Fluorescent and Bioluminescent Reporter Myxoviruses
Abstract
:1. Introduction
2. Characteristics of Fluorescent and Bioluminescent Proteins
3. Reverse Genetics Systems for Paramyxoviruses and Orthomyxoviruses
4. Design of Reporter Paramyxoviruses and Orthomyxoviruses
4.1. Selection of Fluorescent Proteins
4.2. Brightness
4.3. Photostability
4.4. Maturation Time
4.5. pH Stability
4.6. Oligomerization
4.7. Selection of Luciferase Proteins
4.8. Expression Strategies for Fluorescent and Bioluminescent Labels
5. Applications of Reporter Paramyxoviruses and Orthomyxoviruses
5.1. In Vitro Applications
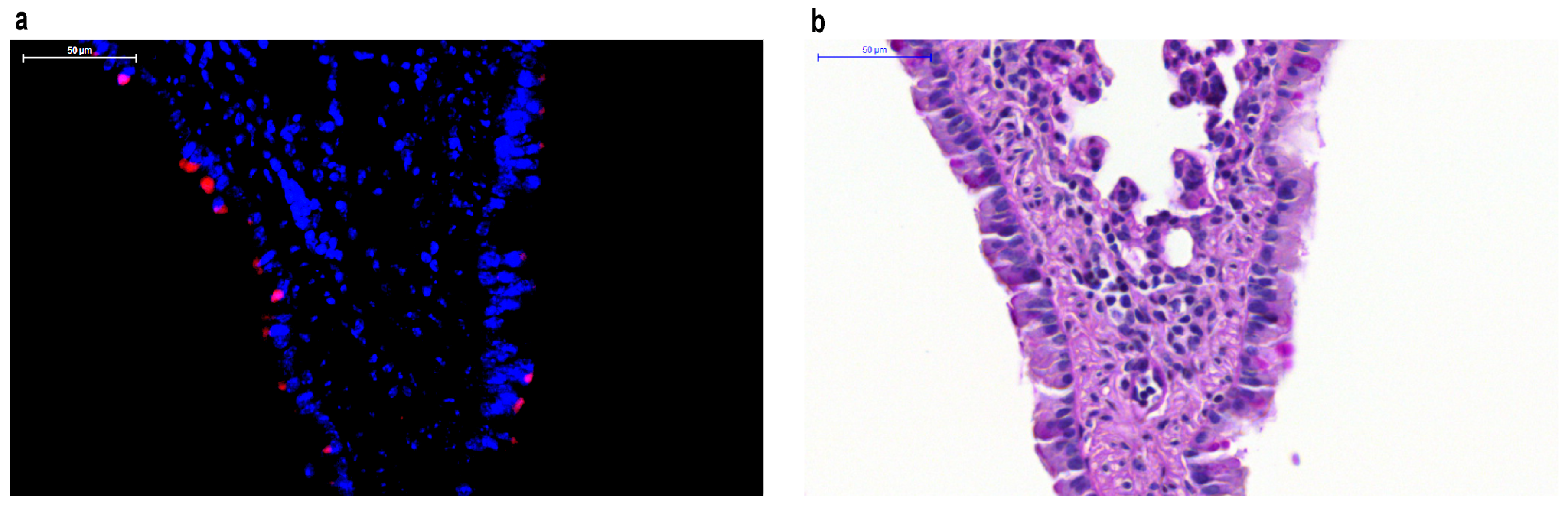
5.2. In Vivo Applications
6. Future Directions
Author Contributions
Conflicts of Interest
References
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Prasher, D.C.; Eckenrode, V.K.; Ward, W.W.; Prendergast, F.G.; Cormier, M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 1992, 111, 229–233. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Luker, K.E.; Luker, G.D. Bioluminescence imaging of reporter mice for studies of infection and inflammation. Antivir. Res. 2010, 86, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Cody, C.W.; Prasher, D.C.; Westler, W.M.; Prendergast, F.G.; Ward, W.W. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry 1993, 32, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Ormo, M.; Cubitt, A.B.; Kallio, K.; Gross, L.A.; Tsien, R.Y.; Reminqton, S.J. Crystal structure of the Aequorea victoria green fluorescent protein. Science 1996, 273, 1392–1395. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Ando, R.; Hama, H.; Yamamoto-Hino, M.; Mizuno, H.; Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 2002, 99, 12651–12656. [Google Scholar] [CrossRef] [PubMed]
- Shcherbo, D.; Merzlyak, E.M.; Chepurnykh, T.V.; Fradkov, A.F.; Ermakova, G.V.; Solovieva, E.A.; Lukyanov, K.A.; Bogdanova, E.A.; Zaraisky, A.G.; Lukyanov, S.; et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 2007, 4, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Shcherbo, D.; Murphy, C.S.; Ermakova, G.V.; Solovievs, E.A.; Chepurnykh, T.V.; Shcheqlov, A.S.; Verkhusha, W.; Pletnev, V.Z.; Hazelwood, K.L.; Roche, P.M.; et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009, 418, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef] [PubMed]
- Badr, C.E.; Tannous, B.A. Bioluminescence imaging: Progress and applications. Trends Biotechnol. 2011, 29, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Luker, G.D.; Bardill, J.P.; Prior, J.L.; Pica, C.M.; Piwnica-Worms, D.; Leib, D.A. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J. Virol. 2002, 76, 12149–12161. [Google Scholar] [CrossRef] [PubMed]
- Berger, F.; Paulmurugan, R.; Bhaumik, S.; Gambhir, S.S. Uptake kinetics and biodistribution of 14C-D-luciferin—A radiolabeled substrate for the firefly luciferase catalyzed bioluminescence reaction: Impact on bioluminescence based reporter gene imaging. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 2275–2285. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Vally, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.A.; Meliopoulos, V.A.; Savaqe, C.; Livinqston, B.; Mehle, A.; Schultz-Cherry, S. Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nat. Commun. 2015, 6, 6378. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.; Moser, L.A.; Poole, D.S.; Mehle, A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J. Virol. 2013, 87, 13321–13329. [Google Scholar] [CrossRef] [PubMed]
- Schnell, M.J.; Mebatsion, T.; Conzelmann, K.K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994, 13, 4195–4203. [Google Scholar] [PubMed]
- Lawson, N.D.; Stillman, E.A.; Whitt, M.A.; Rose, J.K. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 1995, 92, 4477–4481. [Google Scholar] [CrossRef] [PubMed]
- Radecke, F.; Spieihofer, P.; Schneider, H.; Kaelin, K.; Huber, M.; Dotsch, C.; Christiansen, G.; Billeter, M.A. Rescue of measles viruses from cloned DNA. EMBO J. 1995, 14, 5773–5784. [Google Scholar] [PubMed]
- Garcin, D.; PELET, T.; Calain, P.; Roux, L.; Curran, J.; Kolakofsky, D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: Generation of a novel copy-back nondefective interfering virus. EMBO J. 1995, 14, 6087–6094. [Google Scholar] [PubMed]
- Collins, P.L.; HILL, M.G.; Camargo, E.; Grosfeld, H.; Chanock, R.M.; Murphy, B.R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5’ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 1995, 92, 11563–11567. [Google Scholar] [PubMed]
- Wang, J.; Wang, C.; Feng, N.; Wang, H.; Zheng, X.; Yang, S.; Gao, Y.; Xia, X.; Yi, R.; Liu, X.; et al. Development of a reverse genetics system based on RNA polymerase II for Newcastle disease virus genotype VII. Virus Genes 2015, 50, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Staeheli, P.; Schneider, U. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J. Virol. 2006, 80, 5708–5715. [Google Scholar] [CrossRef] [PubMed]
- Luytjes, W.; Krystal, M.; Enami, M.; Parvin, J.D.; Palese, P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 1989, 59, 1107–1113. [Google Scholar] [CrossRef]
- Enami, M.; Luytjes, W.; Krystal, M.; Palese, P. Introduction of site-specific mutations into the genome of influenza virus. Proc. Natl. Acad. Sci. USA 1990, 87, 3802–3805. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Watanabe, T.; Ito, H.; Watanabe, S.; Goto, H.; Gao, P.; Huqhes, M.; Perez, D.R.; Donis, R.; Hoffmann, E.; et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 1999, 96, 9345–9350. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Fujii, K.; Kino, Y.; Kawaoka, Y. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc. Natl. Acad. Sci. USA 2005, 102, 16825–16829. [Google Scholar] [CrossRef] [PubMed]
- Heim, R.; Cubitt, A.B.; Tsien, R.Y. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Merzlyak, E.M.; Goedhart, J.; Shcherbo, D.; Bulina, M.E.; Shcheglov, A.S.; Fradkov, A.F.; Gaintzeva, A.; Lukyanov, K.A.; Lukyanov, S.; Gadella, T.W.J.; et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 2007, 4, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Z.; Mckeown, M.R.; Ng, H.L.; Aquilera, T.A.; Sjhaner, N.C.; Campbell, R.E.; Adams, S.R.; Gross, L.A.; Ma, W.; Alber, T.; et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem. Biol. 2009, 16, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Lin, M.Z.; Mckeown, M.R.; Steinbach, P.A.; Hazelwood, K.L.; Davidson, M.W.; Tsien, R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 2008, 5, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Verkhusha, V.V.; Otsuna, H.; Awasaki, T.; Oda, H.; Tsukita, S.; Ito, K. An enhanced mutant of red fluorescent protein DsRed for double labeling and developmental timer of neural fiber bundle formation. J. Biol. Chem. 2001, 276, 29621–29624. [Google Scholar] [CrossRef] [PubMed]
- Terskikh, A.; Fradkov, A.; Ermakova, G.; Zaraisky, A.; Tan, P.; Kajava, A.V.; Zhao, X.; Lukyanov, S.; Matz, M.; Kim, S.; et al. “Fluorescent timer”: Protein that changes color with time. Science 2000, 290, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Breen, M.; Nogales, A.; Baker, S.F.; Perez, D.R.; Martinez-Sobrido, L. Replication-competent influenza a and b viruses expressing a fluorescent dynamic timer protein for in vitro and in vivo studies. PLoS ONE 2016, 11, e0147723. [Google Scholar] [CrossRef] [PubMed]
- Ward, W.W. Bioluminescence and Chemiluminescence; Academic Press: New York, USA, 1981; pp. 235–242. [Google Scholar]
- Subach, O.M.; Cranfill, P.J.; Davidson, M.W.; Verkhusha, V.V. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS ONE 2011, 6, e28674. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Pike, D.; Sleat, D.E.; Nanda, V.; Lobel, P. Potential pitfalls and solutions for use of fluorescent fusion proteins to study the lysosome. PLoS ONE 2014, 9, e88893. [Google Scholar] [CrossRef] [PubMed]
- Kneen, M.; Farinas, J.; Li, Y.X.; Verkman, A.S. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 1998, 74, 1591–1599. [Google Scholar] [CrossRef]
- Campbell, R.E.; Tour, O.; Plamer, A.E.; Stienbach, P.A.; Baird, G.S.; Zacharias, D.A.; Taien, R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 2002, 99, 7877–7882. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Plamer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Buckley, S.M.; Howe, S.J.; Rahim, A.A.; Buning, H.; Mclntosh, J.; Wong, S.P.; Baker, A.H.; Nathwani, A.; Thrasher, A.J.; Coutelle, C.; et al. Luciferin detection after intranasal vector delivery is improved by intranasal rather than intraperitoneal luciferin administration. Hum. Gene Ther. 2008, 19, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Rameix-Welti, M.A.; Le Goffic, R.; Herve, P.L.; Sourimant, J.; Remot, A.; Riffault, S.; Yu, Q.; Galloux, M.; Gault, E.; Eleouet, J.F. Visualizing the replication of respiratory syncytial virus in cells and in living mice. Nat. Commun. 2014, 5, 5104. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.K.; Kato, A.; Shioda, T.; Sakai, Y.; Yu, D.; Nagai, Y. Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3’ proximal first locus. J. Gen. Virol. 1997, 78, 2813–2820. [Google Scholar] [CrossRef] [PubMed]
- Duprex, W.P.; McQuaid, S.; Hangartner, L.; Billeter, M.A.; Rima, B.K. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J. Virol. 1999, 73, 9568–9575. [Google Scholar] [PubMed]
- Naim, H.Y.; Ehler, E.; Billeter, M.A. Measles virus matrix protein specifies apical virus release and glycoprotein sorting in epithelial cells. EMBO J. 2000, 19, 3576–3585. [Google Scholar] [CrossRef] [PubMed]
- Burke, C.W.; Mason, J.N.; Surman, S.L.; Jones, B.G.; Dalloneau, E.; Hurwitz, J.L.; Russell, C.J. Illumination of parainfluenza virus infection and transmission in living animals reveals a tissue-specific dichotomy. PLoS Pathog. 2011, 7, e1002134. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Ono, N.; Tatsuo, H.; Minagawa, H.; Takeda, M.; Takeuchi, K.; Yanaqi, Y. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 2002, 76, 6743–6749. [Google Scholar] [CrossRef] [PubMed]
- Hotard, A.L.; Shaikh, F.Y.; Lee, S.; Yan, D.; Teng, M.N.; Plemper, R.K.; Crowe, J.E., Jr.; Moore, M.L. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology 2012, 434, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, M.; Guillauma, V.; Ikeda, F.; Sakuma, Y.; Sato, H.; Wild, T.F.; Kai, C. Establishment of a Nipah virus rescue system. Proc. Natl. Acad. Sci. USA 2006, 103, 16508–16513. [Google Scholar] [CrossRef] [PubMed]
- Burke, C.W.; Bridges, O.; Brown, S.; Rahija, R.; Russell, C.J. Mode of parainfluenza virus transmission determines the dynamics of primary infection and protection from reinfection. PLoS Pathog. 2013, 9, e1003786. [Google Scholar] [CrossRef] [PubMed]
- Marsh, G.A.; Virtue, E.R.; Smith, I.S.; Todd, S.; Arinstall, R.; Frazer, L.; Monaghan, P.; Smith, G.A.; Broder, C.C.; Middleton, D.; et al. Recombinant Hendra viruses expressing a reporter gene retain pathogenicity in ferrets. Virol. J. 2013, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, M.; Herfst, S.; Schrauwen, E.J.A.; van den Hoohen, B.G.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. An improved plaque reduction virus neutralization assay for human metapneumovirus. J. Virol. Methods 2007, 143, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Fujino, M.; Takeuchi, K.; Iwata, S.; Nakayama, T. A new method for the detection of neutralizing antibodies against mumps virus. PLoS ONE 2013, 8, e65281. [Google Scholar] [CrossRef] [PubMed]
- Rennick, L.J.; de Vries, R.D.; Carsillo, T.J.; Lemon, K.; van Amerongen, G.; Ludlow, M.; Nquyen, D.T.; Yuksel, S.; Verburqh, R.J.; Haddock, P.; et al. Live-attenuated measles virus vaccine targets dendritic cells and macrophages in muscle of nonhuman primates. J. Virol. 2015, 89, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, Y.; Ma, X.; Seki, F.; Suzuki, T.; Iwasaki, M.; Yanagi, Y.; Komase, K.; Takeda, M. Intracellular transport of the measles virus ribonucleoprotein complex is mediated by Rab11A-positive recycling endosomes and drives virus release from the apical membrane of polarized epithelial cells. J. Virol. 2013, 87, 4683–4693. [Google Scholar] [CrossRef] [PubMed]
- Van Remmerden, Y.; Xu, F.; van Eldik, M.; Heldens, J.G.; Huyisman, W.; Widjojoatmodjo, M.N. An improved respiratory syncytial virus neutralization assay based on the detection of green fluorescent protein expression and automated plaque counting. Virol. J. 2012, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Avilov, S.V.; Mosiy, D.; Munier, S.; Schraidt, O.; Naffakh, N.; Cusack, S. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. J. Virol. 2012, 86, 1433–1448. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Leyva-Grado, V.H.; Tan, G.S.; Eqqink, D.; Hai, R.; Palese, P. In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies. J. Virol. 2013, 87, 8272–8281. [Google Scholar] [CrossRef] [PubMed]
- Fulton, B.O.; Palese, P.; Heaton, N.S. Replication-Competent Influenza B Reporter Viruses as Tools for Screening Antivirals and Antibodies. J. Virol. 2015, 89, 12226–12231. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Dong, Z.; Li, F.; Meng, W.; Feng, L.; Niu, X.; Li, C.; Luo, Q.; Li, Z.; Sun, C.; et al. Visualizing influenza virus infection in living mice. Nat. Commun. 2013, 4, 2369. [Google Scholar] [CrossRef] [PubMed]
- Sutton, T.C.; Obadan, A.; Laviqne, J.; Chan, H.; Li, W.; Perez, D.R. Genome rearrangement of influenza virus for anti-viral drug screening. Virus Res. 2014, 189, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Manicassamy, B.; Manicassamy, S.; Belicha-Villanueva, A.; Pisanelli, G.; Pulendran, B.; Garcia-Sastre, A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. USA 2010, 107, 11531–11536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duprex, W.P.; Collins, F.M.; Rima, B.K. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 2002, 76, 7322–7328. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, S.; Kastura, H.; Zhao, D.; Ozawa, M.; Ando, T.; Shoemaker, J.E.; Ishikawa, I.; Yamada, S.; Neumann, G.; Watanabe, S.; et al. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nat. Commun. 2015, 6, 6600. [Google Scholar] [CrossRef] [PubMed]
- Techaarpornkul, S.; Barretto, N.; Peeples, M.E. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 2001, 75, 6825–6834. [Google Scholar] [CrossRef] [PubMed]
- Batonick, M.; Oomens, A.G.; Wertz, G.W. Human respiratory syncytial virus glycoproteins are not required for apical targeting and release from polarized epithelial cells. J. Virol. 2008, 82, 8664–8672. [Google Scholar] [CrossRef] [PubMed]
- Von Messling, V.; Milosevic, D.; Cattaneo, R. Tropism illuminated: lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proc. Natl. Acad. Sci. USA 2004, 101, 14216–14221. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.F.; Noqales, A.; Finch, C.; Tuffy, K.M.; Domm, W.; Perez, D.R.; Topham, D.J.; Martinez-Sobrido, L. Influenza A and B virus intertypic reassortment through compatible viral packaging signals. J. Virol. 2014, 88, 10778–10791. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.L.; Manicassamy, B.; Lamb, R.A. Influenza A virus uses intercellular connections to spread to neighboring cells. J. Virol. 2015, 89, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Lee, S.; Hotard, A.L.; Moore, M.L. Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. MBio 2014, 5, e01704–e01714. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Crim, R.L.; Beeler, J.; Teng, M.N.; Golding, H.; Khurana, S. Development of a simple, rapid, sensitive, high-throughput luciferase reporter based microneutralization test for measurement of virus neutralizing antibodies following Respiratory Syncytial Virus vaccination and infection. Vaccine 2013, 31, 3987–3994. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.F.; Nogales, A.; Santiago, F.W.; Topham, D.J.; Martinez-Sobrido, L. Competitive detection of influenza neutralizing antibodies using a novel bivalent fluorescence-based microneutralization assay (BiFMA). Vaccine 2015, 33, 3562–3570. [Google Scholar] [CrossRef] [PubMed]
- Fujino, M.; Yoshida, N.; Kimura, K.; Zhou, J.; Motegi, Y.; Komase, K.; Nakayama, T. Development of a new neutralization test for measles virus. J. Virol. Methods 2007, 142, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Kwanten, L.; De Clerck, B.; Roymans, D. A fluorescence-based high-throughput antiviral compound screening assay against respiratory syncytial virus. Methods Mol. Biol. 2013, 1030, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Lo, M.K.; Nichol, S.T.; Spiropoulou, C.F. Evaluation of luciferase and GFP-expressing Nipah viruses for rapid quantitative antiviral screening. Antivir. Res. 2014, 106, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Krumm, S.A.; Sun, A.; Steinhuaer, D.A.; Luo, M.; Moore, M.L.; Plemper, R.K. Dual myxovirus screen identifies a small-molecule agonist of the host antiviral response. J. Virol. 2013, 87, 11076–11087. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Weisshaar, M.; Lamb, K.; Chung, H.K.; Lin, M.Z.; Plemper, R.K. Replication-Competent Influenza Virus and Respiratory Syncytial Virus Luciferase Reporter Strains Engineered for Co-Infections Identify Antiviral Compounds in Combination Screens. Biochemistry 2015, 54, 5589–5604. [Google Scholar] [CrossRef] [PubMed]
- Duprex, W.P.; McQuaid, S.; Rima, B.K. Measles virus-induced disruption of the glial-fibrillary-acidic protein cytoskeleton in an astrocytoma cell line (U-251). J. Virol. 2000, 74, 3874–3880. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.W.; Frenzke, M.; Myers, R.; Soeffker, D.; Harvery, M.; Greiner, S.; Galanis, E.; Cattaneo, R.; Federspiel, M.J.; Russell, S.J. Biodistribution of oncolytic measles virus after intraperitoneal administration into Ifnar-CD46Ge transgenic mice. Hum. Gene Ther. 2003, 14, 1565–1577. [Google Scholar] [CrossRef] [PubMed]
- Baertsch, M.A.; Leber, M.F.; Bossow, S.; Singh, M.; Engeland, C.E.; Alber, J.; Grossardt, C.; Japer, D.; von Kalle, C.; Ungerechts, G. MicroRNA-mediated multi-tissue detargeting of oncolytic measles virus. Cancer Gene Ther. 2014, 21, 373–380. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rostad, C.A.; Currier, M.C.; Moore, M.L. Fluorescent and Bioluminescent Reporter Myxoviruses. Viruses 2016, 8, 214. https://doi.org/10.3390/v8080214
Rostad CA, Currier MC, Moore ML. Fluorescent and Bioluminescent Reporter Myxoviruses. Viruses. 2016; 8(8):214. https://doi.org/10.3390/v8080214
Chicago/Turabian StyleRostad, Christina A., Michael C. Currier, and Martin L. Moore. 2016. "Fluorescent and Bioluminescent Reporter Myxoviruses" Viruses 8, no. 8: 214. https://doi.org/10.3390/v8080214




