A Phylogenetic Survey on the Structure of the HIV-1 Leader RNA Domain That Encodes the Splice Donor Signal
Abstract
:1. Introduction
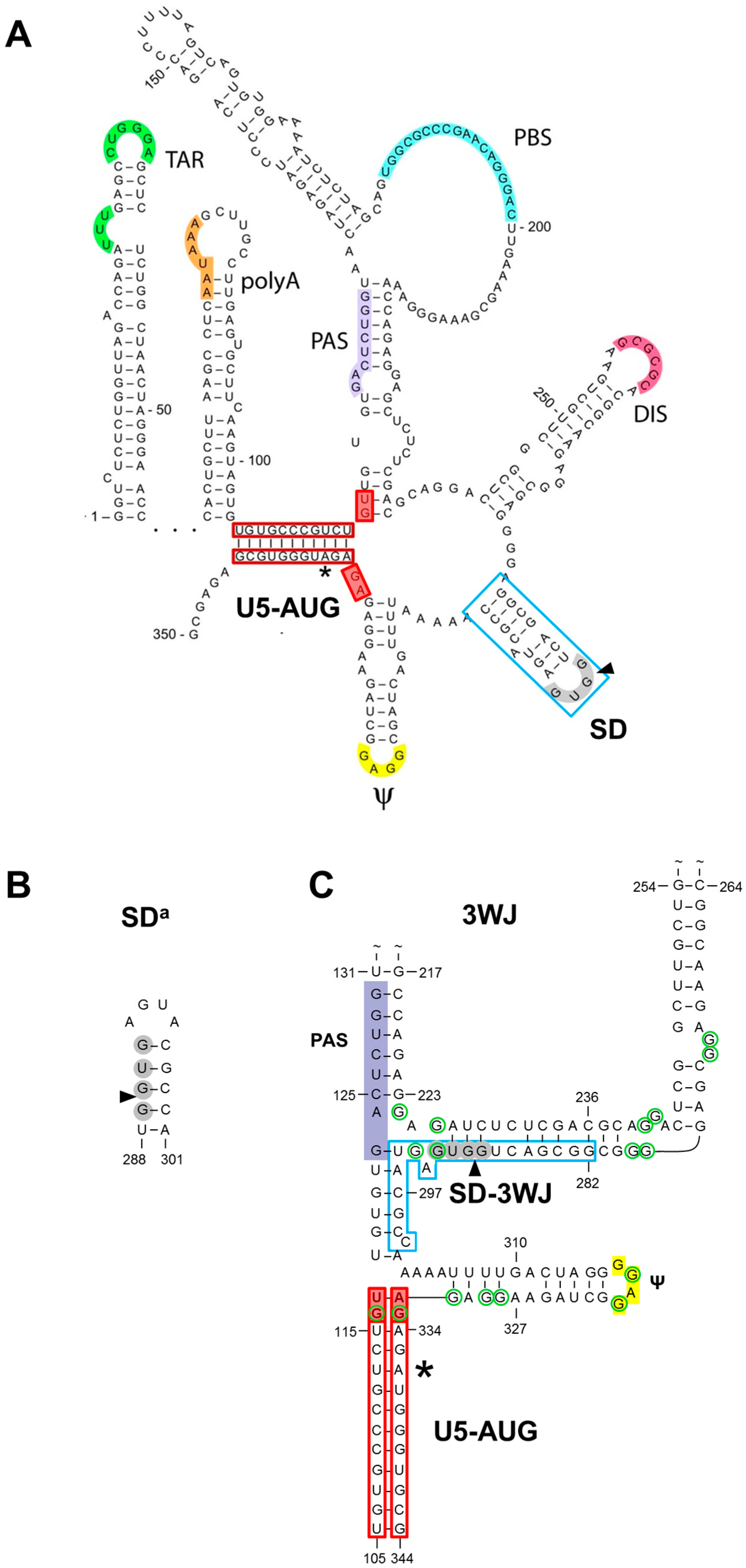
1.1. The SD Hairpin
1.2. The Alternative SDa Hairpin
1.3. The 3WJ Structure
1.4. In Search for Phylogenetic Support for SD, SDa or 3WJ
2. Materials and Methods
3. Results
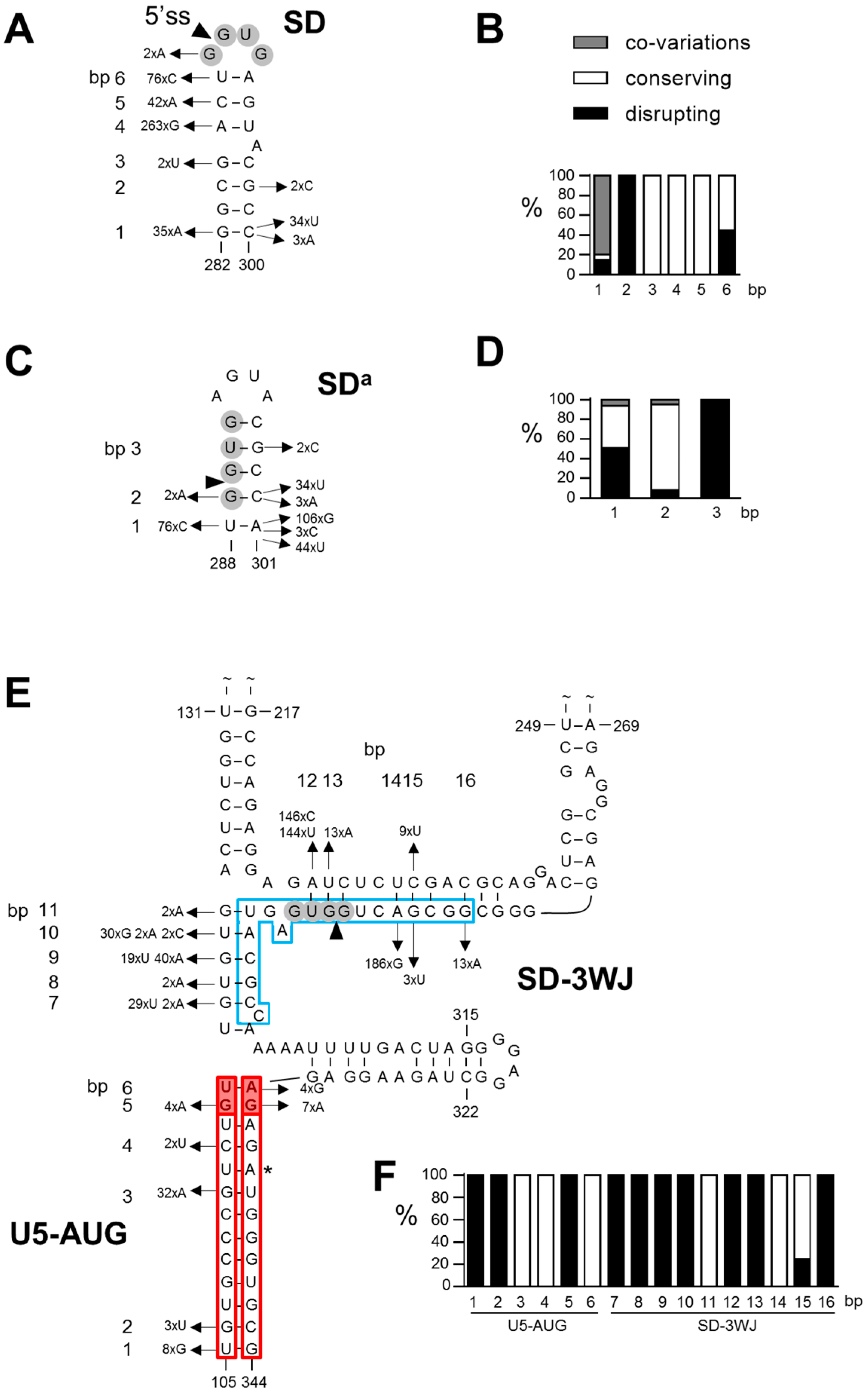
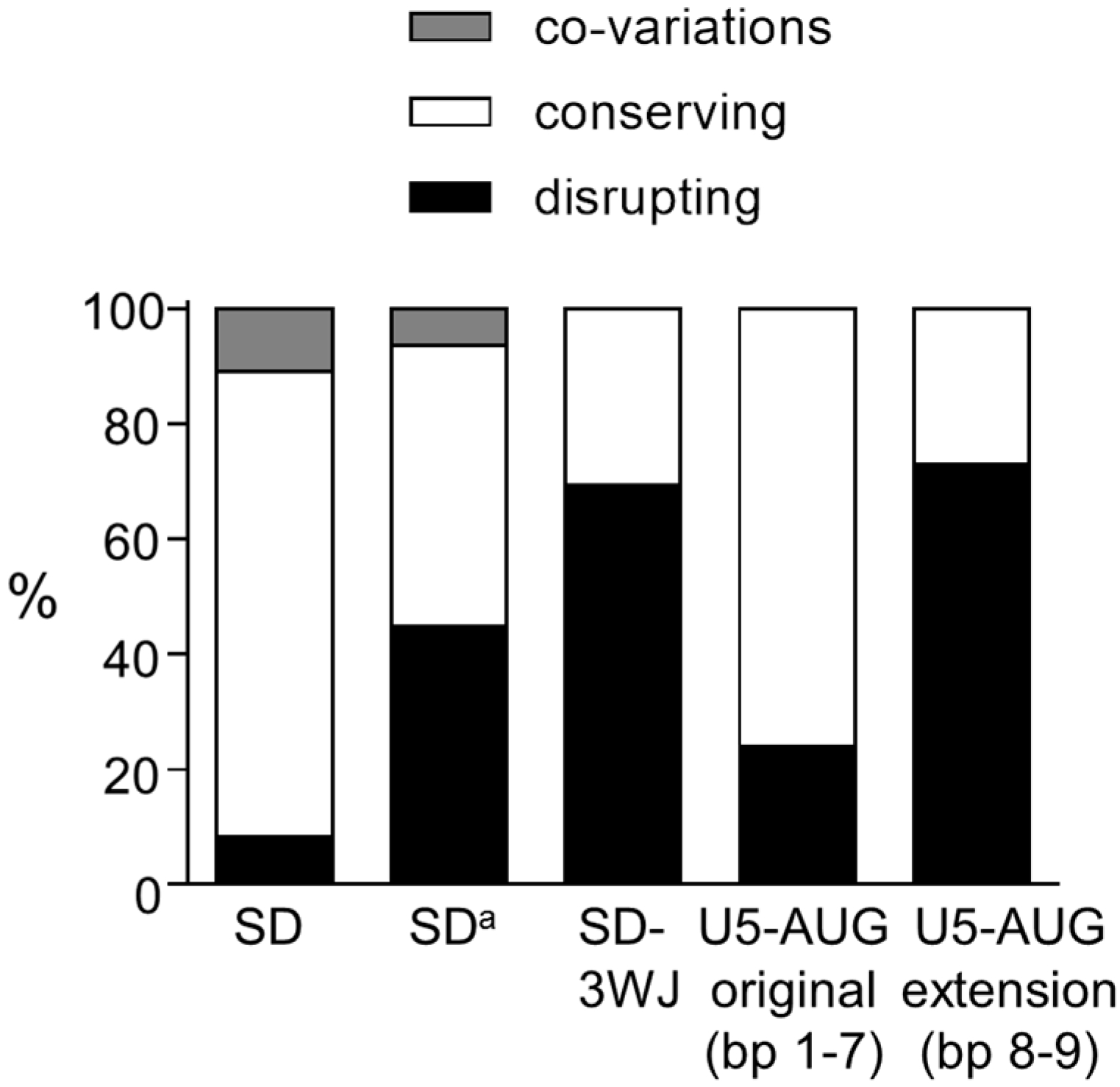
3.1. SD Phylogeny
3.2. SDa Phylogeny
3.3. 3WJ Phylogeny
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflict of Interest
References
- Huthoff, H.; Berkhout, B. Two alternating structures of the HIV-1 leader RNA. RNA 2001, 7, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Van Bel, N.; Das, A.T.; Berkhout, B. In vivo SELEX of single-stranded domains in the HIV-1 leader RNA. J. Virol. 2014, 88, 1870–1880. [Google Scholar] [CrossRef] [PubMed]
- Van Bel, N.; Das, A.T.; Cornelissen, M.; Abbink, T.E.; Berkhout, B. A short sequence motif in the 5′ leader of the HIV-1 genome modulates extended RNA dimer formation and virus replication. J. Biol. Chem. 2014, 289, 35061–35074. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Lavender, C.A.; Gorelick, R.J.; Weeks, K.M. Structure-Based Alignment and Consensus Secondary Structures for Three HIV-Related RNA Genomes. PLoS Comput. Biol. 2015, 11, e1004230. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, K.; Zambrano, N.; Baldwin, E.T.; Shapiro, B.A.; Erickson, J.W.; Omichinski, J.G.; Clore, G.M.; Gronenborn, A.M.; Appella, E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 5219–5223. [Google Scholar] [CrossRef] [PubMed]
- Harrison, G.P.; Miele, G.; Hunter, E.; Lever, A.M. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 1998, 72, 5886–5896. [Google Scholar] [PubMed]
- Pappalardo, L.; Kerwood, D.J.; Pelczer, I.; Borer, P.N. Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J. Mol. Biol. 1998, 282, 801–818. [Google Scholar] [CrossRef] [PubMed]
- Clever, J.; Sassetti, C.; Parslow, T.G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 1995, 69, 2101–2109. [Google Scholar] [PubMed]
- Amarasinghe, G.K.; De Guzman, R.N.; Turner, R.B.; Summers, M.F. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 2000, 299, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Pollom, E.; Dang, K.K.; Potter, E.L.; Gorelick, R.J.; Burch, C.L.; Weeks, K.M.; Swanstrom, R. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 2013, 9, e1003294. [Google Scholar] [CrossRef] [PubMed]
- Klaver, B.; Berkhout, B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994, 13, 2650–2659. [Google Scholar] [PubMed]
- Berkhout, B.; Schoneveld, I. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 1993, 21, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.; Berkhout, B. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J. Virol. 2008, 82, 3090–3098. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.; van Bel, N.; Berkhout, B.; Das, A.T. HIV-1 splicing at the major splice donor site is restricted by RNA structure. Virology 2014, 468–470, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.; Berkhout, B.; Das, A.T. HIV-1 splicing is controlled by local RNA structure and binding of splicing regulatory proteins at the major 5′ splice site. J. Gen. Virol. 2015, 96, 1906–1917. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.; Klaver, B.; Berkhout, B.; Das, A.T. Human immunodeficiency virus type 1 splicing at the major splice donor site is controlled by highly conserved RNA sequence and structural elements. J. Gen. Virol. 2015, 96, 3389–3395. [Google Scholar] [CrossRef] [PubMed]
- Solnick, D. Alternative splicing caused by RNA secondary structure. Cell. 1985, 43, 667–676. [Google Scholar] [CrossRef]
- Eperon, L.P.; Graham, I.R.; Griffiths, A.D.; Eperon, I.C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: Is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988, 54, 393–401. [Google Scholar] [CrossRef]
- Goguel, V.; Wang, Y.; Rosbash, M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol. Cell. Biol. 1993, 13, 6841–6848. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Goodall, G.J.; Kole, R.; Filipowicz, W. Effects of secondary structure on pre-mRNA splicing: Hairpins sequestering the 5′ but not the 3′ splice site inhibit intron processing in Nicotiana plumbaginifolia. EMBO J. 1995, 14, 377–388. [Google Scholar] [PubMed]
- Buratti, E.; Baralle, F.E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004, 24, 10505–10514. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Singh, R.N.; Androphy, E.J. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007, 35, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Shepard, P.J.; Hertel, K.J. Conserved RNA secondary structures promote alternative splicing. RNA 2008, 14, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Warf, M.B.; Berglund, J.A. Role of RNA structure in regulating pre-mRNA splicing. Trends. Biochem. Sci. 2010, 35, 169–178. [Google Scholar] [PubMed]
- McManus, C.J.; Graveley, B.R. RNA structure and the mechanisms of alternative splicing. Curr. Opin. Genet. Dev. 2011, 21, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Houlden, H.; Baker, M.; Adamson, J.; Lewis, J.; Prihar, G.; Pickering-Brown, S.; Duff, K.; Hutton, M. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 1999, 274, 15134–15143. [Google Scholar] [CrossRef] [PubMed]
- Hutton, M.; Lendon, C.L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A.; et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998, 393, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Murrell, J.R.; Goedert, M.; Farlow, M.R.; Klug, A.; Ghetti, B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. USA 1998, 95, 7737–7741. [Google Scholar] [CrossRef] [PubMed]
- Asang, C.; Erkelenz, S.; Schaal, H. The HIV-1 major splice donor D1 is activated by splicing enhancer elements within the leader region and the p17-inhibitory sequence. Virology 2012, 432, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Harrison, G.P.; Lever, A.M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 1992, 66, 4144–4153. [Google Scholar] [PubMed]
- Baudin, F.; Marquet, R.; Isel, C.; Darlix, J.L.; Ehresmann, B.; Ehresmann, C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 1993, 229, 382–397. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.; Berkhout, B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, C.K.; Andersen, E.S.; Knudsen, B.; Gorodkin, J.; Kjems, J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004, 336, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.; Ooms, M.; Haasnoot, P.C.; Berkhout, B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 2005, 44, 9058–9066. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Magrum, L.J.; Gupta, R.; Siegel, R.B.; Stahl, D.A.; Kop, J.; Crawford, N.; Brosius, J.; Gutell, R.; Hogan, J.J.; et al. Secondary structure model for bacterial 16S ribosomal RNA: Phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980, 8, 2275–2293. [Google Scholar] [CrossRef] [PubMed]
- Noller, H.F.; Kop, J.; Wheaton, V.; Brosius, J.; Gutell, R.R.; Kopylov, A.M.; Dohme, F.; Herr, W.; Stahl, D.A.; Gupta, R.; et al. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981, 9, 6167–6189. [Google Scholar] [CrossRef] [PubMed]
- Noller, H.F.; Woese, C.R. Secondary structure of 16S ribosomal RNA. Science 1981, 212, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Michel, F.; Jacquier, A.; Dujon, B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie 1982, 64, 867–881. [Google Scholar] [CrossRef]
- James, B.D.; Olsen, G.J.; Liu, J.S.; Pace, N.R. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988, 52, 19–26. [Google Scholar] [CrossRef]
- Michel, F.; Umesono, K.; Ozeki, H. Comparative and functional anatomy of group II catalytic introns—A review. Gene 1989, 82, 5–30. [Google Scholar] [CrossRef]
- Glotz, C.; Zwieb, C.; Brimacombe, R.; Edwards, K.; Kossel, H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981, 9, 3287–3306. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.C.; Prestwood, L.J.; Le Grice, S.F.; Lever, A.M. In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res. 2013, 41, e174. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Ooms, M.; Beerens, N.; Huthoff, H.; Southern, E.; Verhoef, K. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 2002, 277, 19967–19975. [Google Scholar] [CrossRef] [PubMed]
- Huthoff, H.; Berkhout, B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001, 29, 2594–2600. [Google Scholar] [CrossRef] [PubMed]
- Van Bel, N.; Ghabri, A.; Das, A.T.; Berkhout, B. The HIV-1 leader RNA is exquisitely sensitive to structural changes. Virology 2015, 483, 236–252. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B. Structural features in TAR RNA of human and simian immunodeficiency viruses: A phylogenetic analysis. Nucleic Acids Res. 1992, 20, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Vrolijk, M.M.; Ooms, M.; Harwig, A.; Das, A.T.; Berkhout, B. Destabilization of the TAR hairpin affects the structure and function of the HIV-1 leader RNA. Nucleic Acids Res. 2008, 36, 4352–4363. [Google Scholar] [CrossRef] [PubMed]
- Das, A.T.; Klaver, B.; Berkhout, B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 1999, 73, 81–91. [Google Scholar] [PubMed]
- Berkhout, B.; Klaver, B.; Das, A.T. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 1997, 25, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Klasens, B.I.; Das, A.T.; Berkhout, B. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 1998, 26, 1870–1876. [Google Scholar] [CrossRef] [PubMed]
- Huthoff, H.; Girard, F.; Wijmenga, S.S.; Berkhout, B. Evidence for a base triple in the free HIV-1 TAR RNA. RNA 2004, 10, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Erkelenz, S.; Theiss, S.; Otte, M.; Widera, M.; Peter, J.O.; Schaal, H. Genomic HEXploring allows landscaping of novel potential splicing regulatory elements. Nucleic Acids Res. 2014, 42, 10681–10697. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mueller, N.; Das, A.T.; Berkhout, B. A Phylogenetic Survey on the Structure of the HIV-1 Leader RNA Domain That Encodes the Splice Donor Signal. Viruses 2016, 8, 200. https://doi.org/10.3390/v8070200
Mueller N, Das AT, Berkhout B. A Phylogenetic Survey on the Structure of the HIV-1 Leader RNA Domain That Encodes the Splice Donor Signal. Viruses. 2016; 8(7):200. https://doi.org/10.3390/v8070200
Chicago/Turabian StyleMueller, Nancy, Atze T. Das, and Ben Berkhout. 2016. "A Phylogenetic Survey on the Structure of the HIV-1 Leader RNA Domain That Encodes the Splice Donor Signal" Viruses 8, no. 7: 200. https://doi.org/10.3390/v8070200




