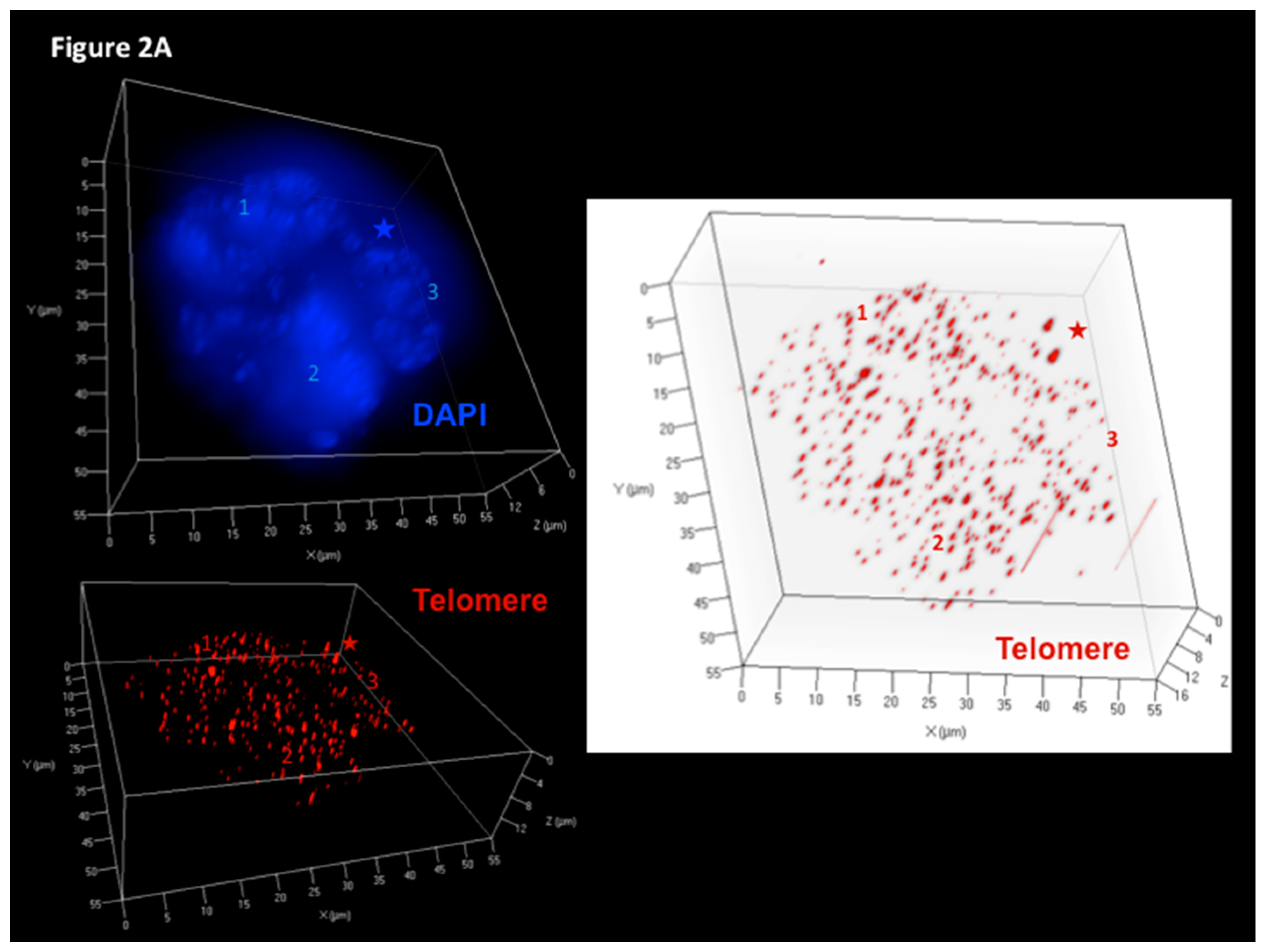
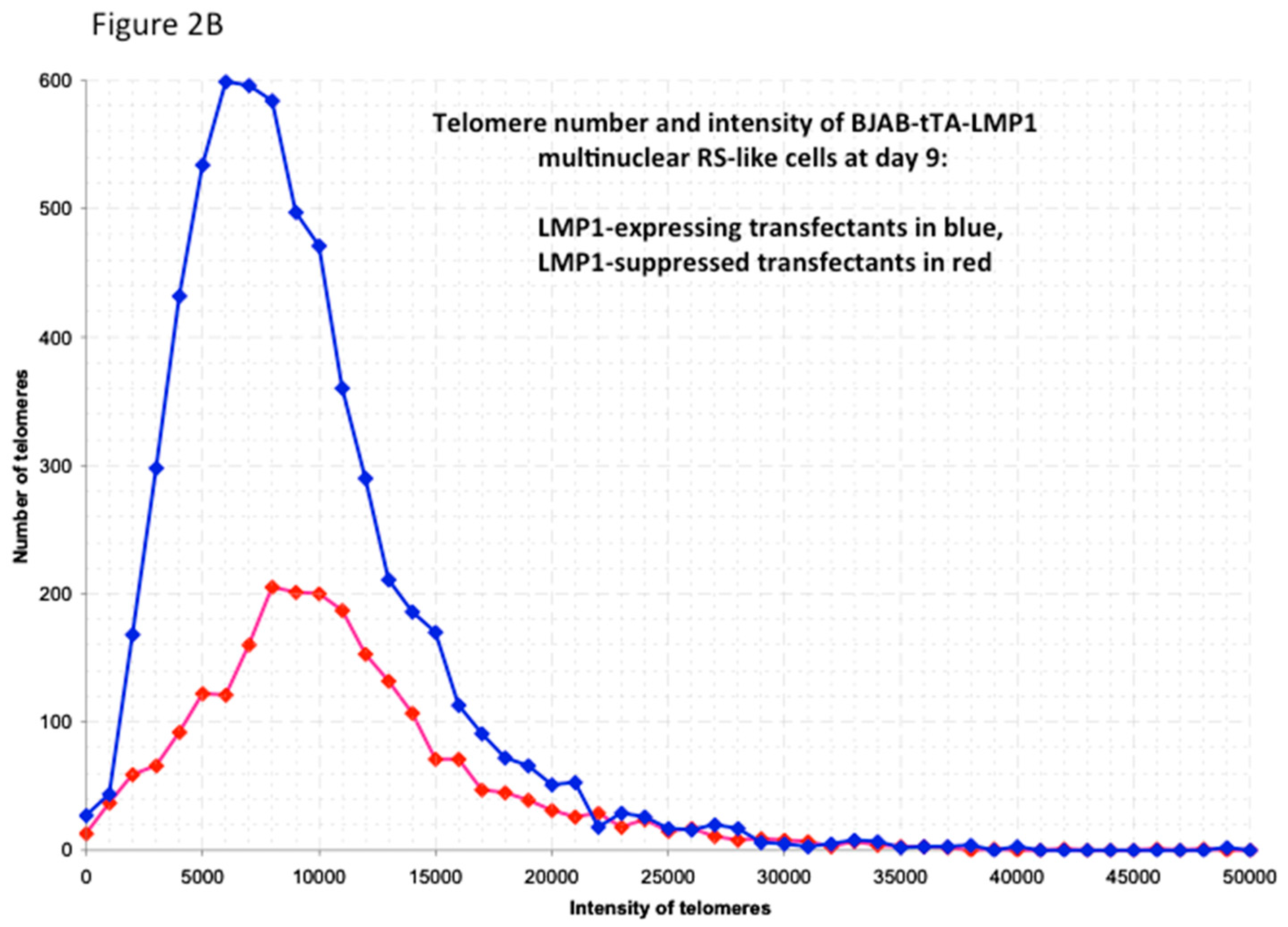
3. Assessment of 3D Telomere Dynamics
4. In Vitro Model for EBV-Associated Classical Hodgkin’s Lymphoma
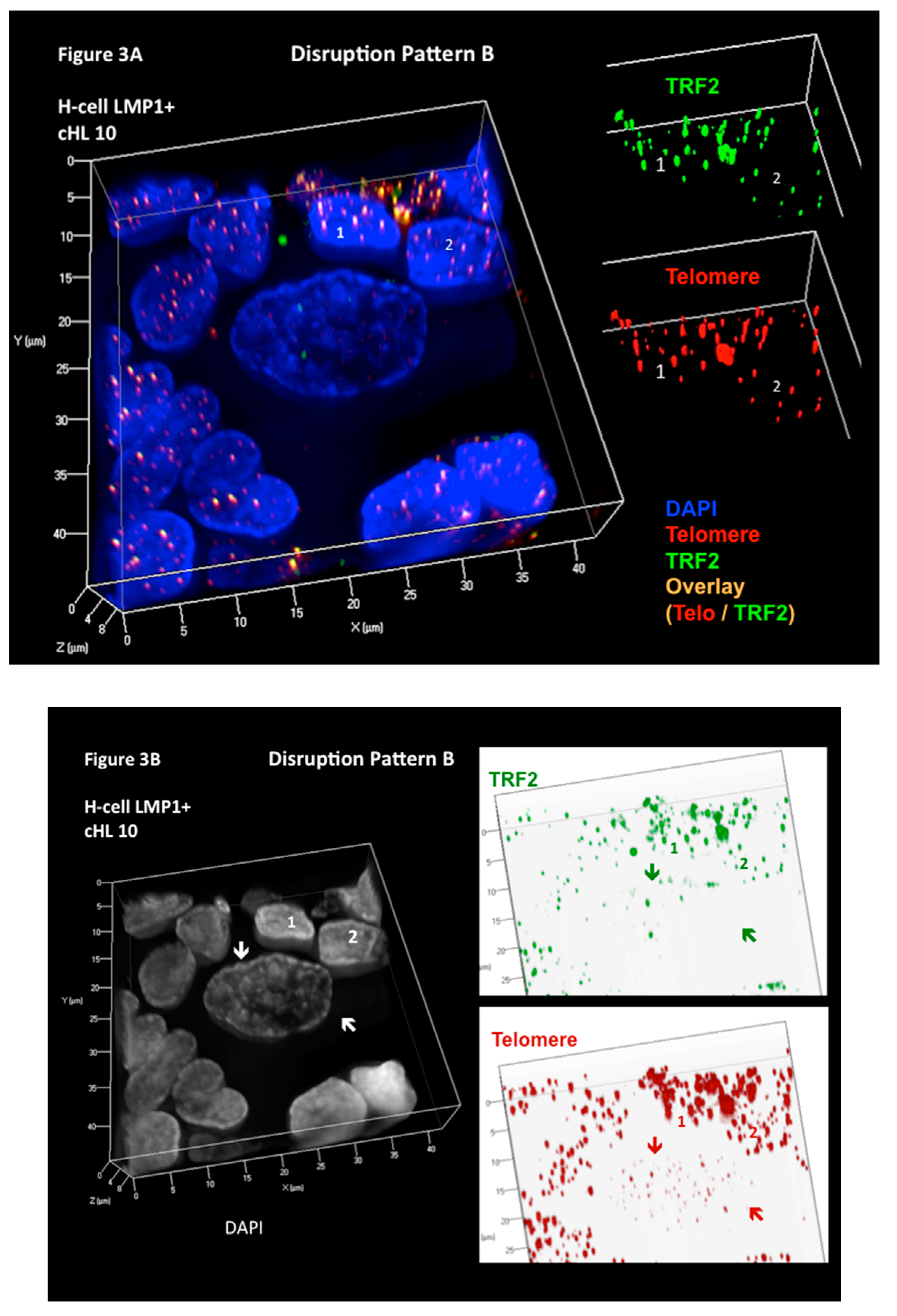
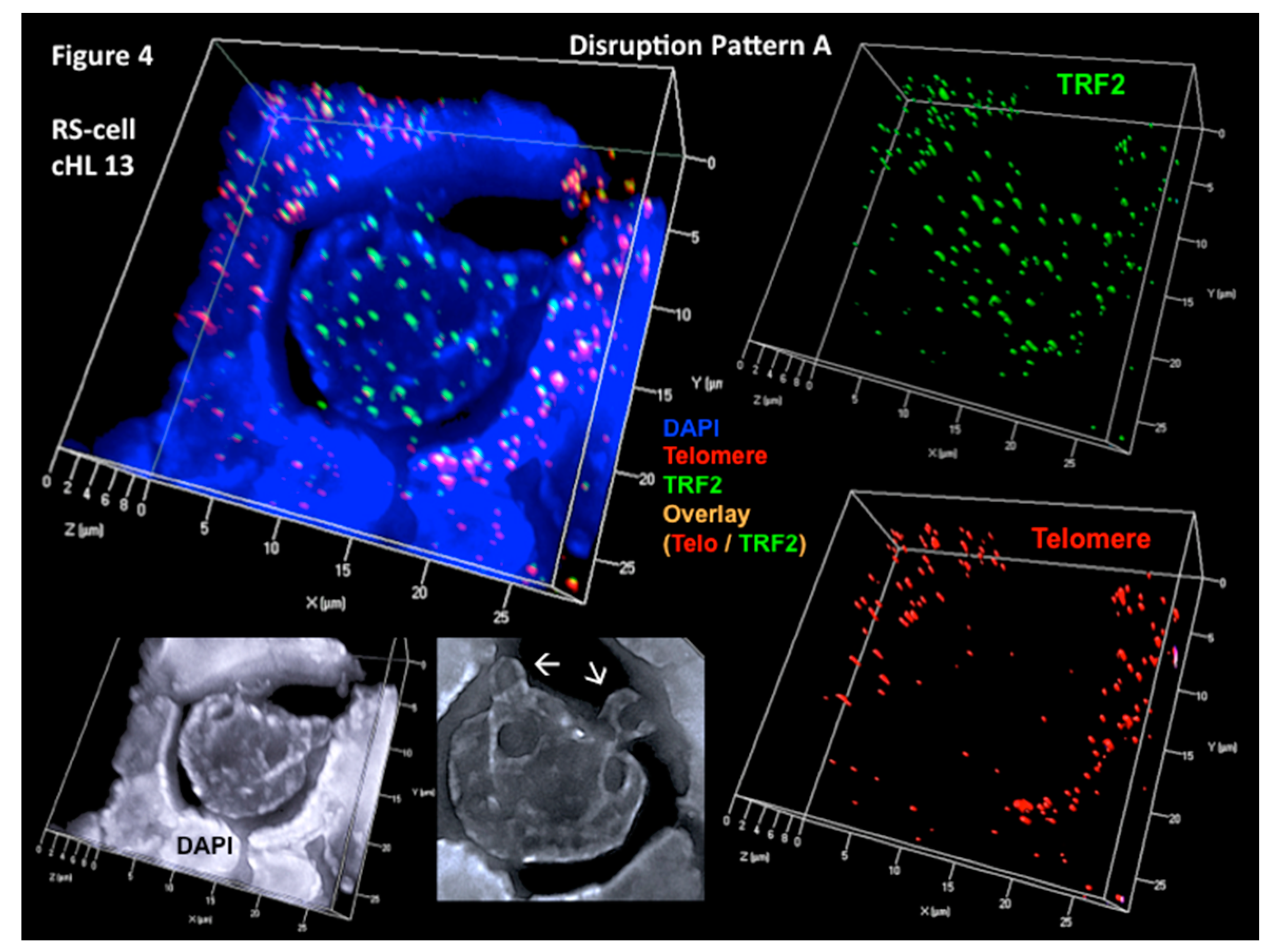
5. Combined 3D Immuno TRF2/Telo-Q-FISH of Primary H and RS Cells
6. Summary
Author Contributions
Conflicts of Interest
References
- Fennewald, S.; van Santen, V.; Kieff, E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J. Virol. 1984, 51, 411–419. [Google Scholar] [PubMed]
- Wang, D.; Liebowitz, D.; Kieff, E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 1985, 43, 831–840. [Google Scholar] [CrossRef]
- Knecht, H.; Berger, C.; Rothenberger, S.; Odermatt, B.F.; Brousset, P. The role of Epstein-Barr virus in neoplastic transformation. Oncology 2001, 60, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Kieser, A.; Sterz, K.R. The Latent Membrane Protein 1 (LMP1). Curr. Top. Microbiol. Immunol. 2015, 391, 119–149. [Google Scholar] [PubMed]
- Minarovits, J.; Niller, H.H. Current Trends and Alternative Scenarios in EBV Research. Methods Mol. Biol. 2017, 1532, 1–32. [Google Scholar] [PubMed]
- Deng, Z.; Lezina, L.; Chen, C.J.; Shtivelband, S.; So, W.; Lieberman, P.M. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 2002, 9, 493–503. [Google Scholar] [CrossRef]
- Deng, Z.; Atanasiu, C.; Burg, J.S.; Broccoli, D.; Lieberman, P.M. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 2003, 77, 11992–12001. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Wang, Z.; Lieberman, P.M. Telomeres and viruses: Common themes of genome maintenance. Front. Oncol. 2012, 2, 201. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; Righolt, C.; Mai, S. Genomic instability: The driving force behind refractory/relapsing Hodgkin’s lymphoma. Cancers 2013, 5, 714–725. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Hug, N.; Lingner, J. Telomere length homeostasis. Chromosoma 2006, 115, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Londoño-Vallejo, J.A.; Wellinger, R.J. Telomeres and telomerase dance to the rhythm of the cell cycle. Trends Biochem. Sci. 2012, 37, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Armanios, M.; Blackburn, E.H. The telomere syndromes. Nat. Rev. Genet. 2012, 13, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.; Choi, J.W.; Kim, Y.S. Prevalence and prognostic significance of Epstein-Barr virus infection in classical Hodgkin’s lymphoma: A meta-analysis. Arch. Med. Res. 2014, 45, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Claviez, A.; Tiemann, M.; Lüders, H.; Krams, M.; Parwaresch, R.; Schellong, G.; Dörffel, W. Impact of latent Epstein-Barr virus infection on outcome in children and adolescents with Hodgkin’s lymphoma. J. Clin. Oncol. 2005, 23, 4048–4056. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.A.; Glaser, S.L.; Dorfman, R.F.; Mann, R.; DiGiuseppe, J.A.; Prehn, A.W.; Ambinder, R.F. Epstein-Barr virus and survival after Hodgkin disease in a population-based series of women. Cancer 2001, 91, 1579–1587. [Google Scholar] [CrossRef]
- Schwarzer, R.; Jundt, F. Notch and NF-κB signaling pathways in the biology of classical Hodgkin lymphoma. Curr. Mol. Med. 2011, 11, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, E.; Younes, A. Lymphomagenesis in Hodgkin lymphoma. Semin. Cancer Biol. 2015, 34, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, K. Human B-lymphoid cell lines. Hum. Cell 1992, 5, 25–41. [Google Scholar] [PubMed]
- Satoh, M.; Yasuda, T.; Higaki, T.; Goto, M.; Tanuma, S.; Ide, T.; Furuichi, Y.; Sugimoto, M. Innate apoptosis of human B lymphoblasts transformed by Epstein-Barr virus: Modulation by cellular immortalization and senescence. Cell Struct. Funct. 2003, 28, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Schmidt, S.C.; Jiang, S.; Willox, B.; Bernhardt, K.; Liang, J.; Johannsen, E.C.; Kharchenko, P.; Gewurz, B.E.; Kieff, E.; et al. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 2015, 17, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Küppers, R. New insights in the biology of Hodgkin lymphoma. Hematol. Am. Soc. Hematol. Educ. Progr. 2012, 2012, 328–334. [Google Scholar]
- Knecht, H.; Brousset, P.; Bachmann, E.; Sandvej, K.; Odermatt, B.F. Latent membrane protein 1: A key oncogene in EBV-related carcinogenesis? Acta Haematol. 1993, 90, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Louissaint, A., Jr.; Ferry, J.A.; Soupir, C.P.; Hasserjian, R.P.; Harris, N.L.; Zukerberg, L.R. Infectious mononucleosis mimicking lymphoma: Distinguishing morphological and immunophenotypic features. Mod. Pathol. 2012, 25, 1149–1159. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; McQuain, C.; Martin, J.; Rothenberger, S.; Drexler, H.G.; Berger, C.; Bachmann, E.; Kittler, E.L.; Odermatt, B.F.; Quesenberry, P.J. Expression of the LMP1 oncoprotein in the EBV negative Hodgkin’s disease cell line L-428 is associated with Reed-Sternberg cell morphology. Oncogene 1996, 13, 947–953. [Google Scholar] [PubMed]
- Knecht, H.; Berger, C.; McQuain, C.; Rothenberger, S.; Bachmann, E.; Martin, J.; Esslinger, C.; Drexler, H.G.; Cai, Y.C.; Quesenberry, P.J.; et al. Latent membrane protein 1 associated signaling pathways are important in tumor cells of Epstein-Barr virus negative Hodgkin’s disease. Oncogene 1999, 18, 7161–7167. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, R.A.; Spitzer, D.; Bar-Am, I.; Sylvester, J.E.; Kaufmann, M.; Wernich, A.; Drexler, H.G. Karyotypic dissection of Hodgkin’s disease cell lines reveals ectopic subtelomeres and ribosomal DNA at sites of multiple jumping translocations and genomic amplification. Leukemia 2000, 14, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Pihan, G.A.; Purohit, A.; Wallace, J.; Knecht, H.; Woda, B.; Quesenberry, P.; Doxsey, S.J. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998, 58, 3974–3985. [Google Scholar] [PubMed]
- Chuang, T.C.; Moshir, S.; Garini, Y.; Chuang, A.Y.; Young, I.T.; Vermolen, B.; van den Doel, R.; Mougey, V.; Perrin, M.; Braun, M.; et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Vermolen, B.J.; Garini, Y.; Mai, S.; Mougey, V.; Fest, T.; Chuang, T.C.; Chuang, A.Y.; Wark, L.; Young, I.T. Characterizing the three-dimensional organization of telomeres. Cytometry Part A 2005, 67, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Mai, S.; Garini, Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J. Cell Biochem. 2006, 97, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Louis, S.F.; Vermolen, B.J.; Garini, Y.; Young, I.T.; Guffei, A.; Lichtensztejn, Z.; Kuttler, F.; Chuang, T.C.; Moshir, S.; Mougey, V.; et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc. Natl. Acad. Sci. USA 2005, 102, 9613–9618. [Google Scholar] [CrossRef] [PubMed]
- Schermelleh, L.; Carlton, P.M.; Haase, S.; Shao, L.; Winoto, L.; Kner, P.; Burke, B.; Cardoso, M.C.; Agard, D.A.; Gustafsson, M.G.; et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 2008, 320, 1332–1336. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Blackburn, E.H. Human cancer cells harbour T-stumps, a distinct class of extremely short telomeres. Mol. Cell 2007, 28, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Lacoste, S.; Wiechec, E.; dos Santos Silva, A.G.; Guffei, A.; Williams, G.; Lowbeer, M.; Benedek, K.; Henriksson, M.; Klein, G.; Mai, S. Chromosomal rearrangements after ex vivo Epstein-Barr virus (EBV) infection of human B cells. Oncogene 2010, 29, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Kamranvar, S.A.; Masucci, M.G. The Epstein-Barr virus nuclear antigen-1 promotes telomere dysfunction via induction of oxidative stress. Leukemia 2011, 25, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Kamranvar, S.A.; Chen, X.; Masucci, M.G. Telomere dysfunction and activation of alternative lengthening of telomeres in B-lymphocytes infected by Epstein-Barr virus. Oncogene 2013, 32, 5522–5530. [Google Scholar] [CrossRef] [PubMed]
- Pallesen, G.; Hamilton-Dutoit, S.J.; Rowe, M.; Young, L.S. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin’s disease. Lancet 1991, 337, 320–322. [Google Scholar] [CrossRef]
- Knecht, H.; Bachmann, E.; Brousset, P.; Sandvej, K.; Nadal, D.; Bachmann, F.; Odermatt, B.F.; Delsol, G.; Pallesen, G. Deletions within the LMP1 oncogene of Epstein-Barr virus are clustered in Hodgkin’s disease and identical to those observed in nasopharyngeal carcinoma. Blood 1993, 82, 2937–2942. [Google Scholar] [PubMed]
- Thorley-Lawson, D.; Gross, A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 2004, 350, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Floettmann, J.E.; Ward, K.; Rickinson, A.B.; Rowe, M. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology 1996, 223, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Glazer, P.M.; Summers, W.C. Oncogene expression in isogenic, EBV-positive and -negative Burkitt lymphoma cell lines. Intervirology 1985, 23, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Wennborg, A.; Aman, P.; Saranath, D.; Pear, W.; Sümegi, J.; Klein, G. Conversion of the lymphoma line “BJAB” by Epstein-Barr virus into phenotypically altered sublines is accompanied by increased c-myc mRNA levels. Int. J. Cancer 1987, 40, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Kalaitzidis, D.; Davis, R.E.; Rosenwald, A.; Staudt, L.M.; Gilmore, T.D. The human B-cell lymphoma cell line RC-K8 has multiple genetic alterations that dysregulate the Rel/NF-κB signal transduction pathway. Oncogene 2002, 21, 8759–8768. [Google Scholar] [CrossRef] [PubMed]
- Ngo, V.N.; Davis, R.E.; Lamy, L.; Yu, X.; Zhao, H.; Lenz, G.; Lam, L.T.; Dave, S.; Yang, L.; Powell, J.; et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 2006, 441, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, V.; Lemieux, B.; Sawan, B.; Lichtensztejn, D.; Lichtensztejn, Z.; Wellinger, R.; Mai, S.; Knecht, H. LMP1 mediates multinuclearity through downregulation of shelterin proteins and formation of telomeric aggregates. Blood 2015, 125, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; Sawan, B.; Lichtensztejn, Z.; Lichtensztejn, D.; Mai, S. 3D Telomere FISH defines LMP1 expressing Reed-Sternberg Cells as End-Stage Cells with Telomere-poor “Ghost” Nuclei and very short Telomeres. Lab. Investig. 2010, 90, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.; Philip, P.S. Functional analysis of exosomes derived from EBV-infected cells. Methods Mol. Biol. 2017, 1532, 159–167. [Google Scholar] [PubMed]
- Schröck, E.; du Manoir, S.; Veldman, T.; Schoell, B.; Wienberg, J.; Ferguson-Smith, M.A.; Ning, Y.; Ledbetter, D.H.; Bar-Am, I.; Soenksen, D.; et al. Multicolor spectral karyotyping of human chromosomes. Science 1996, 273, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Guffei, A.; Sarkar, R.; Klewes, L.; Righolt, C.; Knecht, H.; Mai, S. Dynamic chromosomal rearrangements in Hodgkin’s lymphoma are due to ongoing three-dimensional nuclear remodeling and breakage-bridge-fusion cycles. Haematologica 2010, 95, 2038–2046. [Google Scholar] [CrossRef] [PubMed]
- Feuerhahn, S.; Chen, L.Y.; Luke, B.; Porro, A. No DDRama at chromosome ends: TRF2 takes centre stage. Trends Biochem. Sci. 2015, 40, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; Mai, S. The use of 3D telomere FISH for the characterization of the nuclear architecture in EBV-positive Hodgkin’s lymphoma. Methods Mol. Biol. 2017, 1532, 93–104. [Google Scholar] [PubMed]
- Knecht, H.; Johnson, N.A.; Haliotis, T.; Lichtensztejn, D.; Mai, S. Disruption of direct 3D Telomere-TRF2 interaction through two molecularly disparate mechanisms is a hallmark of primary Hodgkin and Reed-Sternberg cells. Lab. Investig. 2017. [Google Scholar] [CrossRef] [PubMed]
- Righolt, C.; Guffei, A.; Knecht, H.; Young, I.T.; Stallinga, S.; van Vliet, L.J.; Mai, S. Differences in nuclear DNA organization between lymphocytes, Hodgkin and Reed-Sternberg cells revealed by structured illumination microscopy. J. Cell Biochem. 2014, 115, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Celli, G.B.; de Lange, T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005, 7, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Denchi, E.L.; de Lange, T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Lazzerini Denchi, E.; Celli, G.; De Lange, T. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev. 2006, 20, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; Denchi, E.L.; de Lange, T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 2010, 141, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; Sawan, B.; Lichtensztejn, D.; Lemieux, B.; Wellinger, R.; Mai, S. The 3D nuclear organization of telomeres marks the transition from Hodgkin to Reed-Sternberg cells. Leukemia 2009, 23, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Benarroch-Popivker, D.; Pisano, S.; Mendez-Bermudez, A.; Lototska, L.; Kaur, P.; Bauwens, S.; Djerbi, N.; Latrick, C.M.; Fraisier, V.; Pei, B.; et al. TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Mol. Cell 2016, 61, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.M.; Rendtlew Danielsen, J.M.; Lucas, C.A.; Rice, E.L.; Scalzo, D.; Shimi, T.; Goldman, R.D.; Smith, E.D.; Le Beau, M.M.; Kosak, S.T. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun. 2014, 5, 5467. [Google Scholar] [CrossRef] [PubMed]
- Bronshtein, I.; Kepten, E.; Kanter, I.; Berezin, S.; Lindner, M.; Redwood, A.B.; Mai, S.; Gonzalo, S.; Foisner, R.; Shav-Tal, Y.; et al. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat. Commun. 2015, 6, 8044. [Google Scholar] [CrossRef] [PubMed]
- Nera, B.; Huang, H.S.; Lai, T.; Xu, L. Elevated levels of TRF2 induce telomeric ultrafine anaphase bridges and rapid telomere deletions. Nat. Commun. 2015, 6, 10132. [Google Scholar] [CrossRef] [PubMed]
- Xavier de Carvalho, A.; Maiato, H.; Maia, A.F.; Ribeiro, S.A.; Pontes, P.; Bickmore, W.; Earnshaw, W.C.; Sambade, C. Reed-Sternberg cells form by abscission failure in the presence of functional Aurora B kinase. PLoS ONE 2015, 10, e0124629. [Google Scholar] [CrossRef] [PubMed]
- Newcom, S.R.; Kadin, M.E.; Phillips, C. L-428 Reed-Sternberg cells and mononuclear Hodgkin’s cells arise from a single cloned mononuclear cell. Int. J. Cell Cloning 1988, 6, 417–431. [Google Scholar] [CrossRef] [PubMed]
- Küppers, R.; Bräuninger, A.; Müschen, M.; Distler, V.; Hansmann, M.L.; Rajewsky, K. Evidence that Hodgkin and Reed-Sternberg cells in Hodgkin disease do not represent cell fusions. Blood 2001, 97, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Benharroch, D.; Einav, I.; Feldman, A.; Levy, A.; Ariad, S.; Gopas, J. Apoptosis of Hodgkin-Reed-Sternberg cells in classical Hodgkin lymphoma revisited. APMIS 2010, 118, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.; Miralles Fusté, J.; Simavorian, T.; Bartocci, C.; Tsai, J.; Karlseder, J.; Lazzerini Denchi, E. TZAP: A telomere-associated protein involved in telomere length control. Science 2017, 355, 638–641. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knecht, H.; Mai, S. LMP1 and Dynamic Progressive Telomere Dysfunction: A Major Culprit in EBV-Associated Hodgkin’s Lymphoma. Viruses 2017, 9, 164. https://doi.org/10.3390/v9070164
Knecht H, Mai S. LMP1 and Dynamic Progressive Telomere Dysfunction: A Major Culprit in EBV-Associated Hodgkin’s Lymphoma. Viruses. 2017; 9(7):164. https://doi.org/10.3390/v9070164
Chicago/Turabian StyleKnecht, Hans, and Sabine Mai. 2017. "LMP1 and Dynamic Progressive Telomere Dysfunction: A Major Culprit in EBV-Associated Hodgkin’s Lymphoma" Viruses 9, no. 7: 164. https://doi.org/10.3390/v9070164





