Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors
Abstract
:1. Introduction
2. Trafficking of the RSV Gag Polyprotein
3. Determinants of the RSV RNA Sequence Required for Packaging
3.1. The Psi Packaging Sequence
3.2. Mechanisms Governing Gag–vRNA Interaction
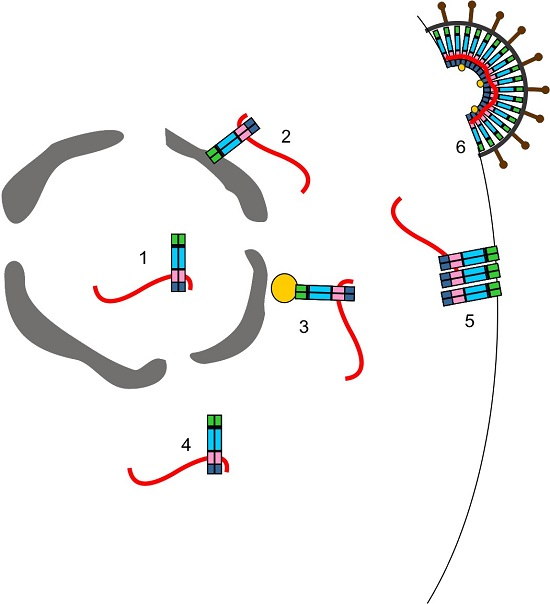
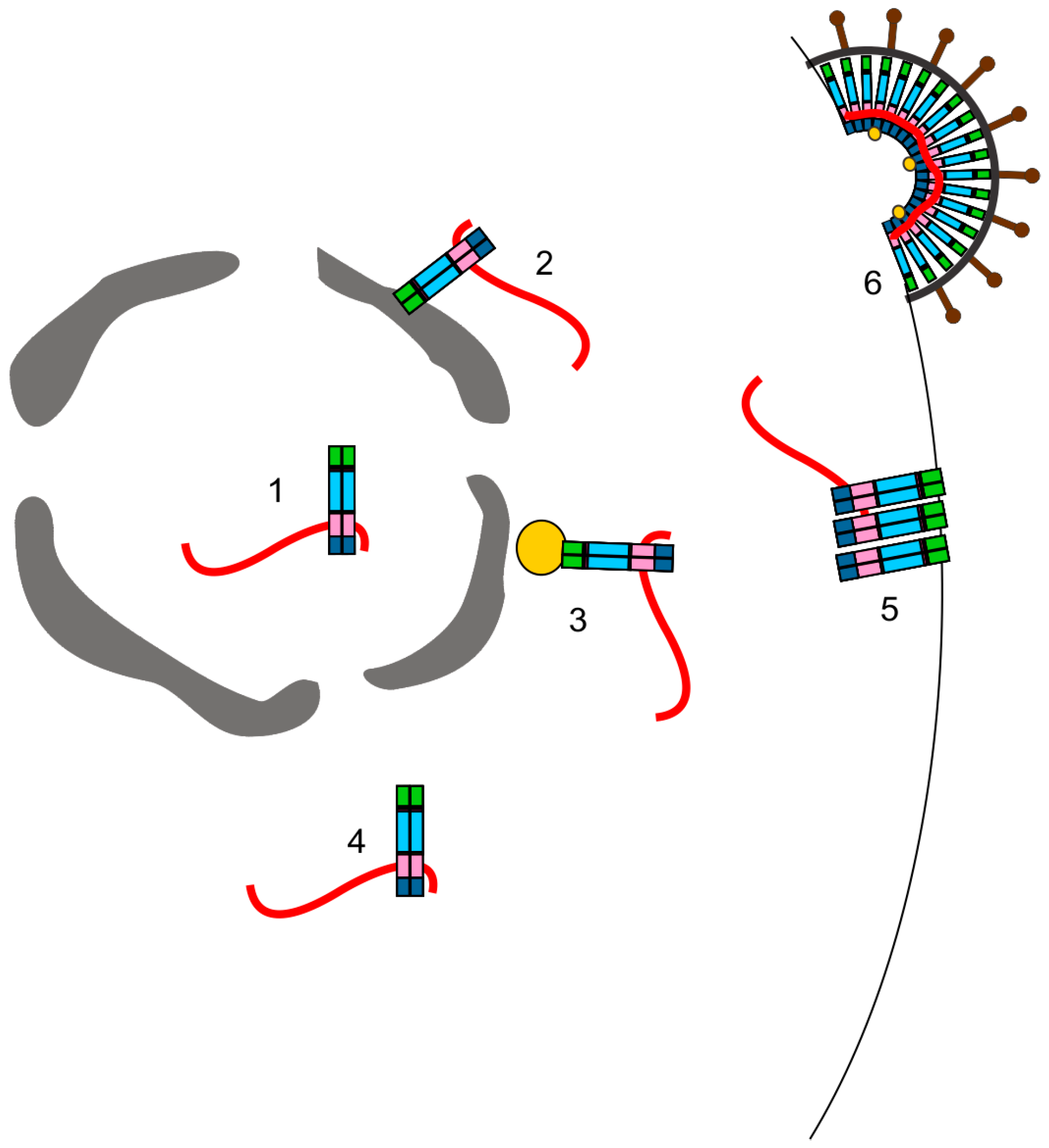
3.3. Where in the Cell Is gRNA Packaging Initiated?
3.4. Impact of RSV Gag Nuclear Trafficking on gRNA Packaging
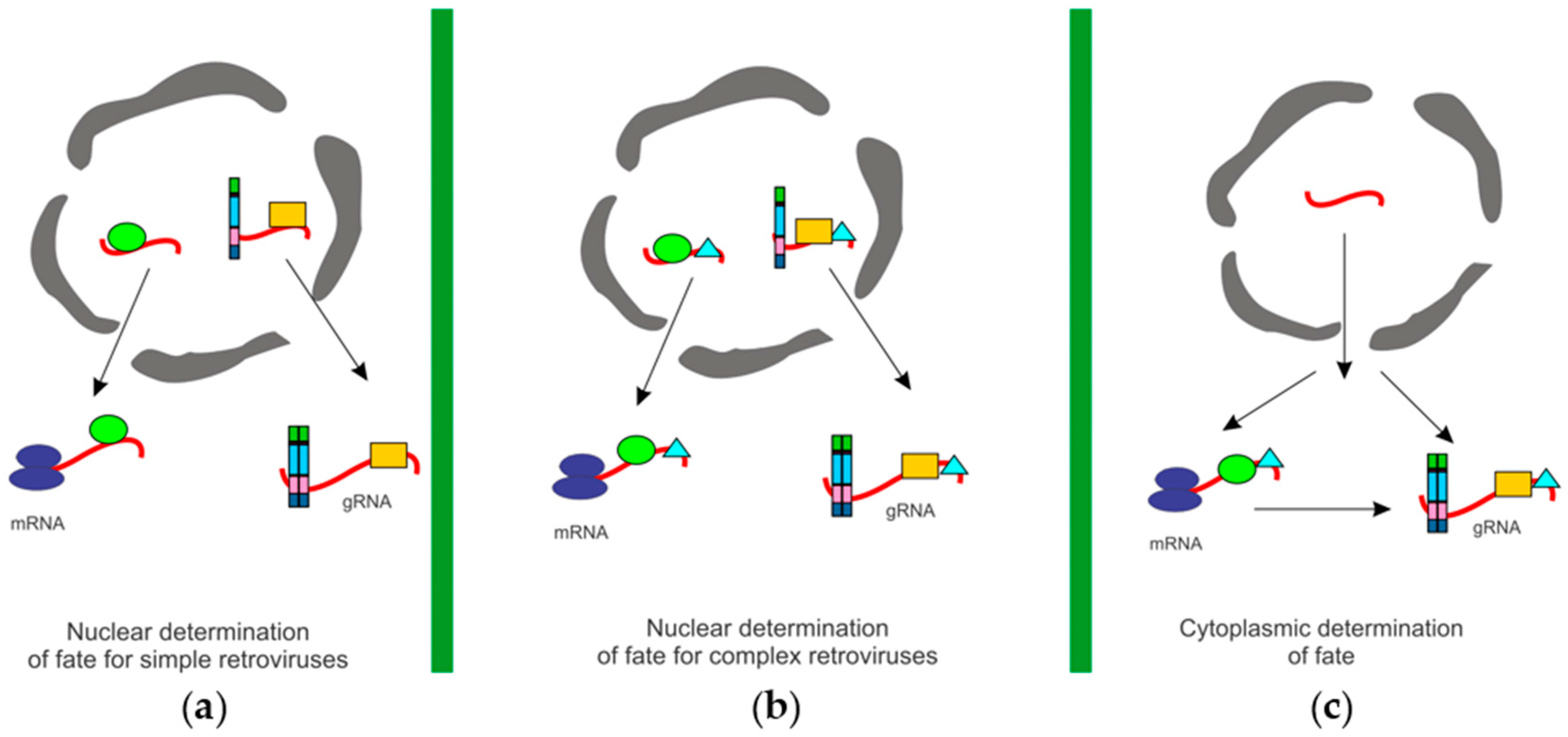
3.5. Determining the Cytoplasmic Fates of Unspliced Retroviral RNA
3.6. Role of vRNA Structure in Selection of gRNA for Packaging
3.7. Contribution of Host Factors to vRNA Sorting
3.8. Unspliced Retroviral RNA Nuclear Export
3.9. Nuclear Trafficking of Other Retroviral Gag Proteins
3.10. Conclusions and Remaining Questions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Giorgi, C.; Moore, M.J. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 2007, 18, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Reed, R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003, 15, 326–331. [Google Scholar] [CrossRef]
- Trcek, T.; Singer, R.H. The cytoplasmic fate of an mRNP is determined cotranscriptionally: Exception or rule? Genes Dev. 2010, 24, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, C.M. Synthesis and processing of avian sarcoma retrovirus RNA. Adv. Virus Res. 1988, 35, 1–38. [Google Scholar] [PubMed]
- Coffin, J.M.; Hughes, S.H.; Varmus, H.E. The interactions of retroviruses and their hosts. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor: New York, NY, USA, 1997. [Google Scholar]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- Butterfield-Gerson, K.L.; Scheifele, L.Z.; Ryan, E.P.; Hopper, A.K.; Parent, L.J. Importin-beta family members mediate alpharetrovirus Gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 2006, 80, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Verderame, M.F.; Nelle, T.D.; Wills, J.W. The membrane-binding domain of the Rous sarcoma virus Gag protein. J. Virol. 1996, 70, 2664–2668. [Google Scholar] [PubMed]
- Wills, J.W.; Cameron, C.E.; Wilson, C.B.; Xiang, Y.; Bennett, R.P.; Leis, J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 1994, 68, 6605–6618. [Google Scholar] [PubMed]
- Xiang, Y.; Cameron, C.E.; Wills, J.W.; Leis, J. Fine mapping and characterization of the Rous sarcoma virus pr76gag late assembly domain. J. Virol. 1996, 70, 5695–5700. [Google Scholar] [PubMed]
- Freed, E.O. Viral late domains. J. Virol. 2002, 76, 4679–4687. [Google Scholar] [CrossRef] [PubMed]
- Scheifele, L.Z.; Kenney, S.P.; Cairns, T.M.; Craven, R.C.; Parent, L.J. Overlapping roles of the Rous sarcoma virus Gag p10 domain in nuclear export and virion core morphology. J. Virol. 2007, 81, 10718–10728. [Google Scholar] [CrossRef] [PubMed]
- Scheifele, L.Z.; Ryan, E.P.; Parent, L.J. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 2005, 79, 8732–8741. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Vogt, V.M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: Identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 1997, 71, 4425–4435. [Google Scholar] [PubMed]
- Craven, R.C.; Parent, L.J. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immunol. 1996, 214, 65–94. [Google Scholar] [PubMed]
- Nandhagopal, N.; Simpson, A.A.; Johnson, M.C.; Francisco, A.B.; Schatz, G.W.; Rossmann, M.G.; Vogt, V.M. Dimeric Rous sarcoma virus capsid protein structure relevant to immature Gag assembly. J. Mol. Biol. 2004, 335, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.M.; Murray, P.S.; Murray, D.; Vogt, V.M. A molecular switch required for retrovirus assembly participates in the hexagonal immature lattice. EMBO J. 2008, 27, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.W.; Johnson, M.C.; Vogt, V.M. Mutations in the spacer peptide and adjoining sequences in Rous sarcoma virus Gag lead to tubular budding. J. Virol. 2008, 82, 6788–6797. [Google Scholar] [CrossRef] [PubMed]
- Lochmann, T.L.; Bann, D.V.; Ryan, E.P.; Beyer, A.R.; Mao, A.; Cochrane, A.; Parent, L.J. NC-mediated nucleolar localization of retroviral Gag proteins. Virus Res. 2013, 171, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Bowzard, J.B.; Bennett, R.P.; Krishna, N.K.; Ernst, S.M.; Rein, A.; Wills, J.W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 1998, 72, 9034–9044. [Google Scholar] [PubMed]
- Oertle, S.; Spahr, P.F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J. Virol. 1990, 64, 5757–5763. [Google Scholar] [PubMed]
- Stewart, L.; Schatz, G.; Vogt, V.M. Properties of avian retrovirus particles defective in viral protease. J. Virol. 1990, 64, 5076–5092. [Google Scholar] [PubMed]
- Canaani, E.; Helm, K.V.; Duesberg, P. Evidence for 30–40 S RNA as precursor of the 60–70 S RNA of Rous sarcoma virus. Proc. Natl. Acad. Sci. USA 1973, 70, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Korb, J.; Travnicek, M.; Riman, J. The oncornavirus maturation process: Quantitative correlation between morphological changes and conversion of genomic virion RNA. Intervirology 1976, 7, 211–224. [Google Scholar] [PubMed]
- Cheung, K.S.; Smith, R.E.; Stone, M.P.; Joklik, W.K. Comparison of immature (rapid harvest) and mature Rous sarcoma virus particles. Virology 1972, 50, 851–864. [Google Scholar] [CrossRef]
- Stoltzfus, C.M.; Snyder, P.N. Structure of b77 sarcoma virus RNA: Stabilization of RNA after packaging. J. Virol. 1975, 16, 1161–1170. [Google Scholar] [PubMed]
- Dupraz, P.; Oertle, S.; Meric, C.; Damay, P.; Spahr, P.F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J. Virol. 1990, 64, 4978–4987. [Google Scholar] [PubMed]
- Dupraz, P.; Spahr, P.F. Specificity of Rous sarcoma virus nucleocapsid protein in genomic rna packaging. J. Virol. 1992, 66, 4662–4670. [Google Scholar] [PubMed]
- Meric, C.; Darlix, J.L.; Spahr, P.F. It is Rous sarcoma virus protein p12 and not p19 that binds tightly to Rous sarcoma virus RNA. J. Mol. Biol. 1984, 173, 531–538. [Google Scholar] [CrossRef]
- Meric, C.; Spahr, P.F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J. Virol. 1986, 60, 450–459. [Google Scholar] [PubMed]
- Banks, J.D.; Kealoha, B.O.; Linial, M.L. An MPsi-containing heterologous RNA, but not Env mRNA, is efficiently packaged into avian retroviral particles. J. Virol. 1999, 73, 8926–8933. [Google Scholar] [PubMed]
- Giles, K.E.; Caputi, M.; Beemon, K.L. Packaging and reverse transcription of snRNAs by retroviruses may generate pseudogenes. RNA 2004, 10, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Eckwahl, M.J.; Telesnitsky, A.; Wolin, S.L. Host RNA packaging by retroviruses: A newly synthesized story. MBio 2016, 7, e02025-15. [Google Scholar] [CrossRef] [PubMed]
- Eckwahl, M.J.; Sim, S.; Smith, D.; Telesnitsky, A.; Wolin, S.L. A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev. 2015, 29, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, R.; Fisher, J.; Goff, S.P. RNA packaging. Curr. Top. Microbiol. Immunol. 1996, 214, 177–218. [Google Scholar] [PubMed]
- Bishop, J.M.; Levinson, W.E.; Sullivan, D.; Fanshier, L.; Quintrell, N.; Jackson, J. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 1970, 42, 927–937. [Google Scholar] [CrossRef]
- Bishop, J.M.; Levinson, W.E.; Quintrell, N.; Sullivan, D.; Fanshier, L.; Jackson, J. The low molecular weight RNAs of Rous sarcoma virus. I. The 4 S RNA. Virology 1970, 42, 182–195. [Google Scholar] [CrossRef]
- Onafuwa-Nuga, A.A.; Telesnitsky, A.; King, S.R. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA 2006, 12, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Onafuwa-Nuga, A.A.; King, S.R.; Telesnitsky, A. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J. Virol. 2005, 79, 13528–13537. [Google Scholar] [CrossRef] [PubMed]
- Cen, S.; Javanbakht, H.; Kim, S.; Shiba, K.; Craven, R.; Rein, A.; Ewalt, K.; Schimmel, P.; Musier-Forsyth, K.; Kleiman, L. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J. Virol. 2002, 76, 13111–13115. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Mak, J.; Ladha, A.; Cohen, E.; Klein, M.; Rovinski, B.; Kleiman, L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 1993, 67, 3246–3253. [Google Scholar] [PubMed]
- Zaitseva, L.; Myers, R.; Fassati, A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 2006, 4, e332. [Google Scholar] [CrossRef] [PubMed]
- Houzet, L.; Paillart, J.C.; Smagulova, F.; Maurel, S.; Morichaud, Z.; Marquet, R.; Mougel, M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007, 35, 2695–2704. [Google Scholar] [CrossRef] [PubMed]
- Pavon-Eternod, M.; Wei, M.; Pan, T.; Kleiman, L. Profiling non-lysyl tRNAs in HIV-1. RNA 2010, 16, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Schopman, N.C.; van Montfort, T.; Willemsen, M.; Knoepfel, S.A.; Pollakis, G.; van Kampen, A.; Sanders, R.W.; Haasnoot, J.; Berkhout, B. Selective packaging of cellular miRNAs in HIV-1 particles. Virus Res. 2012, 169, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Brameier, M.; Ibing, W.; Hofer, K.; Montag, J.; Stahl-Hennig, C.; Motzkus, D. Mapping the small RNA content of simian immunodeficiency virions (SIV). PLoS ONE 2013, 8, e75063. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Wang, T.; Zhang, W.; Yu, X.F. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 2007, 35, 7288–7302. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.J.; Lee, P.; Levine, K.L.; Sang, J.S.; Shah, S.A.; Yang, O.O.; Shank, P.R.; Linial, M.L. Molecular cloning and characterization of the RNA packaging-defective retrovirus SE21Q1b. J. Virol. 1992, 66, 204–216. [Google Scholar] [PubMed]
- Gallis, B.; Linial, M.; Eisenman, R. An avian oncovirus mutant deficient in genomic RNA: Characterization of the packaged RNA as cellular messenger RNA. Virology 1979, 94, 146–161. [Google Scholar] [CrossRef]
- Kawai, S.; Koyama, T. Characterization of a Rous sarcoma virus mutant defective in packaging its own genomic RNA: Biological properties of mutant TK15 and mutant-induced transformants. J. Virol. 1984, 51, 147–153. [Google Scholar] [PubMed]
- Koyama, T.; Harada, F.; Kawai, S. Characterization of a Rous sarcoma virus mutant defective in packaging its own genomic RNA: Biochemical properties of mutant TK15 and mutant-induced transformants. J. Virol. 1984, 51, 154–162. [Google Scholar] [PubMed]
- Linial, M.; Medeiros, E.; Hayward, W.S. An avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: Biological and biochemical characterization. Cell 1978, 15, 1371–1381. [Google Scholar] [CrossRef]
- Nishizawa, M.; Koyama, T.; Kawai, S. Unusual features of the leader sequence of Rous sarcoma virus packaging mutant TK15. J. Virol. 1985, 55, 881–885. [Google Scholar] [PubMed]
- Shank, P.R.; Linial, M. Avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: Characterization of a deletion in the provirus. J. Virol. 1980, 36, 450–456. [Google Scholar] [PubMed]
- Katz, R.A.; Terry, R.W.; Skalka, A.M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J. Virol. 1986, 59, 163–167. [Google Scholar] [PubMed]
- Aronoff, R.; Linial, M. Specificity of retroviral RNA packaging. J. Virol. 1991, 65, 71–80. [Google Scholar] [PubMed]
- Aronoff, R.; Hajjar, A.M.; Linial, M.L. Avian retroviral RNA encapsidation: Reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J. Virol. 1993, 67, 178–188. [Google Scholar] [PubMed]
- Banks, J.D.; Yeo, A.; Green, K.; Cepeda, F.; Linial, M.L. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 1998, 72, 6190–6194. [Google Scholar] [PubMed]
- Banks, J.D.; Linial, M.L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 2000, 74, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Stoker, A.W.; Bissell, M.J. Development of avian sarcoma and leukosis virus-based vector-packaging cell lines. J. Virol. 1988, 62, 1008–1015. [Google Scholar] [PubMed]
- Zhou, J.; Bean, R.L.; Vogt, V.M.; Summers, M. Solution structure of the Rous sarcoma virus nucleocapsid protein: MuPsi RNA packaging signal complex. J. Mol. Biol. 2007, 365, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; McAllen, J.K.; Tailor, Y.; Summers, M.F. High affinity nucleocapsid protein binding to the muPsi RNA packaging signal of Rous sarcoma virus. J. Mol. Biol. 2005, 349, 976–988. [Google Scholar] [CrossRef] [PubMed]
- Hackett, P.B.; Dalton, M.W.; Johnson, D.P.; Petersen, R.B. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1991, 19, 6929–6934. [Google Scholar] [CrossRef] [PubMed]
- Doria-Rose, N.A.; Vogt, V.M. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J. Virol. 1998, 72, 8073–8082. [Google Scholar] [PubMed]
- Knight, J.B.; Si, Z.H.; Stoltzfus, C.M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J. Virol. 1994, 68, 4493–4502. [Google Scholar] [PubMed]
- Bolognesi, D.P.; Montelaro, R.C.; Frank, H.; Schafer, W. Assembly of type C oncornaviruses: A model. Science 1978, 199, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Fleissner, E.; Tress, E. Isolation of a ribonucleoprotein structure from oncornaviruses. J. Virol. 1973, 12, 1612–1615. [Google Scholar] [PubMed]
- Davis, N.L.; Rueckert, R.R. Properties of a ribonucleoprotein particle isolated from Nonidet P-40-treated Rous sarcoma virus. J. Virol. 1972, 10, 1010–1020. [Google Scholar] [PubMed]
- Copeland, T.D.; Morgan, M.A.; Oroszlan, S. Complete amino acid sequence of the basic nucleic acid binding protein of feline leukemia virus. Virology 1984, 133, 137–145. [Google Scholar] [CrossRef]
- Covey, S.N. Amino acid sequence homology in Gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986, 14, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Bowles, N.E.; Damay, P.; Spahr, P.F. Effect of rearrangements and duplications of the Cys-His motifs of Rous sarcoma virus nucleocapsid protein. J. Virol. 1993, 67, 623–631. [Google Scholar] [PubMed]
- Meric, C.; Gouilloud, E.; Spahr, P.F. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): Deletions of Cys-His boxes. J. Virol. 1988, 62, 3328–3333. [Google Scholar] [PubMed]
- Lee, E.; Yeo, A.; Kraemer, B.; Wickens, M.; Linial, M.L. The Gag domains required for avian retroviral RNA encapsidation determined by using two independent assays. J. Virol. 1999, 73, 6282–6292. [Google Scholar] [PubMed]
- Lee, E.G.; Linial, M.L. Basic residues of the retroviral nucleocapsid play different roles in Gag-Gag and Gag-Psi RNA interactions. J. Virol. 2004, 78, 8486–8495. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.G.; Alidina, A.; May, C.; Linial, M.L. Importance of basic residues in binding of Rous sarcoma virus nucleocapsid to the RNA packaging signal. J. Virol. 2003, 77, 2010–2020. [Google Scholar] [CrossRef]
- Lee, E.G.; Linial, M.L. Yeast three-hybrid screening of Rous sarcoma virus mutants with randomly mutagenized minimal packaging signals reveals regions important for Gag interactions. J. Virol. 2000, 74, 9167–9174. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.F.; Telesnitsky, A. Retroviral RNA dimerization and packaging: The what, how, when, where, and why. PLoS Pathog. 2010, 6, e1001007. [Google Scholar] [CrossRef] [PubMed]
- Lear, A.L.; Haddrick, M.; Heaphy, S. A study of the dimerization of Rous sarcoma virus RNA in vitro and in vivo. Virology 1995, 212, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Garbitt-Hirst, R.; Kenney, S.P.; Parent, L.J. Genetic evidence for a connection between Rous sarcoma virus Gag nuclear trafficking and genomic RNA packaging. J. Virol. 2009, 83, 6790–6797. [Google Scholar] [CrossRef] [PubMed]
- Garbitt, R.A.; Albert, J.A.; Kessler, M.D.; Parent, L.J. Trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 2001, 75, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Parent, L.J.; Cairns, T.M.; Albert, J.A.; Wilson, C.B.; Wills, J.W.; Craven, R.C. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 2000, 74, 164–172. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, V.; Summers, M.F. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 2004, 431, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.A.; Telesnitsky, A. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology 2006, 344, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Maurel, S.; Mougel, M. Murine leukemia virus RNA dimerization is coupled to transcription and splicing processes. Retrovirology 2010, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Rein, A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993, 67, 5443–5449. [Google Scholar] [PubMed]
- Moore, M.D.; Nikolaitchik, O.A.; Chen, J.; Hammarskjold, M.L.; Rekosh, D.; Hu, W.S. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009, 5, e1000627. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Clerte, C.; Chamontin, C.; Basyuk, E.; Laine, S.; Hottin, J.; Bertrand, E.; Margeat, E.; Mougel, M. Imaging HIV-1 RNA dimerization in cells by multicolor super-resolution and fluctuation microscopies. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Hu, W.S. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 2009, 11, 91–102. [Google Scholar] [PubMed]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA 2009, 106, 19114–19119. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.A.; Reed, J.C.; Geary, C.D.; Swain, J.V.; Lingappa, J.R. A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells. J. Virol. 2014, 88, 5718–5741. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.R.; Ma, Y.M.; Vogt, V.M.; Webb, W.W. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J. Cell Biol. 2003, 162, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.J.; Beemon, K.L. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J. Virol. 2004, 78, 5139–5146. [Google Scholar] [CrossRef] [PubMed]
- Sonstegard, T.S.; Hackett, P.B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus pr76gag. J. Virol. 1996, 70, 6642–6652. [Google Scholar] [PubMed]
- Gudleski, N.; Flanagan, J.M.; Ryan, E.P.; Bewley, M.C.; Parent, L.J. Directionality of nucleocytoplasmic transport of the retroviral Gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 9358–9363. [Google Scholar] [CrossRef] [PubMed]
- Kenney, S.P.; Lochmann, T.L.; Schmid, C.L.; Parent, L.J. Intermolecular interactions between retroviral Gag proteins in the nucleus. J. Virol. 2008, 82, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Rice, B.L.; Kaddis, R.J.; Stake, M.S.; Lochmann, T.L.; Parent, L.J. Interplay between the alpharetroviral Gag protein and SR proteins SF2 and SC35 in the nucleus. Front. Microbiol. 2015, 6, 925. [Google Scholar] [CrossRef] [PubMed]
- Butsch, M.; Boris-Lawrie, K. Destiny of unspliced retroviral RNA: Ribosome and/or virion? J. Virol. 2002, 76, 3089–3094. [Google Scholar] [CrossRef] [PubMed]
- Balvay, L.; Lopez Lastra, M.; Sargueil, B.; Darlix, J.L.; Ohlmann, T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007, 5, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.G.; Grimley, P.M.; Ramseur, J.M.; Berezesky, I.K. Deficiency of 60 to 70 S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 1974, 14, 152–161. [Google Scholar] [PubMed]
- Levin, J.G.; Rosenak, M.J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: Evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA 1976, 73, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Dorman, N.; Lever, A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 2000, 74, 11413–11417. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.F.; Lever, A.M. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 1999, 73, 3023–3031. [Google Scholar] [PubMed]
- Anderson, E.C.; Lever, A.M. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J. Virol. 2006, 80, 10478–10486. [Google Scholar] [CrossRef] [PubMed]
- Butsch, M.; Boris-Lawrie, K. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected t cells. J. Virol. 2000, 74, 11531–11537. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, C.M.; Dimock, K.; Horikami, S.; Ficht, T.A. Stabilities of avian sarcoma virus RNAs: Comparison of subgenomic and genomic species with cellular mRNAs. J. Gen. Virol. 1983, 64, 2191–2202. [Google Scholar] [CrossRef] [PubMed]
- Donze, O.; Damay, P.; Spahr, P.F. The first and third uORFs in RSV leader RNA are efficiently translated: Implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995, 23, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Donze, O.; Spahr, P.F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992, 11, 3747–3757. [Google Scholar] [PubMed]
- Moustakas, A.; Sonstegard, T.S.; Hackett, P.B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J. Virol. 1993, 67, 4337–4349. [Google Scholar] [PubMed]
- Heng, X.; Kharytonchyk, S.; Garcia, E.L.; Lu, K.; Divakaruni, S.S.; LaCotti, C.; Edme, K.; Telesnitsky, A.; Summers, M.F. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012, 417, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jun, H.J.; Jang, S.I.; You, J.C. The determination of importance of sequences neighboring the Psi sequence in lentiviral vector transduction and packaging efficiency. PLoS ONE 2012, 7, e50148. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Skripkin, E.; Ehresmann, B.; Ehresmann, C.; Marquet, R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 2002, 277, 5995–6004. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.; Berkhout, B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef] [PubMed]
- Ooms, M.; Huthoff, H.; Russell, R.; Liang, C.; Berkhout, B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004, 78, 10814–10819. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.; Ooms, M.; Haasnoot, P.C.; Berkhout, B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 2005, 44, 9058–9066. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Kemler, I.; Barraza, R.; Poeschla, E.M. Mapping the encapsidation determinants of feline immunodeficiency virus. J. Virol. 2002, 76, 11889–11903. [Google Scholar] [CrossRef] [PubMed]
- Browning, M.T.; Mustafa, F.; Schmidt, R.D.; Lew, K.A.; Rizvi, T.A. Sequences within the Gag gene of feline immunodeficiency virus (FIV) are important for efficient RNA encapsidation. Virus Res. 2003, 93, 199–209. [Google Scholar] [CrossRef]
- Browning, M.T.; Mustafa, F.; Schmidt, R.D.; Lew, K.A.; Rizvi, T.A. Delineation of sequences important for efficient packaging of feline immunodeficiency virus RNA. J. Gen. Virol. 2003, 84, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Kemler, I.; Azmi, I.; Poeschla, E.M. The critical role of proximal Gag sequences in feline immunodeficiency virus genome encapsidation. Virology 2004, 327, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, F.; Ghazawi, A.; Jayanth, P.; Phillip, P.S.; Ali, J.; Rizvi, T.A. Sequences intervening between the core packaging determinants are dispensable for maintaining the packaging potential and propagation of feline immunodeficiency virus transfer vector RNAs. J. Virol. 2005, 79, 13817–13821. [Google Scholar] [CrossRef] [PubMed]
- Ghazawi, A.; Mustafa, F.; Phillip, P.S.; Jayanth, P.; Ali, J.; Rizvi, T.A. Both the 5′ and 3′ LTRs of FIV contain minor RNA encapsidation determinants compared to the two core packaging determinants within the 5′ untranslated region and Gag. Microbes Infect. 2006, 8, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.C.; Ghazawi, A.; Cheung, W.K.; Phillip, P.S.; Rizvi, T.A.; Lever, A.M. The secondary structure of the 5′ end of the FIV genome reveals a long-range interaction between R/U5 and Gag sequences, and a large, stable stem-loop. RNA 2008, 14, 2597–2608. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, T.A.; Kenyon, J.C.; Ali, J.; Aktar, S.J.; Phillip, P.S.; Ghazawi, A.; Mustafa, F.; Lever, A.M. Optimal packaging of FIV genomic RNA depends upon a conserved long-range interaction and a palindromic sequence within Gag. J. Mol. Biol. 2010, 403, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Vile, R.G.; Ali, M.; Hunter, E.; McClure, M.O. Identification of a generalised packaging sequence for D-type retroviruses and generation of a D-type retroviral vector. Virology 1992, 189, 786–791. [Google Scholar] [CrossRef]
- Harrison, G.P.; Hunter, E.; Lever, A.M. Secondary structure model of the Mason-Pfizer monkey virus 5′ leader sequence: Identification of a structural motif common to a variety of retroviruses. J. Virol. 1995, 69, 2175–2186. [Google Scholar] [PubMed]
- Guesdon, F.M.; Greatorex, J.; Rhee, S.R.; Fisher, R.; Hunter, E.; Lever, A.M. Sequences in the 5′ leader of Mason-Pfizer monkey virus which affect viral particle production and genomic RNA packaging: Development of MPMV packaging cell lines. Virology 2001, 288, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Jaballah, S.A.; Aktar, S.J.; Ali, J.; Phillip, P.S.; Al Dhaheri, N.S.; Jabeen, A.; Rizvi, T.A. A G-C-rich palindromic structural motif and a stretch of single-stranded purines are required for optimal packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA. J. Mol. Biol. 2010, 401, 996–1014. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.D.; Mustafa, F.; Lew, K.A.; Browning, M.T.; Rizvi, T.A. Sequences within both the 5′ untranslated region and the Gag gene are important for efficient encapsidation of Mason-Pfizer monkey virus RNA. Virology 2003, 309, 166–178. [Google Scholar] [CrossRef]
- Aktar, S.J.; Jabeen, A.; Ali, L.M.; Vivet-Boudou, V.; Marquet, R.; Rizvi, T.A. SHAPE analysis of the 5′ end of the Mason-Pfizer monkey virus (MPMV) genomic RNA reveals structural elements required for genome dimerization. RNA 2013, 19, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Kalloush, R.M.; Vivet-Boudou, V.; Ali, L.M.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA depends upon conserved long-range interactions (LRIs) between U5 and Gag sequences. RNA 2016, 22, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Aktar, S.J.; Vivet-Boudou, V.; Ali, L.M.; Jabeen, A.; Kalloush, R.M.; Richer, D.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Structural basis of genomic RNA (gRNA) dimerization and packaging determinants of mouse mammary tumor virus (MMTV). Retrovirology 2014, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, F.; Al Amri, D.; Al Ali, F.; Al Sari, N.; Al Suwaidi, S.; Jayanth, P.; Philips, P.S.; Rizvi, T.A. Sequences within both the 5′ UTR and Gag are required for optimal in vivo packaging and propagation of mouse mammary tumor virus (MMTV) genomic RNA. PLoS ONE 2012, 7, e47088. [Google Scholar] [CrossRef] [PubMed]
- Salmons, B.; Moritz-Legrand, S.; Garcha, I.; Gunzburg, W.H. Construction and characterization of a packaging cell line for MMTV-based conditional retroviral vectors. Biochem. Biophys Res. Commun. 1989, 159, 1191–1198. [Google Scholar] [CrossRef]
- Rizvi, T.A.; Ali, J.; Phillip, P.S.; Ghazawi, A.; Jayanth, P.; Mustafa, F. Role of a heterologous retroviral transport element in the development of genetic complementation assay for mouse mammary tumor virus (MMTV) replication. Virology 2009, 385, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Farina, K.L.; Singer, R.H. The nuclear connection in RNA transport and localization. Trends Cell Biol. 2002, 12, 466–472. [Google Scholar] [CrossRef]
- Stake, M.; Singh, D.; Singh, G.; Marcela Hernandez, J.; Kaddis Maldonado, R.; Parent, L.J.; Boris-Lawrie, K. HIV-1 and two avian retroviral 5′ untranslated regions bind orthologous human and chicken RNA binding proteins. Virology 2015, 486, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Kula, A.; Guerra, J.; Knezevich, A.; Kleva, D.; Myers, M.P.; Marcello, A. Characterization of the HIV-1 RNA associated proteome identifies matrin 3 as a nuclear cofactor of rev function. Retrovirology 2011, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, C.Y.; Yedavalli, V.S.; Jeang, K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 2013, 4, e00596-12. [Google Scholar] [CrossRef] [PubMed]
- Budhiraja, S.; Liu, H.; Couturier, J.; Malovannaya, A.; Qin, J.; Lewis, D.E.; Rice, A.P. Mining the human complexome database identifies RBM14 as an XPO1-associated protein involved in HIV-1 Rev function. J. Virol. 2015, 89, 3557–3567. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.L.; Zhou, Z.; Magni, K.; Christoforides, C.; Rappsilber, J.; Mann, M.; Reed, R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 2001, 413, 644–647. [Google Scholar] [CrossRef] [PubMed]
- Sommer, P.; Nehrbass, U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr. Opin. Cell Biol. 2005, 17, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Fasken, M.B.; Corbett, A.H. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009, 6, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Saguez, C.; Olesen, J.R.; Jensen, T.H. Formation of export-competent mRNP: Escaping nuclear destruction. Curr. Opin. Cell Biol. 2005, 17, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Greco, A.; Pierotti, M.A.; Bongarzone, I.; Pagliardini, S.; Lanzi, C.; Della Porta, G. TRK-T1 is a novel oncogene formed by the fusion of TPR and TRK genes in human papillary thyroid carcinomas. Oncogene 1992, 7, 237–242. [Google Scholar] [PubMed]
- Park, M.; Dean, M.; Cooper, C.S.; Schmidt, M.; O’Brien, S.J.; Blair, D.G.; Vande Woude, G.F. Mechanism of Met oncogene activation. Cell 1986, 45, 895–904. [Google Scholar] [CrossRef]
- Soman, N.R.; Correa, P.; Ruiz, B.A.; Wogan, G.N. The TPR-Met oncogenic rearrangement is present and expressed in human gastric carcinoma and precursor lesions. Proc. Natl. Acad. Sci. USA 1991, 88, 4892–4896. [Google Scholar] [CrossRef] [PubMed]
- Bangs, P.; Burke, B.; Powers, C.; Craig, R.; Purohit, A.; Doxsey, S. Functional analysis of TPR: Identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J. Cell. Biol. 1998, 143, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Ben-Efraim, I.; Frosst, P.D.; Gerace, L. Karyopherin binding interactions and nuclear import mechanism of nuclear pore complex protein TPR. BMC Cell Biol. 2009, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Hauber, J.; Le, S.Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 dead box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Zolotukhin, A.S.; Michalowski, D.; Bear, J.; Smulevitch, S.V.; Traish, A.M.; Peng, R.; Patton, J.; Shatsky, I.N.; Felber, B.K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell Biol. 2003, 23, 6618–6630. [Google Scholar] [CrossRef] [PubMed]
- Ajamian, L.; Abrahamyan, L.; Milev, M.; Ivanov, P.V.; Kulozik, A.E.; Gehring, N.H.; Mouland, A.J. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 2008, 14, 914–927. [Google Scholar] [CrossRef] [PubMed]
- Coyle, J.H.; Bor, Y.C.; Rekosh, D.; Hammarskjold, M.L. The TPR protein regulates export of mRNAs with retained introns that traffic through the NXF1 pathway. RNA 2011, 17, 1344–1356. [Google Scholar] [CrossRef] [PubMed]
- Gruter, P.; Tabernero, C.; von Kobbe, C.; Schmitt, C.; Saavedra, C.; Bachi, A.; Wilm, M.; Felber, B.K.; Izaurralde, E. Tap, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1998, 1, 649–659. [Google Scholar] [CrossRef]
- Teplova, M.; Wohlbold, L.; Khin, N.W.; Izaurralde, E.; Patel, D.J. Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor Tap. Nat. Struct. Mol. Biol. 2011, 18, 990–998. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.; Prasad, S.; Dubay, J.W.; Hunter, E.; Jeang, K.T.; Rekosh, D.; Hammarskjold, M.L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 1994, 91, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.J.; Uddowla, S.; Abraham, B.; Clatterbuck, S.; Beemon, K.L. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced rous sarcoma virus RNA. Virology 2007, 363, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Ogert, R.A.; Lee, L.H.; Beemon, K.L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 1996, 70, 3834–3843. [Google Scholar] [PubMed]
- Stake, M.S.; Bann, D.V.; Kaddis, R.J.; Parent, L.J. Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication. Viruses 2013, 5, 2767–2795. [Google Scholar] [CrossRef] [PubMed]
- Nash, M.A.; Meyer, M.K.; Decker, G.L.; Arlinghaus, R.B. A subset of pr65gag is nucleus associated in murine leukemia virus-infected cells. J. Virol. 1993, 67, 1350–1356. [Google Scholar] [PubMed]
- Royer, M.; Cerutti, M.; Gay, B.; Hong, S.S.; Devauchelle, G.; Boulanger, P. Functional domains of HIV-1 Gag-polyprotein expressed in baculovirus-infected cells. Virology 1991, 184, 417–422. [Google Scholar] [CrossRef]
- Poole, E.; Strappe, P.; Mok, H.P.; Hicks, R.; Lever, A.M. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic 2005, 6, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.L.; Lee, S.H.; Lee, E.S.; You, J.C. HIV-1 nucleocapsid protein localizes efficiently to the nucleus and nucleolus. Virology 2016, 492, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Crumpacker, C.S. Human immunodeficiency virus type 1 nucleocapsid protein nuclear localization mediates early viral mRNA expression. J. Virol. 2002, 76, 10444–10454. [Google Scholar] [CrossRef] [PubMed]
- Kemler, I.; Saenz, D.; Poeschla, E. Feline immunodeficiency virus Gag is a nuclear shuttling protein. J. Virol. 2012, 86, 8402–8411. [Google Scholar] [CrossRef] [PubMed]
- Baluyot, M.F.; Grosse, S.A.; Lyddon, T.D.; Janaka, S.K.; Johnson, M.C. CRM1-dependent trafficking of retroviral Gag proteins revisited. J. Virol. 2012, 86, 4696–4700. [Google Scholar] [CrossRef] [PubMed]
- Grewe, B.; Hoffmann, B.; Ohs, I.; Blissenbach, M.; Brandt, S.; Tippler, B.; Grunwald, T.; Uberla, K. Cytoplasmic utilization of human immunodeficiency virus type 1 genomic RNA is not dependent on a nuclear interaction with Gag. J. Virol. 2012, 86, 2990–3002. [Google Scholar] [CrossRef] [PubMed]
- Kemler, I.; Meehan, A.; Poeschla, E.M. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J. Virol. 2010, 84, 6352–6366. [Google Scholar] [CrossRef] [PubMed]
- Flugel, R.M.; Pfrepper, K.I. Proteolytic processing of foamy virus Gag and Pol proteins. Curr. Top. Microbiol. Immunol. 2003, 277, 63–88. [Google Scholar] [PubMed]
- Schliephake, A.W.; Rethwilm, A. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 1994, 68, 4946–4954. [Google Scholar] [PubMed]
- Tobaly-Tapiero, J.; Bittoun, P.; Lehmann-Che, J.; Delelis, O.; Giron, M.L.; de The, H.; Saib, A. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 2008, 9, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Mullers, E.; Stirnnagel, K.; Kaulfuss, S.; Lindemann, D. Prototype foamy virus Gag nuclear localization: A novel pathway among retroviruses. J. Virol. 2011, 85, 9276–9285. [Google Scholar] [CrossRef] [PubMed]
- Renault, N.; Tobaly-Tapiero, J.; Paris, J.; Giron, M.L.; Coiffic, A.; Roingeard, P.; Saib, A. A nuclear export signal within the structural Gag protein is required for prototype foamy virus replication. Retrovirology 2011, 8, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Checkley, M.A.; Mitchell, J.A.; Eizenstat, L.D.; Lockett, S.J.; Garfinkel, D.J. Ty1 Gag enhances the stability and nuclear export of Ty1 mRNA. Traffic 2013, 14, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Dang, V.D.; Levin, H.L. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol. Cell. Biol. 2000, 20, 7798–7812. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaddis Maldonado, R.J.; Parent, L.J. Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors. Viruses 2016, 8, 257. https://doi.org/10.3390/v8090257
Kaddis Maldonado RJ, Parent LJ. Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors. Viruses. 2016; 8(9):257. https://doi.org/10.3390/v8090257
Chicago/Turabian StyleKaddis Maldonado, Rebecca J., and Leslie J. Parent. 2016. "Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors" Viruses 8, no. 9: 257. https://doi.org/10.3390/v8090257