Arms Race between Enveloped Viruses and the Host ERAD Machinery
Abstract
:1. Enveloped Viruses
2. Virus Envelope Glycoproteins
3. Glycosylation and Unfolded Protein Response
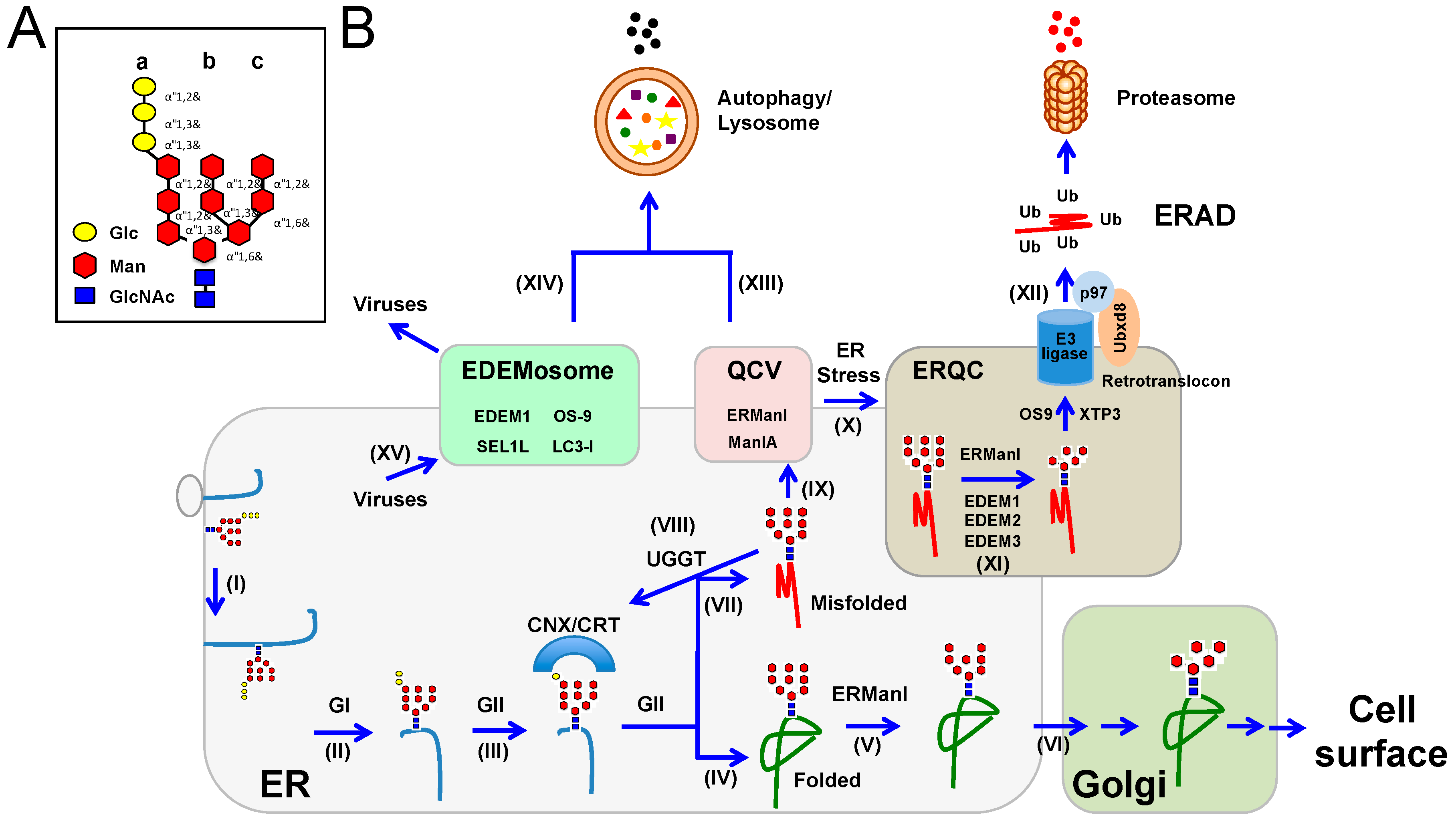
4. ER-Associated Protein Degradation
5. Viruses and UPR
6. Roles of ERAD in Promotion of Virus Replication
7. Roles of ERAD in Inhibition of Virus Replication
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Knipe, D.M.; Howley, P.M. Fields Virology, 6th ed.; Wolters Kluwer/Lippincott Williams & Wilkins Health: Philadelphia, PA, USA, 2013. [Google Scholar]
- Dimitrov, D.S. Virus entry: Molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004, 2, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.; Marsh, M. The cell biology of receptor-mediated virus entry. J. Cell Biol. 2011, 195, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Wu, L.; Bohks, S.M.; KewalRamani, V.N.; Unutmaz, D.; Hope, T.J. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 2003, 300, 1295–1297. [Google Scholar] [CrossRef] [PubMed]
- Tsai, B. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell dev. Biol. 2007, 23, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Podbilewicz, B. Virus and cell fusion mechanisms. Annu. Rev. Cell dev. Biol. 2014, 30, 111–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Leang, S.K.; Davidson, A.; Lobigs, M. Both E protein glycans adversely affect dengue virus infectivity but are beneficial for virion release. J. Virol. 2010, 84, 5171–5180. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Nie, J.; Prochnow, C.; Truong, C.; Jia, Z.; Wang, S.; Chen, X.S.; Wang, Y. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology 2013, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Tate, M.D.; Job, E.R.; Deng, Y.M.; Gunalan, V.; Maurer-Stroh, S.; Reading, P.C. Playing hide and seek: How glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 2014, 6, 1294–1316. [Google Scholar] [CrossRef] [PubMed]
- Helle, F.; Duverlie, G.; Dubuisson, J. The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses 2011, 3, 1909–1932. [Google Scholar] [CrossRef] [PubMed]
- Lennemann, N.J.; Rhein, B.A.; Ndungo, E.; Chandran, K.; Qiu, X.; Maury, W. Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio 2014, 5, e00862-00813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gaschen, B.; Blay, W.; Foley, B.; Haigwood, N.; Kuiken, C.; Korber, B. Tracking global patterns of n-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 2004, 14, 1229–1246. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, K.L.; Fried, V.A.; Ando, M.; Webster, R.G. Glycosylation affects cleavage of an H5N2 influenza virus hemagglutinin and regulates virulence. Proc. Natl. Acad. Sci. USA 1987, 84, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Klenk, H.D.; Wagner, R.; Heuer, D.; Wolff, T. Importance of hemagglutinin glycosylation for the biological functions of influenza virus. Virus Res. 2002, 82, 73–75. [Google Scholar] [CrossRef]
- Wagner, R.; Wolff, T.; Herwig, A.; Pleschka, S.; Klenk, H.D. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: A study by reverse genetics. J. Virol. 2000, 74, 6316–6323. [Google Scholar] [CrossRef] [PubMed]
- Doores, K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015, 282, 4679–4691. [Google Scholar] [CrossRef] [PubMed]
- Vigerust, D.J.; Shepherd, V.L. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007, 15, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef] [PubMed]
- Lamriben, L.; Graham, J.B.; Adams, B.M.; Hebert, D.N. N-glycan-based ER molecular chaperone and protein quality control system: The calnexin binding cycle. Traffic 2016, 17, 308–326. [Google Scholar] [CrossRef] [PubMed]
- Schallus, T.; Jaeckh, C.; Feher, K.; Palma, A.S.; Liu, Y.; Simpson, J.C.; Mackeen, M.; Stier, G.; Gibson, T.J.; Feizi, T.; et al. Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 2008, 19, 3404–3414. [Google Scholar] [CrossRef] [PubMed]
- Slominska-Wojewodzka, M.; Sandvig, K. The role of lectin-carbohydrate interactions in the regulation of ER-associated protein degradation. Molecules 2015, 20, 9816–9846. [Google Scholar] [CrossRef] [PubMed]
- Meusser, B.; Hirsch, C.; Jarosch, E.; Sommer, T. ERAD: The long road to destruction. Nat. Cell Biol. 2005, 7, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Benyair, R.; Ogen-Shtern, N.; Mazkereth, N.; Shai, B.; Ehrlich, M.; Lederkremer, G.Z. Mammalian ER mannosidase I resides in quality control vesicles, where it encounters its glycoprotein substrates. Mol. Biol. Cell 2015, 26, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Herscovics, A.; Romero, P.A.; Tremblay, L.O. The specificity of the yeast and human class I ER alpha 1,2-mannosidases involved in ER quality control is not as strict previously reported. Glycobiology 2002, 12, 14G–15G. [Google Scholar] [PubMed]
- Frenkel, Z.; Gregory, W.; Kornfeld, S.; Lederkremer, G.Z. Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6-5GlcNAc2. J. Biol. Chem. 2003, 278, 34119–34124. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Martinon, F.; Rodriguez, D.; Glimcher, L.H. The unfolded protein response: Integrating stress signals through the stress sensor IRE1alpha. Physiol. Rev. 2011, 91, 1219–1243. [Google Scholar] [CrossRef] [PubMed]
- Gardner, B.M.; Pincus, D.; Gotthardt, K.; Gallagher, C.M.; Walter, P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 2013, 5, a013169. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BIP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [PubMed]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Gardner, B.M.; Walter, P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.H.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Romisch, K. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell dev. Biol. 2005, 21, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Lemus, L.; Goder, V. Regulation of endoplasmic reticulum-associated protein degradation (ERAD) by ubiquitin. Cells 2014, 3, 824–847. [Google Scholar] [CrossRef] [PubMed]
- Needham, P.G.; Brodsky, J.L. How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: The early history of erad. Biochim. Biophys. Acta 2013, 1833, 2447–2457. [Google Scholar] [CrossRef] [PubMed]
- Foresti, O.; Ruggiano, A.; Hannibal-Bach, H.K.; Ejsing, C.S.; Carvalho, P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife 2013, 2, e00953. [Google Scholar] [CrossRef] [PubMed]
- Wojcikiewicz, R.J.; Pearce, M.M.; Sliter, D.A.; Wang, Y. When worlds collide: Ip(3) receptors and the ERAD pathway. Cell Calcium 2009, 46, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, H.; Kang, H.; Lee, S.Y.; Lee, S.J.; Back, S.H.; Lee, S.H.; Kim, M.S.; Lee, J.E.; Park, J.Y.; et al. Regulation of diacylglycerol acyltransferase 2 protein stability by gp78-associated endoplasmic-reticulum-associated degradation. FEBS J. 2014, 281, 3048–3060. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.H.; Chiang, M.T.; Chau, L.Y. Ubiquitin-proteasome system mediates heme oxygenase-1 degradation through endoplasmic reticulum-associated degradation pathway. Biochim. Biophys. Acta 2008, 1783, 1826–1834. [Google Scholar] [CrossRef] [PubMed]
- Vij, N.; Fang, S.; Zeitlin, P.L. Selective inhibition of endoplasmic reticulum-associated degradation rescues DeltaF508-cystic fibrosis transmembrane regulator and suppresses interleukin-8 levels: Therapeutic implications. J. Biol. Chem. 2006, 281, 17369–17378. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, C.J.; Brodsky, J.L. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 2012, 92, 537–576. [Google Scholar] [CrossRef] [PubMed]
- Hebert, D.N.; Molinari, M. Flagging and docking: Dual roles for N-glycans in protein quality control and cellular proteostasis. Trends Biochem. Sci. 2012, 37, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Mast, S.W.; Moremen, K.W. Family 47 alpha-mannosidases in N-glycan processing. Methods Enzymol. 2006, 415, 31–46. [Google Scholar] [PubMed]
- Ninagawa, S.; Okada, T.; Sumitomo, Y.; Kamiya, Y.; Kato, K.; Horimoto, S.; Ishikawa, T.; Takeda, S.; Sakuma, T.; Yamamoto, T.; et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 2014, 206, 347–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirao, K.; Natsuka, Y.; Tamura, T.; Wada, I.; Morito, D.; Natsuka, S.; Romero, P.; Sleno, B.; Tremblay, L.O.; Herscovics, A.; et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 2006, 281, 9650–9658. [Google Scholar] [CrossRef] [PubMed]
- Aikawa, J.; Matsuo, I.; Ito, Y. In vitro mannose trimming property of human ER alpha-1,2 mannosidase I. Glycoconj. J. 2012, 29, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hauri, H.P.; Kappeler, F.; Andersson, H.; Appenzeller, C. ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 2000, 113 Pt 4, 587–596. [Google Scholar] [PubMed]
- Avezov, E.; Frenkel, Z.; Ehrlich, M.; Herscovics, A.; Lederkremer, G.Z. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol. Biol. Cell 2008, 19, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; You, Z.; Tremblay, L.O.; Nagata, K.; Herscovics, A. Stimulation of ERAD of misfolded null hong kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem. Biophys. Res. Commun. 2007, 362, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Ogen-Shtern, N.; Avezov, E.; Shenkman, M.; Benyair, R.; Lederkremer, G.Z. Mannosidase IA is in quality control vesicles and participates in glycoprotein targeting to ERAD. J. Mol. Biol. 2016, 428, 3194–3205. [Google Scholar] [CrossRef] [PubMed]
- Olivari, S.; Cali, T.; Salo, K.E.; Paganetti, P.; Ruddock, L.W.; Molinari, M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 2006, 349, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Olivari, S.; Molinari, M. Glycoprotein folding and the role of EDEM1, EDEM2 and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 2007, 581, 3658–3664. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Munro, S. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr. Biol. 2001, 11, R499–R501. [Google Scholar] [CrossRef]
- Garza, R.M.; Sato, B.K.; Hampton, R.Y. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J. Biol. Chem. 2009, 284, 14710–14722. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Otsuka, T.; Ohsaki, Y.; Cheng, J.; Taniguchi, T.; Hashimoto, H.; Taniguchi, H.; Fujimoto, T. Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol. Biol. Cell 2012, 23, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Forster, F. Structure of the native SEC61 protein-conducting channel. Nat. Commun. 2015, 6, 8403. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, M.L.; Romisch, K. Proteasome 19s rp binding to the sec61 channel plays a key role in erad. PLoS ONE 2015, 10, e0117260. [Google Scholar] [CrossRef] [PubMed]
- Lilley, B.N.; Ploegh, H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 2004, 429, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Shibata, Y.; Yun, C.; Ron, D.; Rapoport, T.A. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 2004, 429, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, E.J.; Olzmann, J.A.; Kopito, R.R. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2011, 18, 1147–1152. [Google Scholar]
- Claessen, J.H.; Kundrat, L.; Ploegh, H.L. Protein quality control in the ER: Balancing the ubiquitin checkbook. Trends Cell Biol. 2012, 22, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, C.; Gauss, R.; Horn, S.C.; Neuber, O.; Sommer, T. The ubiquitylation machinery of the endoplasmic reticulum. Nature 2009, 458, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Hampton, R.Y.; Gardner, R.G.; Rine, J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 1996, 7, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, R.D.; Rapoport, T.A. Autoubiquitination of the HRD1 ligase triggers protein retrotranslocation in ERAD. Cell 2016, 166, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Stein, A.; Ruggiano, A.; Carvalho, P.; Rapoport, T.A. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 2014, 158, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Shibata, E.; Abbas, T.; Huang, X.; Wohlschlegel, J.A.; Dutta, A. Selective ubiquitylation of p21 and Cdt1 by UBCH8 and UBE2G ubiquitin-conjugating enzymes via the CRL4Cdt2 ubiquitin ligase complex. Mol. Cell. Biol. 2011, 31, 3136–3145. [Google Scholar] [CrossRef] [PubMed]
- Gardner, R.G.; Swarbrick, G.M.; Bays, N.W.; Cronin, S.R.; Wilhovsky, S.; Seelig, L.; Kim, C.; Hampton, R.Y. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of HRD1P by HRD3P. J. Cell Biol. 2000, 151, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Wada, I. Association of the SEL1L protein transmembrane domain with HRD1 ubiquitin ligase regulates ERAD-L. FEBS J. 2016, 283, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.C.; Hanna, J.; Hirsch, C.; Volkwein, C.; Schutz, A.; Heinemann, U.; Sommer, T.; Jarosch, E. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol. Cell 2009, 36, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Standera, S.; Buerger, E.; Kikkert, M.; van Voorden, S.; Wiertz, E.; Koning, F.; Kloetzel, P.M.; Seeger, M. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J. Mol. Biol. 2005, 354, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Kreft, S.G.; Wang, L.; Hochstrasser, M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-IV). J. Biol. Chem. 2006, 281, 4646–4653. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Herr, R.A.; Rabelink, M.; Hoeben, R.C.; Wiertz, E.J.; Hansen, T.H. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J. Cell Biol. 2009, 187, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Burr, M.L.; Cano, F.; Svobodova, S.; Boyle, L.H.; Boname, J.M.; Lehner, P.J. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. USA 2011, 108, 2034–2039. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.; Goder, V.; Rapoport, T.A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 2006, 126, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Habeck, G.; Ebner, F.A.; Shimada-Kreft, H.; Kreft, S.G. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J. Cell Biol. 2015, 209, 621. [Google Scholar] [CrossRef] [PubMed]
- Jakob, C.A.; Bodmer, D.; Spirig, U.; Battig, P.; Marcil, A.; Dignard, D.; Bergeron, J.J.; Thomas, D.Y.; Aebi, M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001, 2, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.; Lipari, F.; Igdoura, S.; Herscovics, A. The saccharomyces cerevisiae processing alpha 1,2-mannosidase is localized in the endoplasmic reticulum, independently of known retrieval motifs. Eur. J. Cell Biol. 1996, 70, 298–305. [Google Scholar] [PubMed]
- Cali, T.; Galli, C.; Olivari, S.; Molinari, M. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem. Biophys. Res. Commun. 2008, 371, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Termine, D.J.; Swulius, M.T.; Moremen, K.W.; Sifers, R.N. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J. Biol. Chem. 2007, 282, 4841–4849. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, R.; Molinari, M. ERAD and ERAD tuning: Disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 2011, 23, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Fujita, E.; Kouroku, Y.; Isoai, A.; Kumagai, H.; Misutani, A.; Matsuda, C.; Hayashi, Y.K.; Momoi, T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: Ubiquitin/proteasome ERAD(i) and autophagy/lysosome ERAD(ii). Hum. Mol. Genet. 2007, 16, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Wada, I.; Hasegawa, K.; Yorihuzi, T.; Tremblay, L.O.; Herscovics, A.; Nagata, K. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001, 2, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Olivari, S.; Galli, C.; Alanen, H.; Ruddock, L.; Molinari, M. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 2005, 280, 2424–2428. [Google Scholar] [CrossRef] [PubMed]
- Benyair, R.; Ogen-Shtern, N.; Lederkremer, G.Z. Glycan regulation of ER-associated degradation through compartmentalization. Semin. Cell Dev. Biol 2015, 41, 99–109. [Google Scholar]
- Termine, D.J.; Moremen, K.W.; Sifers, R.N. The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of er mannosidase i. J. Cell Sci. 2009, 122, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Tsai, B. How viruses use the endoplasmic reticulum for entry, replication, and assembly. Cold Spring Harb. Perspect. Biol. 2013, 5, a013250. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kong, L.; Yu, X. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit. Rev. Microbiol. 2015, 41, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W. Unfolded protein response in hepatitis c virus infection. Front. Microbiol. 2014, 5, 233. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.; Uta, M.; Branza-Nichita, N. Modulation of the unfolded protein response by the human hepatitis B virus. Front. Microbiol. 2014, 5, 433. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014, 5, 296. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.P.; Ng, M.L.; Vasudevan, S.G. Differential unfolded protein response during chikungunya and sindbis virus infection: Chikv nsp4 suppresses eif2alpha phosphorylation. Virol. J. 2013, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Borsa, M.; Ferreira, P.L.; Petry, A.; Ferreira, L.G.; Camargo, M.M.; Bou-Habib, D.C.; Pinto, A.R. HIV infection and antiretroviral therapy lead to unfolded protein response activation. Virol. J. 2015, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.R. Signal integration via PKR. Sci. STKE Signal Trans. Knowl. Environ. 2001, 2001, re2. [Google Scholar] [CrossRef] [PubMed]
- Otto, G.A.; Puglisi, J.D. The pathway of hcv ires-mediated translation initiation. Cell 2004, 119, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Sugden, B. The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood 2008, 111, 2280–2289. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Feng, Z.; He, B. Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERL and mediates eIF-2alpha dephosphorylation by the gamma(1)34.5 protein. J. Virol. 2005, 79, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Moon, A.; Childs, K.; Goodbourn, S.; Dixon, L.K. The african swine fever virus DP71L protein recruits the protein phosphatase 1 catalytic subunit to dephosphorylate eIF2alpha and inhibits CHOP induction but is dispensable for these activities during virus infection. J. Virol. 2010, 84, 10681–10689. [Google Scholar] [CrossRef] [PubMed]
- Pavio, N.; Romano, P.R.; Graczyk, T.M.; Feinstone, S.M.; Taylor, D.R. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK. J. Virol. 2003, 77, 3578–3585. [Google Scholar] [CrossRef] [PubMed]
- Hassan, I.H.; Zhang, M.S.; Powers, L.S.; Shao, J.Q.; Baltrusaitis, J.; Rutkowski, D.T.; Legge, K.; Monick, M.M. Influenza a viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IER1) stress pathway. J. Biol. Chem. 2012, 287, 4679–4689. [Google Scholar] [CrossRef] [PubMed]
- Galindo, I.; Hernaez, B.; Munoz-Moreno, R.; Cuesta-Geijo, M.A.; Dalmau-Mena, I.; Alonso, C. The ATF6 branch of unfolded protein response and apoptosis are activated to promote african swine fever virus infection. Cell Death Dis. 2012, 3, e341. [Google Scholar] [CrossRef]
- Jheng, J.R.; Ho, J.Y.; Horng, J.T. Er stress, autophagy, and rna viruses. Front. Microbiol. 2014, 5, 388. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Benedetti, S.; Gentili, V.; Grigolato, J.; Di Luca, D. Short communication: Activating transcription factor 4 (ATF4) promotes HIV type 1 activation. AIDS Res. Hum. Retrovir. 2012, 28, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Leymarie, O.; Embury-Hyatt, C.; Chevalier, C.; Jouneau, L.; Moroldo, M.; Da Costa, B.; Berhane, Y.; Delmas, B.; Weingartl, H.M.; Le Goffic, R. PB1-F2 attenuates virulence of highly pathogenic avian H5N1 influenza virus in chickens. PLoS ONE 2014, 9, e100679. [Google Scholar] [CrossRef] [PubMed]
- Pena, J.; Harris, E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J. Biol. Chem. 2011, 286, 14226–14236. [Google Scholar] [CrossRef] [PubMed]
- Stahl, S.; Burkhart, J.M.; Hinte, F.; Tirosh, B.; Mohr, H.; Zahedi, R.P.; Sickmann, A.; Ruzsics, Z.; Budt, M.; Brune, W. Cytomegalovirus downregulates IRE1 to repress the unfolded protein response. PLoS Pathog. 2013, 9, e1003544. [Google Scholar] [CrossRef] [PubMed]
- Minakshi, R.; Padhan, K.; Rani, M.; Khan, N.; Ahmad, F.; Jameel, S. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS ONE 2009, 4, e8342. [Google Scholar] [CrossRef] [PubMed]
- van der Wal, F.J.; Kikkert, M.; Wiertz, E. The HCMN gene products US2 and US11 target MHC class I molecules for degradation in the cytosol. Curr. Top. Microbiol. Immunol. 2002, 269, 37–55. [Google Scholar] [PubMed]
- Hsu, J.L.; van den Boomen, D.J.; Tomasec, P.; Weekes, M.P.; Antrobus, R.; Stanton, R.J.; Ruckova, E.; Sugrue, D.; Wilkie, G.S.; Davison, A.J.; et al. Plasma membrane profiling defines an expanded class of cell surface proteins selectively targeted for degradation by HCMV US2 in cooperation with UL141. PLoS Pathog. 2015, 11, e1004811. [Google Scholar] [CrossRef] [PubMed]
- Boname, J.M.; Bloor, S.; Wandel, M.P.; Nathan, J.A.; Antrobus, R.; Dingwell, K.S.; Thurston, T.L.; Smith, D.L.; Smith, J.C.; Randow, F.; et al. Cleavage by signal peptide peptidase is required for the degradation of selected tail-anchored proteins. J. Cell Biol. 2014, 205, 847–862. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.; Lilley, B.N.; Spooner, E.; Noriega, V.; Tortorella, D.; Ploegh, H.L. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature 2006, 441, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Stagg, H.R.; Thomas, M.; van den Boomen, D.; Wiertz, E.J.; Drabkin, H.A.; Gemmill, R.M.; Lehner, P.J. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 2009, 186, 685–692. [Google Scholar] [CrossRef] [PubMed]
- van den Boomen, D.J.; Timms, R.T.; Grice, G.L.; Stagg, H.R.; Skodt, K.; Dougan, G.; Nathan, J.A.; Lehner, P.J. TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc. Natl. Acad. Sci. USA 2014, 111, 11425–11430. [Google Scholar] [CrossRef] [PubMed]
- van den Boomen, D.J.; Lehner, P.J. Identifying the ERAD ubiquitin E3 ligases for viral and cellular targeting of MHC class I. Mol. Immunol. 2015, 68, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Mueller, B.; Klemm, E.J.; Spooner, E.; Claessen, J.H.; Ploegh, H.L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA 2008, 105, 12325–12330. [Google Scholar] [CrossRef] [PubMed]
- Boname, J.M.; Stevenson, P.G. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 2001, 15, 627–636. [Google Scholar] [CrossRef]
- Levesque, K.; Finzi, A.; Binette, J.; Cohen, E.A. Role of CD4 receptor down-regulation during HIV-1 infection. Curr. HIV Res. 2004, 2, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Lindwasser, O.W.; Chaudhuri, R.; Bonifacino, J.S. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr. Mol. Med. 2007, 7, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.L.; Garcia, J.V. HIV-1 Nef: At the crossroads. Retrovirology 2008, 5, 84. [Google Scholar] [CrossRef] [PubMed]
- Margottin, F.; Bour, S.P.; Durand, H.; Selig, L.; Benichou, S.; Richard, V.; Thomas, D.; Strebel, K.; Benarous, R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1998, 1, 565–574. [Google Scholar] [CrossRef]
- Strebel, K. Hiv-1 vpu - an ion channel in search of a job. Biochim. Biophys. Acta 2014, 1838, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Magadan, J.G.; Perez-Victoria, F.J.; Sougrat, R.; Ye, Y.; Strebel, K.; Bonifacino, J.S. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010, 6, e1000869. [Google Scholar] [CrossRef] [PubMed]
- Byun, H.; Halani, N.; Mertz, J.A.; Ali, A.F.; Lozano, M.M.; Dudley, J.P. Retroviral REM protein requires processing by signal peptidase and retrotranslocation for nuclear function. Proc. Natl. Acad. Sci. USA 2010, 107, 12287–12292. [Google Scholar] [CrossRef] [PubMed]
- Surjit, M.; Jameel, S.; Lal, S.K. Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J. Virol. 2007, 81, 3339–3345. [Google Scholar] [CrossRef] [PubMed]
- Neu, U.; Stehle, T.; Atwood, W.J. The polyomaviridae: Contributions of virus structure to our understanding of virus receptors and infectious entry. Virology 2009, 384, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Lilley, B.N.; Gilbert, J.M.; Ploegh, H.L.; Benjamin, T.L. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J. Virol. 2006, 80, 8739–8744. [Google Scholar] [CrossRef] [PubMed]
- Schelhaas, M.; Malmstrom, J.; Pelkmans, L.; Haugstetter, J.; Ellgaard, L.; Grunewald, K.; Helenius, A. Simian virus 40 depends on er protein folding and quality control factors for entry into host cells. Cell 2007, 131, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Monastyrska, I.; Verheije, M.H.; Cali, T.; Ulasli, M.; Bianchi, S.; Bernasconi, R.; de Haan, C.A.; Molinari, M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 2010, 7, 500–508. [Google Scholar]
- Noack, J.; Bernasconi, R.; Molinari, M. How viruses hijack the ERAD tuning machinery. J. Virol. 2014, 88, 10272–10275. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Chertova, E.; Bess, J., Jr.; Lifson, J.D.; Arthur, L.O.; Liu, J.; Taylor, K.A.; Roux, K.H. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. USA 2003, 100, 15812–15817. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Liu, J.; Bess, J., Jr.; Chertova, E.; Lifson, J.D.; Grise, H.; Ofek, G.A.; Taylor, K.A.; Roux, K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 2006, 441, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Forster, F.; Medalia, O.; Zauberman, N.; Baumeister, W.; Fass, D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 2005, 102, 4729–4734. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, K.; Desai, P.; Winkler, D.C.; Heymann, J.B.; Belnap, D.M.; Baumeister, W.; Steven, A.C. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 2003, 302, 1396–1398. [Google Scholar] [CrossRef] [PubMed]
- Loney, C.; Mottet-Osman, G.; Roux, L.; Bhella, D. Paramyxovirus ultrastructure and genome packaging: Cryo-electron tomography of sendai virus. J. Virol. 2009, 83, 8191–8197. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Danev, R.; Nishiyama, K.; Sugawara, K.; Nagayama, K. Zernike phase contrast electron microscopy of ice-embedded influenza A virus. J. Struct. Biol. 2008, 162, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.; Chackerian, B. Why HIV virions have low numbers of envelope spikes: Implications for vaccine development. PLoS Pathog. 2014, 10, e1004254. [Google Scholar] [CrossRef] [PubMed]
- Fennie, C.; Lasky, L.A. Model for intracellular folding of the human immunodeficiency virus type 1 gp120. J. Virol. 1989, 63, 639–646. [Google Scholar] [PubMed]
- Hallenberger, S.; Tucker, S.P.; Owens, R.J.; Bernstein, H.B.; Compans, R.W. Secretion of a truncated form of the human immunodeficiency virus type 1 envelope glycoprotein. Virology 1993, 193, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Willey, R.L.; Bonifacino, J.S.; Potts, B.J.; Martin, M.A.; Klausner, R.D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 1988, 85, 9580–9584. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Han, Y.; Dang, Y.; Wang, X.; Zheng, Y.H. A novel HIV-1 restriction factor that is biologically distinct from APOBEC3 cytidine deaminases in a human T cell line CEM.NKR. Retrovirology 2009, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Dang, Y.; Zheng, Y.H. The mitochondrial translocator protein, TSPO, inhibits HIV-1 envelope glycoprotein biosynthesis via the endoplasmic reticulum-associated protein degradation pathway. J. Virol. 2014, 88, 3474–3484. [Google Scholar] [CrossRef] [PubMed]
- Zamzami, N.; Kroemer, G. The mitochondrion in apoptosis: How pandora's box opens. Nat. Rev. Mol. Cell Biol. 2001, 2, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Gatliff, J.; East, D.; Crosby, J.; Abeti, R.; Harvey, R.; Craigen, W.; Parker, P.; Campanella, M. TSPO interacts with VDAC1 and triggers a ROS-mediated inhibition of mitochondrial quality control. Autophagy 2014, 10, 2279–2296. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Swulius, M.T.; Moremen, K.W.; Sifers, R.N. Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc. Natl. Acad. Sci. USA 2003, 100, 8229–8234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Frabutt, D.A.; Moremen, K.W.; Zheng, Y.H. ERManI (endoplasmic reticulum class i alpha-mannosidase) is required for HIV-1 envelope glycoprotein degradation via endoplasmic reticulum-associated protein degradation pathway. J. Biol. Chem. 2015, 290, 22184–22192. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.S.; Karaveg, K.; Vandersall-Nairn, A.S.; Lal, A.; Moremen, K.W. Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase i, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J. Biol. Chem. 1999, 274, 21375–21386. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, L.O.; Herscovics, A. Cloning and expression of a specific human alpha 1,2-mannosidase that trims Man(9)GlcNAc(2) to Man(8)GlcNAc(2) isomer B during N-glycan biosynthesis. Glycobiology 1999, 9, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Vallee, F.; Karaveg, K.; Herscovics, A.; Moremen, K.W.; Howell, P.L. Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases. J. Biol. Chem. 2000, 275, 41287–41298. [Google Scholar] [CrossRef] [PubMed]
- Lipari, F.; Herscovics, A. Role of the cysteine residues in the alpha1,2-mannosidase involved in N-glycan biosynthesis in saccharomyces cerevisiae. The conserved Cys340 and Cys385 residues form an essential disulfide bond. J. Biol. Chem. 1996, 271, 27615–27622. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, M.A.; Kuss, A.W.; Puettmann, L.; Noor, A.; Ramiah, A.; Ali, G.; Hu, H.; Kerio, N.A.; Xiang, Y.; Garshasbi, M.; et al. Mutations in the alpha 1,2-mannosidase gene, MAN1B1, cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2011, 89, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Fahiminiya, S.; Wang, T.; Dempsey Nunez, L.; Rosenblatt, D.S.; Gibson, W.T.; Gilfix, B.; Bergeron, J.J.; Jerome-Majewska, L.A. Somatic overgrowth associated with homozygous mutations in both MAN1B! and SEC23A. Cold Spring Harb. Mol. Case Stud. 2016, 2, a000737. [Google Scholar] [CrossRef] [PubMed]
- Iannotti, M.J.; Figard, L.; Sokac, A.M.; Sifers, R.N. A golgi-localized mannosidase (MAN1B1) plays a non-enzymatic gatekeeper role in protein biosynthetic quality control. J. Biol. Chem. 2014, 289, 11844–11858. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, T.; Frabutt, D.A.; Zheng, Y.H. HIV-1 Vpr increases env expression by preventing Env from endoplasmic reticulum-associated protein degradation (ERAD). Virology 2016, 496, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.; Macovei, A.; Petrescu, S.; Branza-Nichita, N. Activation of ERAD pathway by human hepatitis B virus modulates viral and subviral particle production. PLoS ONE 2012, 7, e34169. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Suzuki, R.; Watanabe, N.; Masaki, T.; Tomonaga, M.; Muhammad, A.; Kato, T.; Matsuura, Y.; Watanabe, H.; Wakita, T.; et al. Role of the endoplasmic reticulum-associated degradation (ERAD) pathway in degradation of hepatitis C virus envelope proteins and production of virus particles. J. Biol. Chem. 2011, 286, 37264–37273. [Google Scholar] [CrossRef] [PubMed]
- McPherson, S.; Powell, E.E.; Barrie, H.D.; Clouston, A.D.; McGuckin, M.; Jonsson, J.R. No evidence of the unfolded protein response in patients with chronic hepatitis C virus infection. J. Gastroenterol. Hepatol. 2011, 26, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Britt, W.J.; Mach, M. Human cytomegalovirus glycoproteins. Intervirology 1996, 39, 401–412. [Google Scholar] [PubMed]
- Compton, T.; Nepomuceno, R.R.; Nowlin, D.M. Human cytomegalovirus penetrates host cells by ph-independent fusion at the cell surface. Virology 1992, 191, 387–395. [Google Scholar] [CrossRef]
- Zhou, M.; Lanchy, J.M.; Ryckman, B.J. Human cytomegalovirus gH/gL/gO promotes the fusion step of entry into all cell types, whereas gH/gL/UL128-131 broadens virus tropism through a distinct mechanism. J. Virol. 2015, 89, 8999–9009. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, L.; Browne, H.; Wargent, V.; Davis-Poynter, N.; Primorac, S.; Goldsmith, K.; Minson, A.C.; Johnson, D.C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein h (gh) and affects normal folding and surface expression of gH. J. Virol. 1992, 66, 2240–2250. [Google Scholar] [PubMed]
- Gardner, T.J.; Hernandez, R.E.; Noriega, V.M.; Tortorella, D. Human cytomegalovirus gH stability and trafficking are regulated by ER-associated degradation and transmembrane architecture. Sci. Rep. 2016, 6, 23692. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Cormier, J.H.; Hebert, D.N. Characterization of early EDEM1 protein maturation events and their functional implications. J. Biol. Chem. 2011, 286, 24906–24915. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.; Calanca, V.; Galli, C.; Lucca, P.; Paganetti, P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 2003, 299, 1397–1400. [Google Scholar] [CrossRef] [PubMed]
- Cormier, J.H.; Tamura, T.; Sunryd, J.C.; Hebert, D.N. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol. Cell 2009, 34, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Wang, S.; Utama, B.; Huang, L.; Blok, N.; Estes, M.K.; Moremen, K.W.; Sifers, R.N. Golgi localization of ERManI defines spatial separation of the mammalian glycoprotein quality control system. Mol. Biol. Cell 2011, 22, 2810–2822. [Google Scholar] [CrossRef] [PubMed]
| Virus | UPR Pathway | Description | Ref. |
|---|---|---|---|
| HIV-1 | PERK | ATF4 enhances HIV-1 replication synergistically with Tat. | [103] |
| IAV | IRE1 | IAV infection induces IRE1. Treatment with an IRE1 inhibitor reduces viral replication. An alternate splice variant of the PB1 polymerase subunit (PB1-F2) from an avian influenza A strain has been implicated in the induction of IRE1 in chickens. ΔPB1-F2 mutant virus displayed enhanced virulence in chickens. | [100,104] |
| HCV | PERK | HCV E2 glycoprotein binds to PERK as a pseudo-substrate to repress PERK activation. | [99] |
| DENV | PERK, ATF6, IREI | PERK-mediated eIF2α phosphorylation is reversed through the viral-induced expression of GADD34, which works with PP1 to dephosphorylate eIF2α. ATF6 is activated by PERK in a cell-type specific manner. PERK and IREI knockout producer cells have decreased production of virus. | [105] |
| ASFV | PERK, ATF6 | Ectopic DP71L expression dephosphorylates eIF2α in vitro. DP71L mutant viruses lack increased eIF2α phosphorylation, suggesting redundant viral factors. ATF6 activation by virus is implicated in caspase activation and early apoptosis required for viral exit. | [98,101] |
| EBV | PERK, IRE1, ATF6 | LMP1 activates all three UPR sensors through an unknown mechanism. ATF4 is induced by the activation of PERK binding to the LMP1 promoter to stimulate LMP1 expression. | [96] |
| HSV1 | PERK | Viral infection induces PERK and PKR, causing eIF2α phosphorylation. The HSV1 gamma(1)34.5 protein is involved in the dephosphorylation of the eIF2α through an interaction with the phosphatase PP1 | [97] |
| CHIKV | PERK | NSP4, the viral polymerase, reduces PERK-mediated eIF2α phosphorylation. | [92] |
| HCMV | IRE1 | HCMV late protein UL50 down-regulates IRE1 protein expression. | [106] |
| SARS-CoV | PERK | SARS coronavirus protein 3a activates PERK independently of IRE1 and ATF6. | [107] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frabutt, D.A.; Zheng, Y.-H. Arms Race between Enveloped Viruses and the Host ERAD Machinery. Viruses 2016, 8, 255. https://doi.org/10.3390/v8090255
Frabutt DA, Zheng Y-H. Arms Race between Enveloped Viruses and the Host ERAD Machinery. Viruses. 2016; 8(9):255. https://doi.org/10.3390/v8090255
Chicago/Turabian StyleFrabutt, Dylan A., and Yong-Hui Zheng. 2016. "Arms Race between Enveloped Viruses and the Host ERAD Machinery" Viruses 8, no. 9: 255. https://doi.org/10.3390/v8090255






