Seasonal Dynamics of Haptophytes and dsDNA Algal Viruses Suggest Complex Virus-Host Relationship
Abstract
:1. Introduction
2. Results
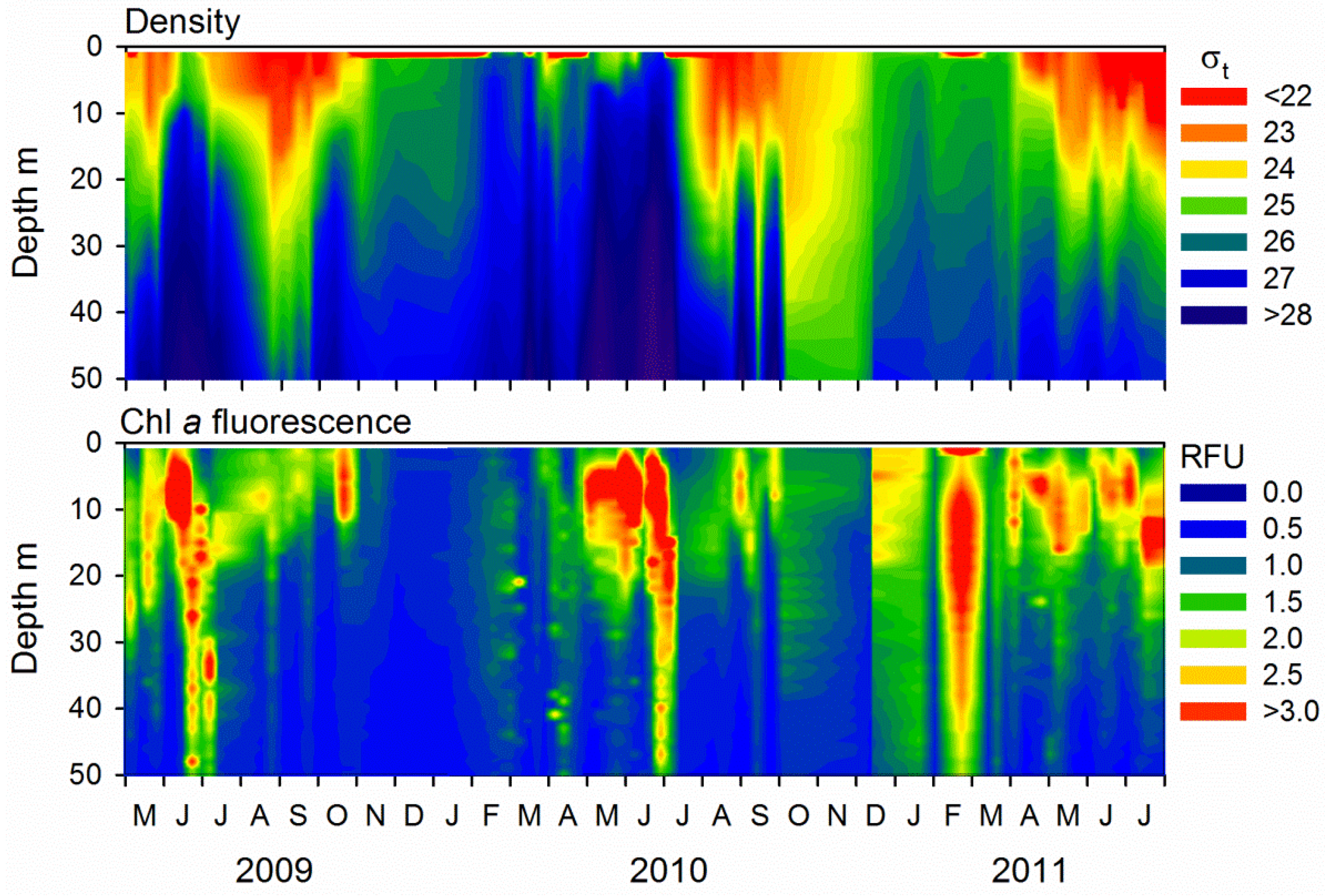
2.1. Microbial Abundance and Abiotic Factors
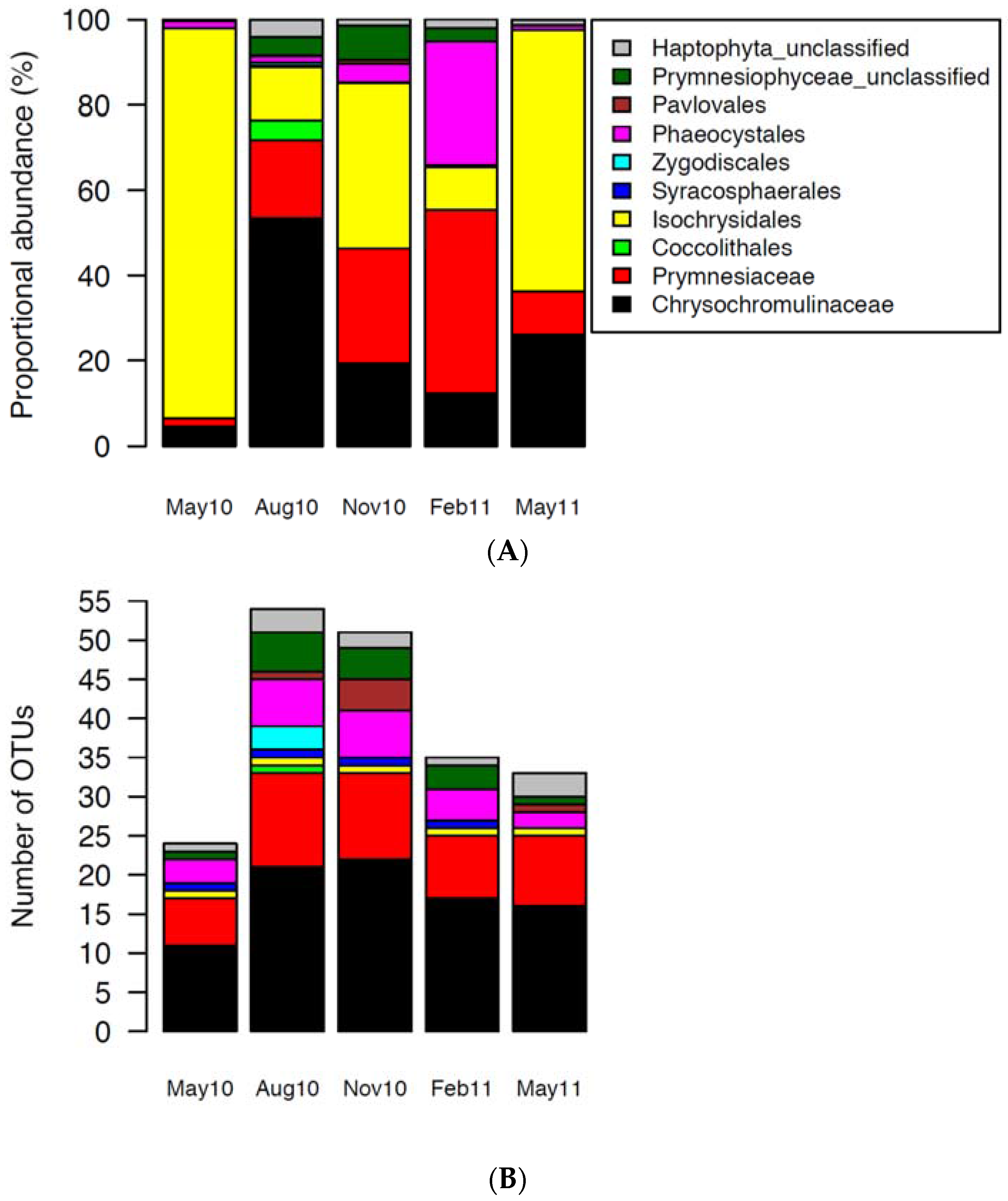
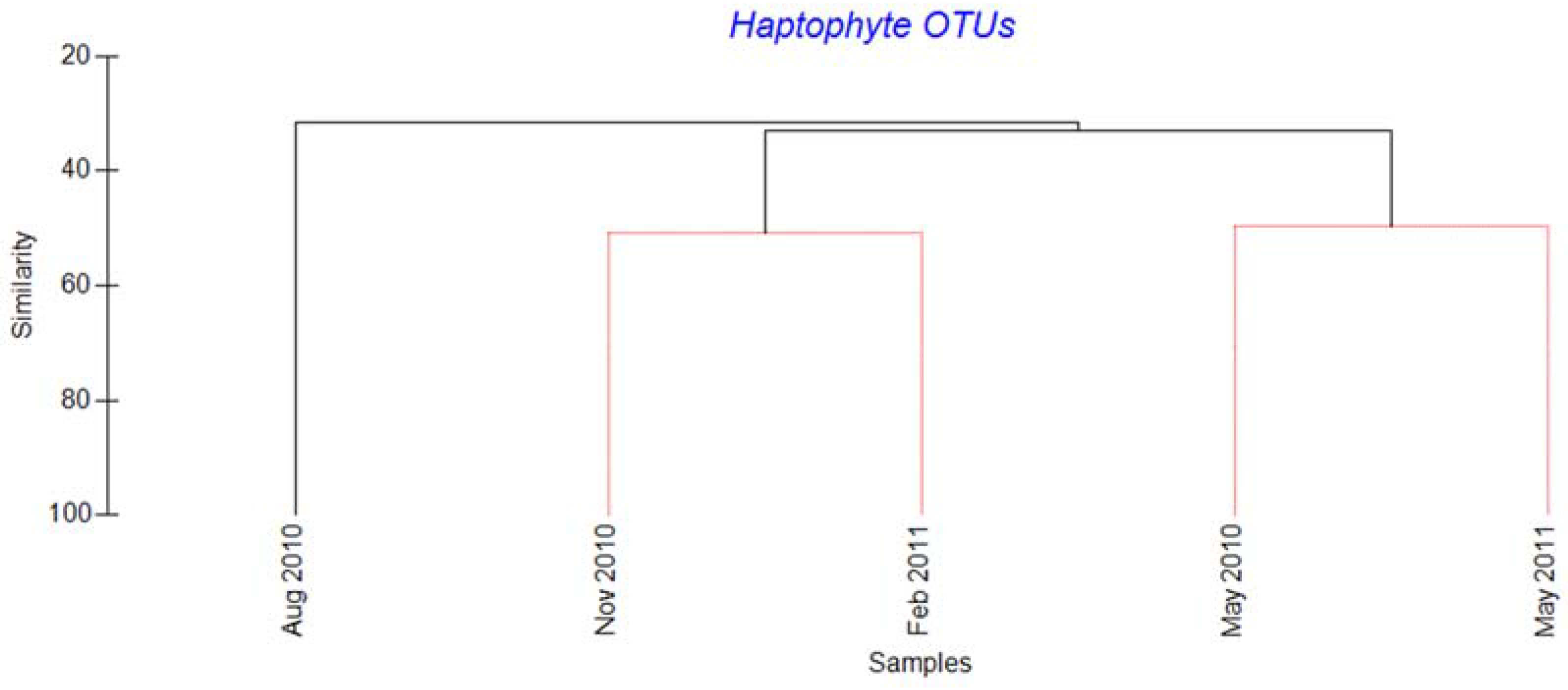
2.2. Haptophytes
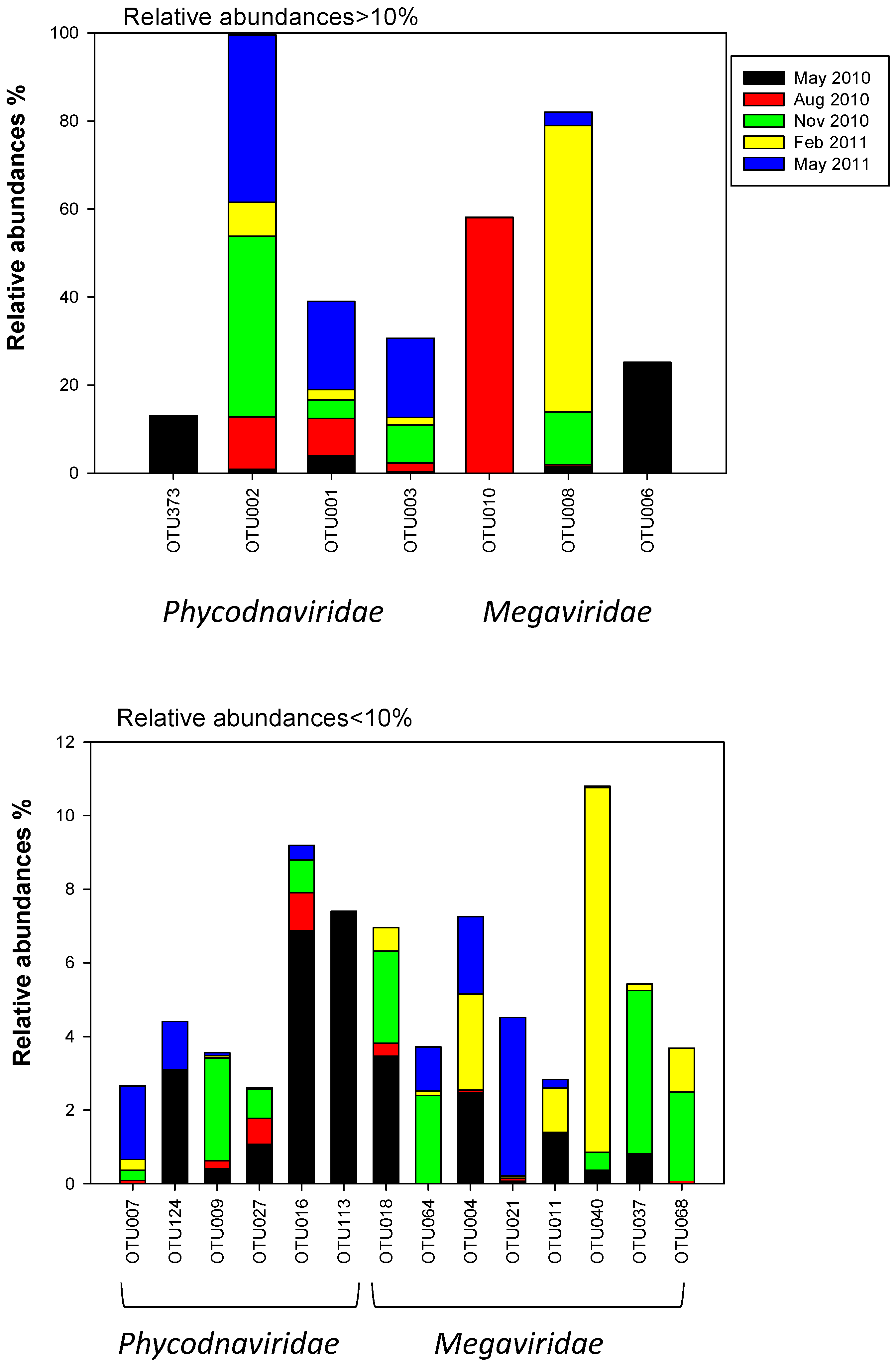
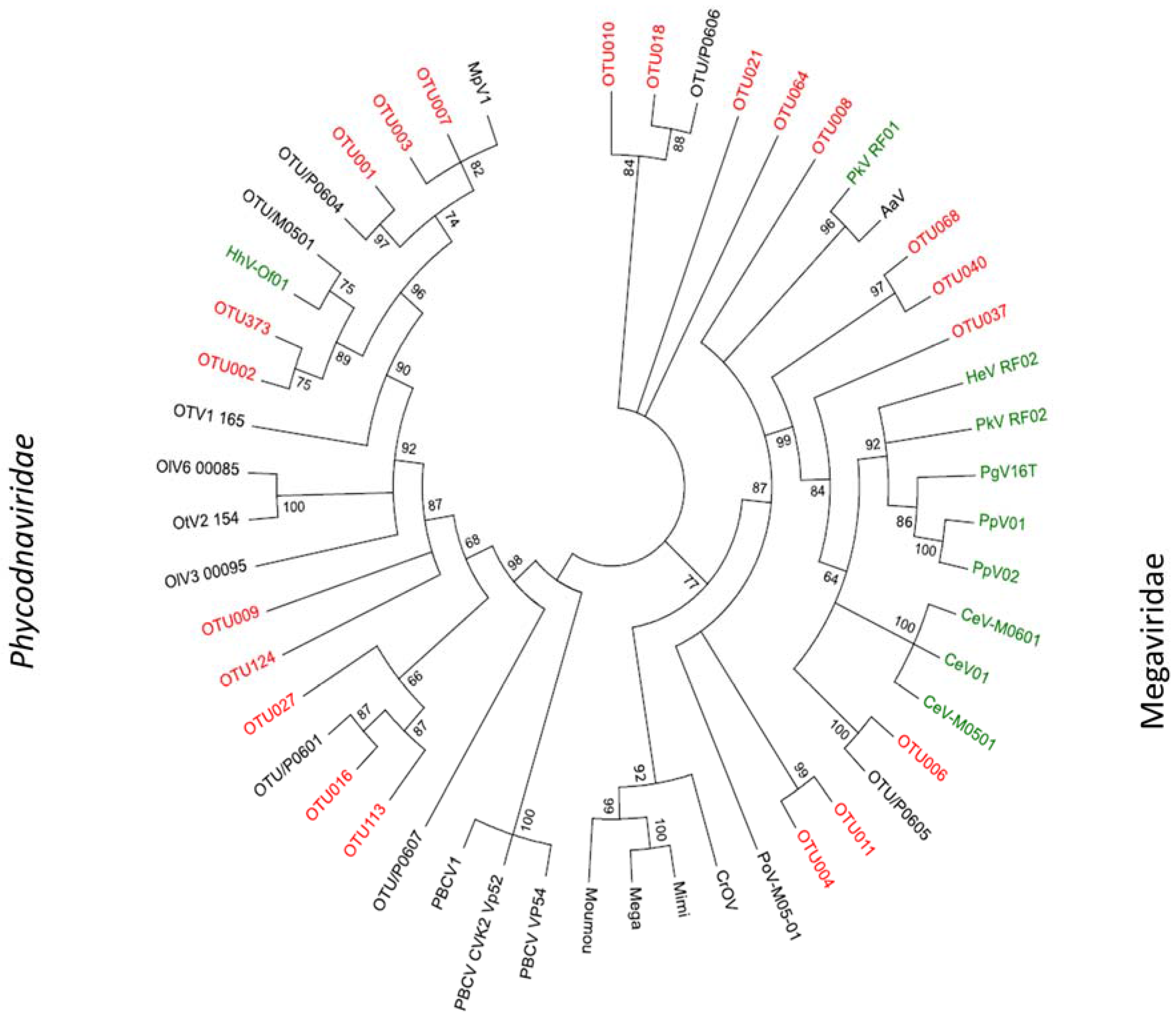
2.3. Megaviridae and Phycodnaviridae
3. Discussion
3.1. Seasonal Patterns in Microbial Dynamics and Environmental Factors
3.2. Succession of Haptophytes and Co-Occurring DNA Viruses
4. Material and Methods
4.1. Sample Collection
4.2. Microbial Abundance Measured by Flow Cytometry (FCM)
4.3. DNA Extraction, PCR and 454-Pyrosequencing
4.4. Sequence Analysis
Supplementary Materials
Data Accessibility
Acknowledgments
Author Contributions
Conflict of Interest
References
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Eikrem, E.; Medlin, L.K.; Henderiks, J.; Rokitta, S.; Rost, B.; Probert, I.; Throndsen, J.; Edvardsen, B. Haptophyta. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Margulis, L., Melkonian, M., Chapman, D.J., Corliss, J.O., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–61. [Google Scholar]
- Unrein, F.; Gasol, J.M.; Not, F.; Forn, I.; Massana, R. Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J. 2014, 8, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Hallegraeff, G.M. Ocean climate change, phytoplankton community responses, and harmful algal blooms: A formidable predictive challenge. J. Phycol. 2010, 46, 220–235. [Google Scholar] [CrossRef]
- Leadbeater, B.S.C. Identification, by means of electron microscopy, of flagellate nanoplankton from the coast of Norway. Sarsia 1972, 49, 107–124. [Google Scholar] [CrossRef]
- Thomsen, H.A.; Buck, K.R.; Chavez, F.P. Haptophytes as components of marine phytoplankton. Syst. Assoc. Spec. Vol. Ser. 1994, 51, 187–208. [Google Scholar]
- Egge, E.S.; Johannessen, T.V.; Andersen, T.; Eikrem, W.; Bittner, L.; Larsen, A.; Sandaa, R.-A.; Edvardsen, B. Seasonal diversity and dynamics of haptophytes in the Skagerrak, Norway, explored by high-throughput sequencing. Mol. Ecol. 2015, 24, 3026–3042. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.; Noble, R. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 1995, 40, 1236–1242. [Google Scholar] [CrossRef]
- Short, S.M. The ecology of viruses that infect eukaryotic algae. Environ. Microbiol. 2012, 14, 2253–2271. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, T.; Del Giorgio, P.A. Key role of selective viral-induced mortality in determining marine bacterial community composition. Envir. Microbiol. 2007, 9, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Castberg, T.; Larsen, A.; Sandaa, R.A.; Brussaard, C.P.D.; Egge, J.K.; Heldal, M.; Thyrhaug, R.; van Hannen, E.J.; Bratbak, G. Microbial population dynamics and diversity during a bloom of the marine coccolithophorid Emiliania huxleyi (Haptophyta). Mar. Ecol. Prog. Ser. 2001, 221, 39–46. [Google Scholar] [CrossRef]
- Brussaard, C.P.D.; Bratbak, G.; Baudoux, A.C.; Ruardij, P. Phaeocystis and its interaction with viruses. Biogeochemistry 2007, 83, 201–215. [Google Scholar] [CrossRef]
- Martinez, J.M.; Schroeder, D.C.; Larsen, A.; Bratbak, G.; Wilson, W.H. Molecular Dynamics of Emiliania huxleyi and Cooccurring Viruses during Two Separate Mesocosm Studies. Appl. Environ. Microbiol. 2007, 73, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Santini, S.; Jeudy, S.; Bartoli, J.; Poirot, O.; Lescot, M.; Abergel, C.; Barbe, V.; Wommack, K.E.; Noordeloos, A.A.M.; Brussaard, C.P.D.; et al. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. USA 2103, 110, 10800–10805. [Google Scholar] [CrossRef] [PubMed]
- Brussaard, C.P.D.; Kuipers, B.; Veldhuis, M.J.W. A mesocosm study of Phaeocystis globosa population dynamics: I. Regulatory role of viruses in bloom control. Harmful Algae 2005, 4, 859–874. [Google Scholar] [CrossRef]
- Tomaru, Y.; Hata, N.; Masuda, T.; Tsuji, M.; Igata, K.; Masuda, Y.; Yamatogi, T.; Sakaguchi, M.; Nagasaki, K. Ecological dynamics of the bivalve-killing dinoflagellate Heterocapsa circularisquama and its infectious viruses in different locations of western Japan. Environ. Microbiol. 2007, 9, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A.; Chan, A.M. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 1994, 60, 3167–3174. [Google Scholar] [PubMed]
- Larsen, A.; Castberg, T.; Sandaa, R.A.; Brussaard, C.P.D.; Egge, J.; Heldal, M.; Paulino, A.; Thyrhaug, R.; van Hannen, E.J.; Bratbak, G. Population dynamics and diversity of phytoplankton, bacteria and viruses in a seawater enclosure. Mar. Ecol. Prog. Ser. 2001, 221, 47–57. [Google Scholar] [CrossRef]
- Cottrell, M.T.; Suttle, C.A. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol. Oceanogr. 1995, 40, 730–739. [Google Scholar] [CrossRef]
- Tarutani, K.; Nagasaki, K.; Itakura, S.; Yamaguchi, M. Isolation of a virus infecting the novel shellfish-killing dinoflagellate Heterocapsa circularisquama. Aquat. Microb. Ecol. 2001, 23, 103–111. [Google Scholar] [CrossRef]
- Thyrhaug, R.; Larsen, A.; Thingstad, T.F.; Bratbak, G. Stable coexistence in marine algal host-virus systems. Mar. Ecol. Prog. Ser. 2003, 254, 27–35. [Google Scholar] [CrossRef]
- Brussaard, C.P.D. Viral control of phytoplankton populations—A review. J. Eukaryot. Microbiol. 2004, 51, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Demory, D.; Arsenieff, L.; Simon, N.; Six, C.; Rigaut-Jalabert, F.; Marie, D.; Ge, P.; Bigeard, E.; Jacquet, S.; Sciandra, A.; et al. Temperature is a key factor in Micromonas-virus interactions. ISME J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sandaa, R.A.; Heldal, M.; Castberg, T.; Thyrhaug, R.; Bratbak, G. Isolation and characterization of two viruses with large genome size infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae). Virology 2001, 290, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, T.V.; Bratbak, G.; Larsen, A.; Ogata, H.; Egge, E.S.; Edvardsen, B.; Eikrem, W.; Sandaa, R.-A. Characterisation of three novel giant viruses reveals huge diversity among viruses infecting Prymnesiales (Haptophyta). Virology 2015, 476, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.; Balaji, S.; Koonin, E.; Aravind, L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006, 117, 156–184. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.H.; Etten, J.L.; Allen, M.J. The Phycodnaviridae: The story of how tiny giants rule the world. In Lesser Known Large dsDNA Viruses; Papers in Plant Pathology; Springer: Heidelberg, Germany, 2009; pp. 1–42. [Google Scholar]
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; de Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.-M.; Raoult, D. A giant virus in amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.G.; Allen, M.J.; Wilson, W.H.; Suttle, C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef] [PubMed]
- Moniruzzaman, M.; LeCleir, G.R.; Brown, C.M.; Gobler, C.J.; Bidle, K.D.; Wilson, W.H.; Wilhelm, S.W. Genome of brown tide virus (AaV), the little giant of the Megaviridae, elucidates NCLDV genome expansion and host-virus coevolution. Virology 2014, 466–467, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Moniruzzaman, M.; Gan, E.R.; LeCleir, G.R.; Kang, Y.; Gobler, C.J.; Wilhelm, S.W. Diversity and dynamics of algal Megaviridae members during a harmful brown tide caused by the pelagophyte, Aureococcus anophagefferens. FEMS Microbiol. Ecol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.B.; Larsen, A.; Bratbak, G.; Sandaa, R.A. Phylogenetic analysis of members of the Phycodnaviridae virus family, using amplified fragments of the major capsid protein gene. Appl. Environ. Microbiol. 2008, 74, 3048–3057. [Google Scholar] [CrossRef] [PubMed]
- Monier, A.; Larsen, J.B.; Sandaa, R.-A.; Bratbak, G.; Claverie, J.M.; Ogata, H. Marine mimivirus relatives are probably large algal viruses. Virol. J. 2008, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, D.M.; Mushegian, A.R.; Dolja, V.V.; Koonin, E.V. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010, 18, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Lee, K.; Lee, Y.S.; Kim, S.W.; Choi, T.J. Detection of diverse marine algal viruses in the South Sea regions of Korea by PCR amplification of the DNA polymerase and major capsid protein genes. Virus Res. 2011, 159, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Short, S.M.; Rusanova, O.; Staniewski, M.A. Novel phycodnavirus genes amplified from Canadian freshwater environments. Aquat. Microb. Ecol. 2011, 63, 61–67. [Google Scholar] [CrossRef]
- Rozon, R.M.; Short, S.M. Complex seasonality observed amongst diverse phytoplankton viruses in the Bay of Quinte, an embayment of Lake Ontario. Freshw. Biol. 2013, 58, 2648–2663. [Google Scholar] [CrossRef]
- Short, S.M.; Suttle, C.A. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 2002, 68, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-N.; Ge, X.-Y.; Wu, Y.-Q.; Yang, X.-L.; Tan, B.; Zhang, Y.-J.; Shi, Z.-L. Genetic diversity and temporal dynamics of phytoplankton viruses in East Lake, China. Virol. Sin. 2015, 30, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.; Flaten, G.A.F.; Sandaa, R.A.; Castberg, T.; Thyrhaug, R.; Erga, S.R.; Jacquet, S.; Bratbak, G. Spring phytoplankton bloom dynamics in Norwegian coastal waters: Microbial community succession and diversity. Limnol. Oceanogr. 2004, 49, 180–190. [Google Scholar] [CrossRef]
- Sandaa, R.A.; Larsen, A. Seasonal variations in viral-host populations in Norwegian coastal waters: Focusing on the cyanophage community infecting marine Synechococcus species. Appl. Environ. Microbiol. 2006, 72, 4610–4618. [Google Scholar] [CrossRef] [PubMed]
- Edvardsen, B.; Egge, E.S.; Vaulot, D. Diversity and distribution of haptophytes revealed by environmental sequencing and metabarcoding—A review. Perspect. Phycol. 2016, 3, 77–91. [Google Scholar] [CrossRef]
- Johannesen, T.V. Marine Virus-phytoplankton Interactions. Ph.D. Thesis, University of Bergen, Bergen, Norway, 2015. [Google Scholar]
- Erga, S.R.; Heimdal, B.R. Ecological studies on the phytoplankton of Korsfjorden, western Norway. The dynamics of a spring bloom seen in relation to hydrographical conditions and light regime. J. Plankton. Res. 1984, 6, 67–90. [Google Scholar] [CrossRef]
- Bratbak, G.; Heldal, M.; Norland, S.; Thingstad, T.F. Viruses as partners in spring bloom microbial trophodynamics. Appl. Environ. Microbiol. 1990, 56, 1400–1405. [Google Scholar] [PubMed]
- Pagarete, A.; Chow, C.-E.T.; Johannessen, T.; Fuhrman, J.A.; Thingstad, T.F.; Sandaa, R.A. Strong Seasonality and Interannual Recurrence in Marine Myovirus Communities. Appl. Environ. Microbiol. 2013, 79, 6253–6259. [Google Scholar] [CrossRef] [PubMed]
- Egge, J.S.; Eikrem, W.; Edvardsen, B. Deep branching novel lineages and high diversity of haptophytes in Skagerak (Norway) uncovered by 454-pyrosequencing. J. Eukaryot. Microbiol. 2015, 62, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Moon-van der Staay, S.Y.; van der Staay, G.W.M.; Guillou, L.; Vaulot, D.; Claustre, H.; Medlin, L.K. Abundance and diversity of prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol. Oceanogr. 2000, 45, 98–109. [Google Scholar] [CrossRef]
- Liu, H.; Probert, I.; Uitz, J.; Claustre, H.; Aris-Brosou, S.; Frada, M.; Not, F.; de Vargas, C. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc. Natl. Acad. Sci. USA 2009, 106, 12803–12808. [Google Scholar] [CrossRef] [PubMed]
- Thingstad, T.F.; Våge, S.; Storesund, J.E.; Sandaa, R.-A.; Giske, J. A theoretical analysis of how strain-specific viruses can control microbial species diversity. Proc. Natl. Acad. Sci. USA 2014, 111, 7813–7818. [Google Scholar] [CrossRef] [PubMed]
- Baudoux, A.; Brussaard, C. Characterization of different viruses infecting the marine harmful algal bloom species Phaeocystis globosa. Virology 2005, 341, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, K. Dinoflagellates, diatoms, and their viruses. J. Microbiol. 2008, 46, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Bratbak, G.; Egge, J.K.; Heldal, M. Viral mortality of the marine alga Emiliania-huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 1993, 93, 39–48. [Google Scholar] [CrossRef]
- Bratbak, G.; Levasseur, M.; Michaud, S.; Cantin, G.; Fernandez, E.; Heimdal, B.R.; Heldal, M. Viral activity in relation to Emiliana huxleyi blooms: A mechanism of DSMP release? Mar. Ecol. Prog. Ser. 1995, 128, 133–142. [Google Scholar] [CrossRef]
- Wilson, W.H.; Tarran, G.A.; Schroeder, D.; Cox, M.; Oke, J.; Malin, G. Isolation of viruses responsible for the demise of an Emiliania huxleyi bloom in the English Channel. J. Mar. Biol. Assoc. UK 2002, 82, 369–377. [Google Scholar] [CrossRef]
- Jacquet, S.; Heldal, M.; Iglesias-Rodriguez, D.; Larsen, A.; Wilson, W.; Bratbak, G. Flow cytometric analysis of an Emiliana huxleyi bloom terminated by viral infection. Aquat. Microb. Ecol. 2002, 27, 111–124. [Google Scholar] [CrossRef]
- Brussaard, C.P.D. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 2004, 70, 1506–1513. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.; Bratbak, G.; Heldal, M. Isolation and characterization of a virus infecting Phaeocystis pouchetii (Prymnesiophyceae). J. Phycol. 1996, 32, 923–927. [Google Scholar] [CrossRef]
- Brussaard, C.P.D.; Short, S.M.; Frederickson, C.M.; Suttle, C.A. Isolation and phylogenetic analysis of novel viruses infecting the phytoplankton Phaeocystis globosa (Prymnesiophyceae). Appl. Environ. Microbiol. 2004, 70, 3700–3705. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, K.; Bratbak, G. Isolation of viruses infecting photosynthetic and nonphotosynthetic protists. In Manual of Aquatic Viral Ecology; Wilhelm, S.W., Weinbauer, M.G., Suttle, C.A., Eds.; ASLO: Waco, TX, USA, 2010; pp. 92–101. [Google Scholar]
- Sandaa, R.-A. Burden or benefit? Virus-host interactions in the marine environment. Res. Microbiol. 2008, 159, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A.; Chen, F. Mechanisms and Rates of Decay of Marine Viruses in Seawater. Appl. Environ. Microbiol. 1992, 58, 3721–3729. [Google Scholar] [PubMed]
- Noble, R.T.; Fuhrman, J.A. Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl. Environ. Microbiol. 2000, 66, 3790–3797. [Google Scholar] [CrossRef] [PubMed]
- Mojica, K.D.A.; Brussaard, C.P.D. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 2014, 89, 495–515. [Google Scholar] [CrossRef] [PubMed]
- Needham, D.M.; Chow, C.E.T.; Cram, J.A.; Sachdeva, R.; Parada, A.; Fuhrman, J.A. Short-term observations of marine bacterial and viral communities: Patterns, connections and resilience. ISME J. 2013, 7, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Ray, J.L.; Haramaty, L.; Thyrhaug, R.; Fredricks, H.F.; Van Mooy, B.A.S.; Larsen, A.; Bidle, K.D.; Sandaa, R.-A. Virus infection of Haptolina ericina and Phaeocystis pouchetii implicates evolutionary conservation of programmed cell death induction in marine haptophyte–virus interactions. J. Plankton Res. 2014, 36, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Rozenn, T.; Grimsley, N.; Escande, M.L.; Subirana, L.; Derelle, E.; Moreau, H. Acquisition and maintenance of resistance to viruses in eukaryotic phytoplankton populations. Environ. Microbiol. 2011, 13, 1412–1420. [Google Scholar]
- Marie, D.; Brussaard, C.P.D.; Thyrhaug, R.; Bratbak, G.; Vaulot, D. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 1999, 65, 45–52. [Google Scholar] [PubMed]
- Edvardsen, B.; Eikrem, W.; Throndsen, J.; Sáez, A.G.; Probert, I.; Medlin, L.K. Ribosomal DNA phylogenies and a morphological revision provide the basis for a revised taxonomy of the Prymnesiales (Haptophyta). Eur. J. Phycol. 2011, 46, 202–228. [Google Scholar] [CrossRef]
- Not, F.; Siano, R.; Kooistra, W.H.C.F.; Simon, N.; Vaulot, D.; Probert, I. Diversity and Ecology of Eukaryotic Marine Phytoplankton. In Genomic Insights into the Biology of Algae; Piganeau, G., Ed.; Academic Press: London, UK, 2012; Volume 64, pp. 1–53. [Google Scholar]
- Egge, E.; Bittner, L.; Andersen, T.; Audic, S.; de Vargas, C.; Edvardsen, B. 454 Pyrosequencing to Describe Microbial Eukaryotic Community Composition, Diversity and Relative Abundance: A Test for Marine Haptophytes. PLoS ONE 2013, 8, e74371. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Lanzen, A.; Davenport, R.J.; Turnbaugh, P.J. Removing Noise From Pyrosequenced Amplicons. BMC Bioinform. 2011, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Caron, D.A.; Countway, P.D. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat. Microb. Ecol. 2009, 57, 227–238. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Figshare. Available online: https://dx.doi.org/10.6084/m9.figshare.2759983.v1 (accessed on 22 March 2017).
- Oksanen, J.; Blanchet, E.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R package version 2.4-1. 2016. Available online: https://CRAN.R-project.org/package=vegan (accessed on 2 March 2017).
- Hall, T. BioEdit: A user friendly biologicla sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- GenomeNet, Tree. Available online: http://www.genome.jp/tools/ete/ (accessed on 18 November 2016).
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johannessen, T.V.; Larsen, A.; Bratbak, G.; Pagarete, A.; Edvardsen, B.; Egge, E.D.; Sandaa, R.-A. Seasonal Dynamics of Haptophytes and dsDNA Algal Viruses Suggest Complex Virus-Host Relationship. Viruses 2017, 9, 84. https://doi.org/10.3390/v9040084
Johannessen TV, Larsen A, Bratbak G, Pagarete A, Edvardsen B, Egge ED, Sandaa R-A. Seasonal Dynamics of Haptophytes and dsDNA Algal Viruses Suggest Complex Virus-Host Relationship. Viruses. 2017; 9(4):84. https://doi.org/10.3390/v9040084
Chicago/Turabian StyleJohannessen, Torill Vik, Aud Larsen, Gunnar Bratbak, António Pagarete, Bente Edvardsen, Elianne D. Egge, and Ruth-Anne Sandaa. 2017. "Seasonal Dynamics of Haptophytes and dsDNA Algal Viruses Suggest Complex Virus-Host Relationship" Viruses 9, no. 4: 84. https://doi.org/10.3390/v9040084





