Ferrets as a Novel Animal Model for Studying Human Respiratory Syncytial Virus Infections in Immunocompetent and Immunocompromised Hosts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Animal Care and Use
- Twice-daily oral dosing with immunosuppressive medication and daily collection of throat- and nose swabs without sedation;
- Inoculation and preparation for anesthesia: sedation with a mixture of ketamine and medetomidine (IM doses of 0.2 and 0.01 mL/kg body weight (bw), respectively). After inoculation the effect of the medetomidine was antagonized with atipamezole (0.005 mL/kg bw) upon completion of the procedure;
- Blood sampling: sedation with ketamine (intra-muscular [IM] dose of 0.2 mL/kg bw);
2.2. Cells and Virus
2.3. Immunosuppressive, Antibiotic, and Antiviral Drugs
2.4. Human Respiratory Syncytial Virus (HRSV) Infections of Cotton Rats
2.5. HRSV Infections of Ferrets: Study Design
2.6. Samples and Assays
3. Results
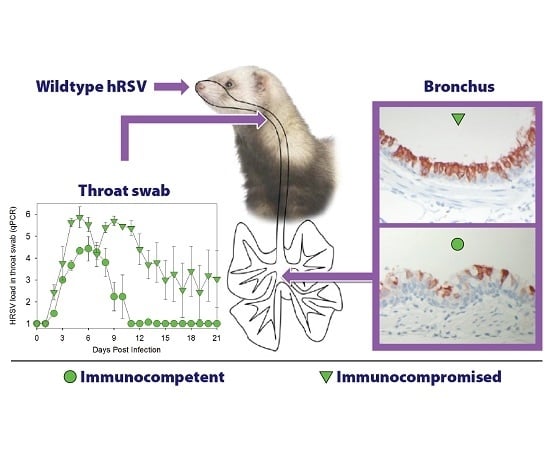
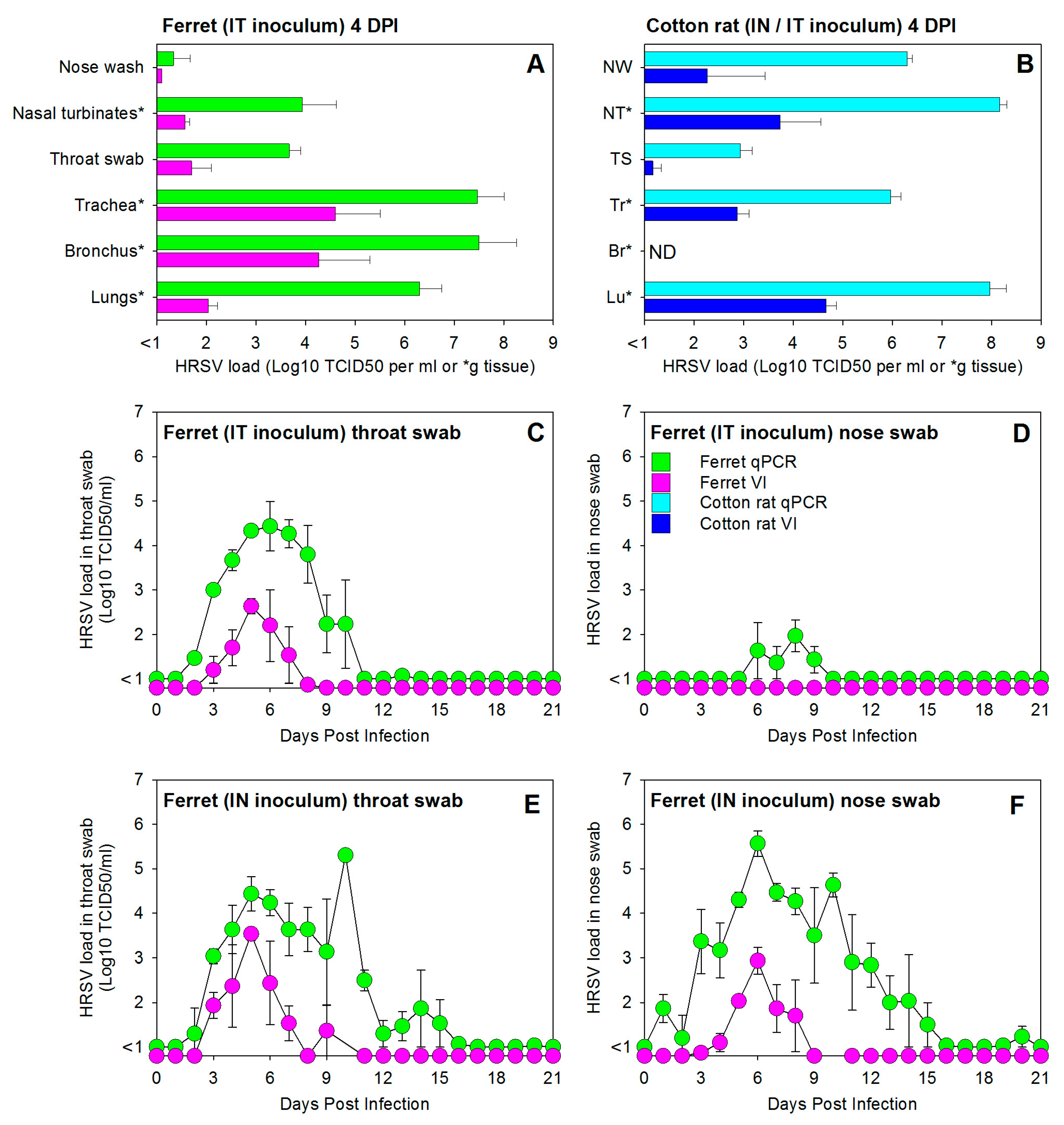
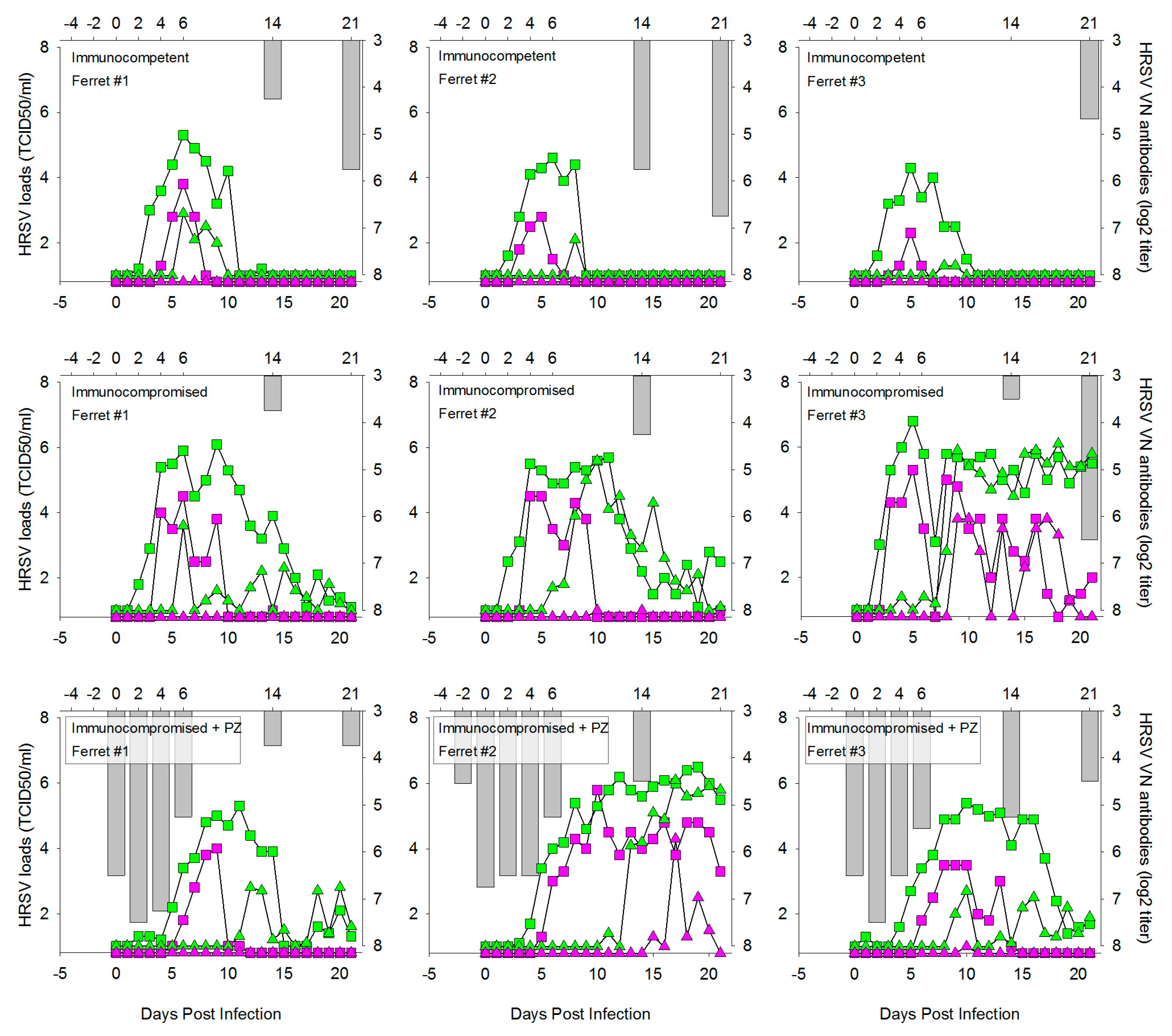
3.1. HRSV Infection of Immunocompetent Ferrets Results in Productive Virus Replication in the Upper and Lower Respiratory Tract
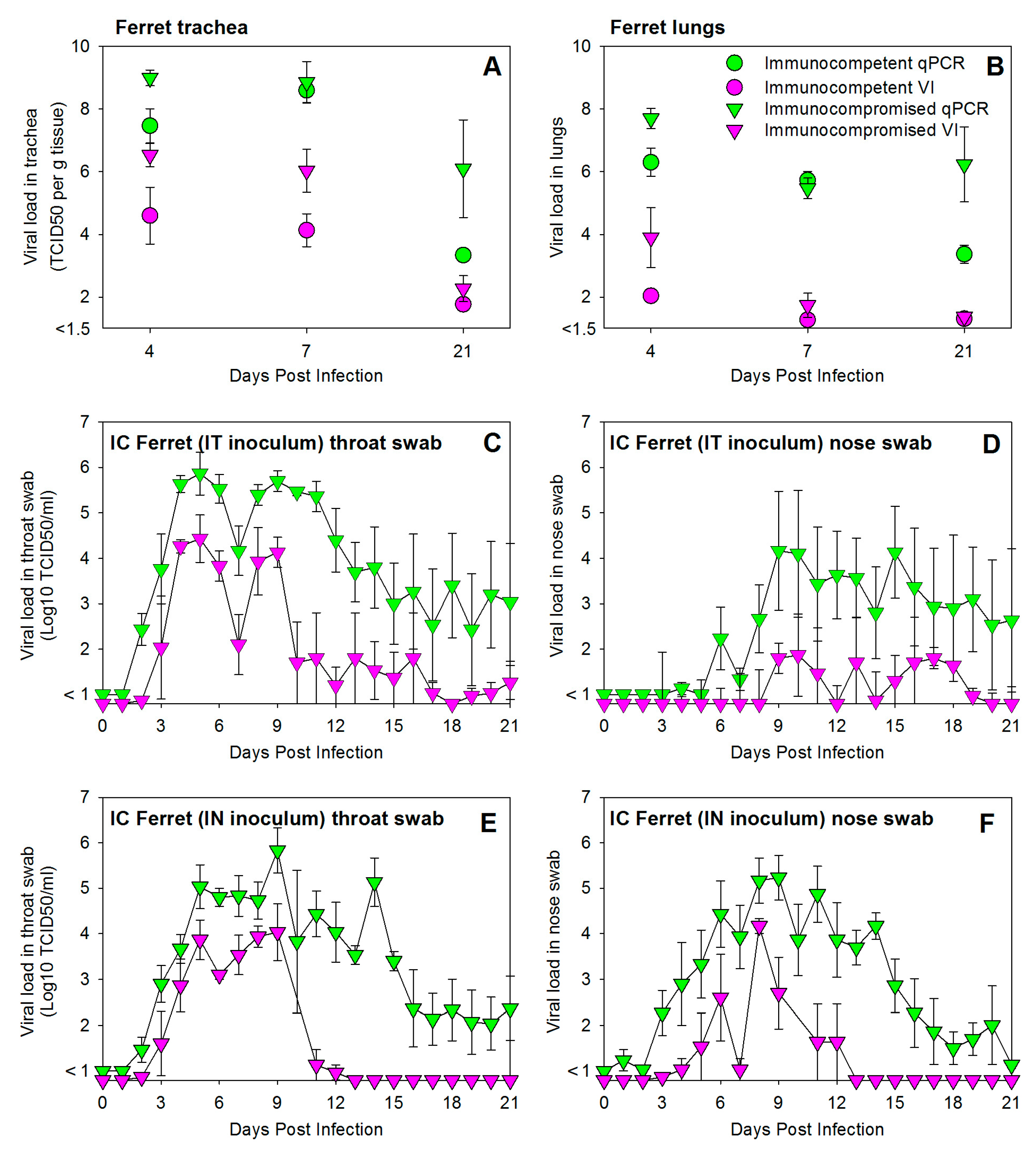
3.2. HRSV Infection of Immunocompromised Ferrets Results in Higher and Prolonged Virus Replication in the Upper and Lower Respiratory Tract
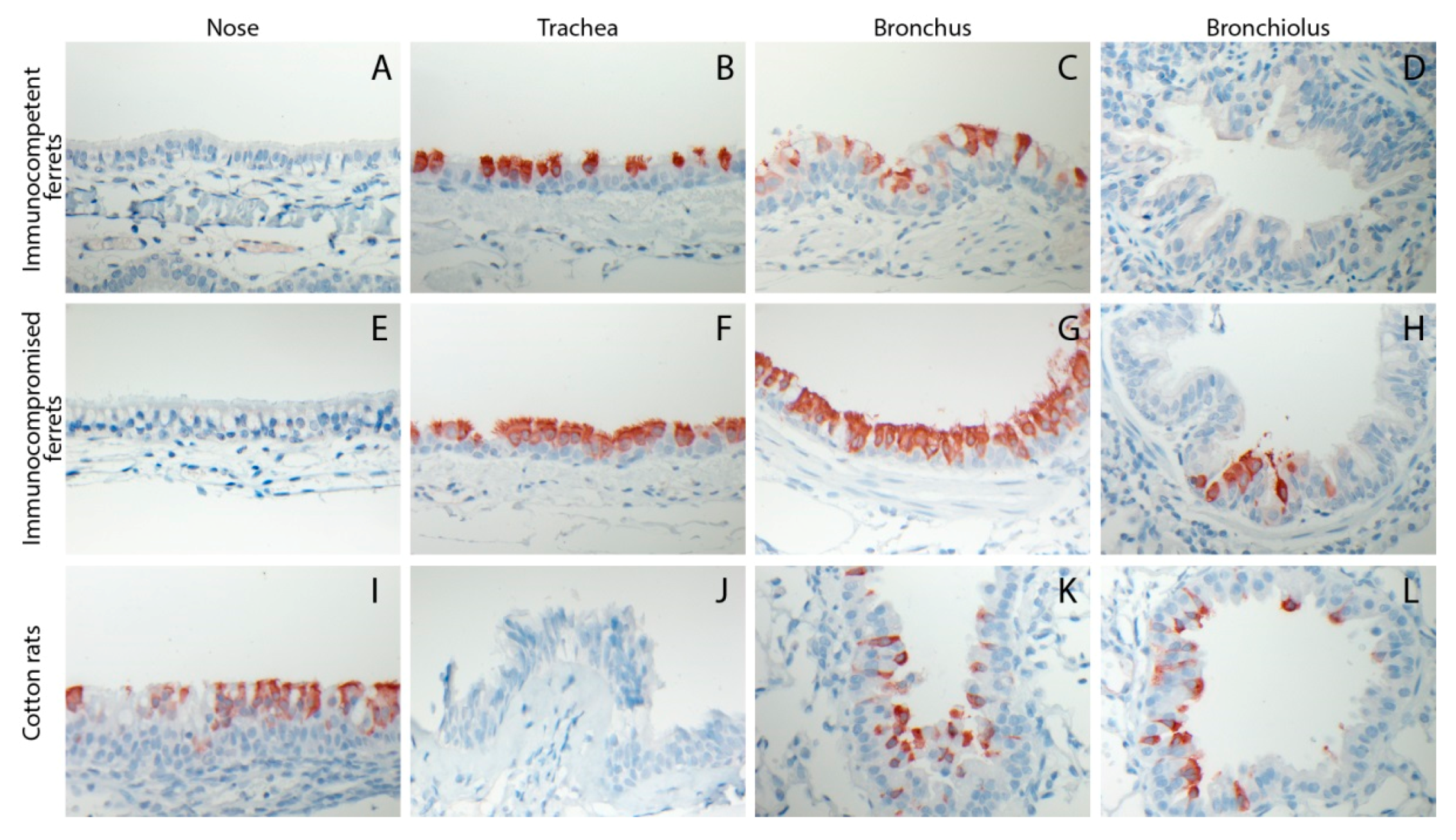
3.3. HRSV Replicates in Respiratory Epithelial Cells of the Ferret
3.4. Prophylactic Treatment of Immunocompromised Ferrets with Palivizumab Results in Temporary Inhibition of HRSV Replication
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Collins, P.L.; Karron, R.A. Respiratory syncytial virus and metapneumovirus. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer|Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 6, pp. 1086–1123. [Google Scholar]
- Mazur, N.I.; Martinon-Torres, F.; Baraldi, E.; Fauroux, B.; Greenough, A.; Heikkinen, T.; Manzoni, P.; Mejias, A.; Nair, H.; Papadopoulos, N.G.; et al. Lower respiratory tract infection caused by respiratory syncytial virus: Current management and new therapeutics. Lancet Respir. Med. 2015, 3, 888–900. [Google Scholar] [CrossRef]
- Falsey, A.R.; McElhaney, J.E.; Beran, J.; van Essen, G.A.; Duval, X.; Esen, M.; Galtier, F.; Gervais, P.; Hwang, S.J.; Kremsner, P.; et al. Respiratory syncytial virus and other respiratory viral infections in older adults with moderate to severe influenza-like illness. J. Infect. Dis. 2014, 209, 1873–1881. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.B.; Powell, K.R.; MacDonald, N.E.; Gala, C.L.; Menegus, M.E.; Suffin, S.C.; Cohen, H.J. Respiratory syncytial viral infection in children with compromised immune function. N. Engl. J. Med. 1986, 315, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Guthrie, K.A.; Waghmare, A.; Walsh, E.E.; Falsey, A.R.; Kuypers, J.; Cent, A.; Englund, J.A.; Boeckh, M. Respiratory syncytial virus in hematopoietic cell transplant recipients: Factors determining progression to lower respiratory tract disease. J. Infect. Dis. 2014, 209, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Englund, J.A.; Sullivan, C.J.; Jordan, M.C.; Dehner, L.P.; Vercellotti, G.M.; Balfour, H.H., Jr. Respiratory syncytial virus infection in immunocompromised adults. Ann. Intern. Med. 1988, 109, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Englund, J.; Feuchtinger, T.; Ljungman, P. Viral infections in immunocompromised patients. Biol. Blood Marrow Transplant. 2011, 17, S2–S5. [Google Scholar] [CrossRef] [PubMed]
- Van der Vries, E.; Stittelaar, K.J.; van Amerongen, G.; Veldhuis Kroeze, E.J.; de Waal, L.; Fraaij, P.L.; Meesters, R.J.; Luider, T.M.; van der Nagel, B.; Koch, B.; et al. Prolonged influenza virus shedding and emergence of antiviral resistance in immunocompromised patients and ferrets. PLoS Pathog. 2013, 9, e1003343. [Google Scholar] [CrossRef] [PubMed]
- De Lima, C.R.; Mirandolli, T.B.; Carneiro, L.C.; Tusset, C.; Romer, C.M.; Andreolla, H.F.; Baethgen, L.F.; Pasqualotto, A.C. Prolonged respiratory viral shedding in transplant patients. Transpl. Infect. Dis. 2014, 16, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Lehners, N.; Tabatabai, J.; Prifert, C.; Wedde, M.; Puthenparambil, J.; Weissbrich, B.; Biere, B.; Schweiger, B.; Egerer, G.; Schnitzler, P. Long-term shedding of influenza virus, parainfluenza virus, respiratory syncytial virus and nosocomial epidemiology in patients with hematological disorders. PLoS ONE 2016, 11, e0148258. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.P.; Ghantoji, S.S.; Ariza-Heredia, E.J.; Shah, J.N.; El Taoum, K.K.; Shah, P.K.; Nesher, L.; Hosing, C.; Rondon, G.; Champlin, R.E.; et al. Immunodeficiency scoring index to predict poor outcomes in hematopoietic cell transplant recipients with RSV infections. Blood 2014, 123, 3263–3268. [Google Scholar] [CrossRef] [PubMed]
- Kmeid, J.; Vanichanan, J.; Shah, D.P.; El Chaer, F.; Azzi, J.; Ariza-Heredia, E.J.; Hosing, C.; Mulanovich, V.; Chemaly, R.F. Outcomes of influenza infections in hematopoietic cell transplant recipients: Application of an immunodeficiency scoring index. Biol. Blood Marrow Transplant. 2016, 22, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Chemaly, R.F.; Shah, D.P.; Boeckh, M.J. Management of respiratory viral infections in hematopoietic cell transplant recipients and patients with hematologic malignancies. Clin. Infect. Dis. 2014, 59 (Suppl. S5), S344–S351. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, J.; Zamora, M.R.; Hodges, T.; Musk, A.W.; Sommerwerk, U.; Dilling, D.; Arcasoy, S.; DeVincenzo, J.; Karsten, V.; Shah, S.; et al. ALN-RSV01 for prevention of bronchiolitis obliterans syndrome after respiratory syncytial virus infection in lung transplant recipients. J. Heart Lung Transplant. 2016, 35, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Marcelin, J.R.; Wilson, J.W.; Razonable, R.R.; Mayo Clinic Hematology/Oncology; Transplant Infectious Diseases Services. Oral ribavirin therapy for respiratory syncytial virus infections in moderately to severely immunocompromised patients. Transpl. Infect. Dis. 2014, 16, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Beaird, O.E.; Freifeld, A.; Ison, M.G.; Lawrence, S.J.; Theodoropoulos, N.; Clark, N.M.; Razonable, R.R.; Alangaden, G.; Miller, R.; Smith, J.; et al. Current practices for treatment of respiratory syncytial virus and other non-influenza respiratory viruses in high-risk patient populations: A survey of institutions in the midwestern respiratory virus collaborative. Transpl. Infect. Dis. 2016, 18, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Simoes, E.A.; DeVincenzo, J.P.; Boeckh, M.; Bont, L.; Crowe, J.E., Jr.; Griffiths, P.; Hayden, F.G.; Hodinka, R.L.; Smyth, R.L.; Spencer, K.; et al. Challenges and opportunities in developing respiratory syncytial virus therapeutics. J. Infect. Dis. 2015, 211 (Suppl. S1), S1–S20. [Google Scholar] [CrossRef] [PubMed]
- Sacco, R.E.; Durbin, R.K.; Durbin, J.E. Animal models of respiratory syncytial virus infection and disease. Curr. Opin. Virol. 2015, 13, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R. Addressing a challenge with a challenge. Investigating respiratory syncytial virus immunity with the human challenge model. Am. J. Respir. Crit. Care Med. 2015, 191, 975–977. [Google Scholar] [CrossRef] [PubMed]
- DeVincenzo, J.P.; Whitley, R.J.; Mackman, R.L.; Scaglioni-Weinlich, C.; Harrison, L.; Farrell, E.; McBride, S.; Lambkin-Williams, R.; Jordan, R.; Xin, Y.; et al. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N. Engl. J. Med. 2014, 371, 711–722. [Google Scholar] [CrossRef] [PubMed]
- DeVincenzo, J.P.; McClure, M.W.; Symons, J.A.; Fathi, H.; Westland, C.; Chanda, S.; Lambkin-Williams, R.; Smith, P.; Zhang, Q.; Beigelman, L.; et al. Activity of oral ALS-008176 in a respiratory syncytial virus challenge study. N. Engl. J. Med. 2015, 373, 2048–2058. [Google Scholar] [CrossRef] [PubMed]
- Prince, G.A.; Porter, D.D. The pathogenesis of respiratory syncytial virus infection in infant ferrets. Am. J. Pathol. 1976, 82, 339–352. [Google Scholar] [PubMed]
- Lemon, K.; Nguyen, D.T.; Ludlow, M.; Rennick, L.J.; Yuksel, S.; van Amerongen, G.; McQuaid, S.; Rima, B.K.; de Swart, R.L.; Duprex, W.P. Recombinant subgroup b human respiratory syncytial virus expressing enhanced green fluorescent protein efficiently replicates in primary human cells and is virulent in cotton rats. J. Virol. 2015, 89, 2849–2856. [Google Scholar] [CrossRef] [PubMed]
- Munster, V.J.; de Wit, E.; van den Brand, J.M.; Herfst, S.; Schrauwen, E.J.; Bestebroer, T.M.; van de Vijver, D.; Boucher, C.A.; Koopmans, M.; Rimmelzwaan, G.F.; et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 2009, 325, 481–483. [Google Scholar] [CrossRef] [PubMed]
- De Waal, L.; Power, U.F.; Yuksel, S.; van Amerongen, G.; Nguyen, T.N.; Niesters, H.G.; de Swart, R.L.; Osterhaus, A.D. Evaluation of BBG2Na in infant macaques: Specific immune responses after vaccination and RSV challenge. Vaccine 2004, 22, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Power, U.F.; Nguyen, T.N.; Rietveld, E.; de Swart, R.L.; Groen, J.; Osterhaus, A.D.; de Groot, R.; Corvaia, N.; Beck, A.; Bouveret-Le-Cam, N.; et al. Safety and immunogenicity of a novel recombinant subunit respiratory syncytial virus vaccine (BBG2Na) in healthy young adults. J. Infect. Dis. 2001, 184, 1456–1460. [Google Scholar] [CrossRef] [PubMed]
- Homaira, N.; Rawlinson, W.; Snelling, T.L.; Jaffe, A. Effectiveness of palivizumab in preventing RSV hospitalization in high risk children: A real-world perspective. Int. J. Pediatr. 2014, 2014, 571609. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Gonzales, R.A.; Olson, S.J.; Wright, P.F.; Graham, B.S. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod. Pathol. 2007, 20, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Stokes, K.L.; Chi, M.H.; Sakamoto, K.; Newcomb, D.C.; Currier, M.G.; Huckabee, M.M.; Lee, S.; Goleniewska, K.; Pretto, C.; Williams, J.V.; et al. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J. Virol. 2011, 85, 5782–5793. [Google Scholar] [CrossRef] [PubMed]
- Villenave, R.; O’Donoghue, D.; Thavagnanam, S.; Touzelet, O.; Skibinski, G.; Heaney, L.G.; McKaigue, J.P.; Coyle, P.V.; Shields, M.D.; Power, U.F. Differential cytopathogenesis of respiratory syncytial virus prototypic and clinical isolates in primary pediatric bronchial epithelial cells. Virol. J. 2011, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Levitz, R.; Wattier, R.; Phillips, P.; Solomon, A.; Lawler, J.; Lazar, I.; Weibel, C.; Kahn, J.S. Induction of IL-6 and CCL5 (rantes) in human respiratory epithelial (A549) cells by clinical isolates of respiratory syncytial virus is strain specific. Virol. J. 2012, 9, 190. [Google Scholar] [CrossRef] [PubMed]
- Bem, R.A.; Domachowske, J.B.; Rosenberg, H.F. Animal models of human respiratory syncytial virus disease. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 301, L148–L156. [Google Scholar] [CrossRef] [PubMed]
- Van der Poel, W.H.M.; Schrijver, R.S.; Middel, W.G.J.; Kramps, J.A.; Brand, A.; Van Oirschot, J.T. Experimental reproduction of respiratory disease in calves with non-cell-culture-passaged bovine respiratory syncytial virus. Vet. Q. 1996, 18, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Woldehiwet, Z. Pathogenesis of bovine respiratory syncytial virus in lambs. Vet. Microbiol. 1990, 23, 267–272. [Google Scholar] [CrossRef]
- Openshaw, P.J.; Tregoning, J.S. Immune responses and disease enhancement during respiratory syncytial virus infection. Clin. Microbiol. Rev. 2005, 18, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Suffin, S.C.; Prince, G.A.; Muck, K.B.; Porter, D.D. Immunoprophylaxis of respiratory syncytial virus infection in the infant ferret. J. Immunol. 1979, 123, 10–14. [Google Scholar] [PubMed]
- Nguyen, D.T.; Louwen, R.; Elberse, K.; van Amerongen, G.; Yuksel, S.; Luijendijk, A.; Osterhaus, A.D.; Duprex, W.P.; de Swart, R.L. Streptococcus pneumoniae enhances human respiratory syncytial virus infection in vitro and in vivo. PLoS ONE 2015, 10, e0127098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridevaux, P.O.; Aubert, J.D.; Soccal, P.M.; Mazza-Stalder, J.; Berutto, C.; Rochat, T.; Turin, L.; Van Belle, S.; Nicod, L.; Meylan, P.; et al. Incidence and outcomes of respiratory viral infections in lung transplant recipients: A prospective study. Thorax 2014, 69, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Weigt, S.S.; Gregson, A.L.; Deng, J.C.; Lynch, J.P., 3rd; Belperio, J.A. Respiratory viral infections in hematopoietic stem cell and solid organ transplant recipients. Semin. Respir. Crit. Care Med. 2011, 32, 471–493. [Google Scholar] [CrossRef] [PubMed]
- Fraaij, P.L.; van der Vries, E.; Osterhaus, A.D. Reply to chan-tack et al. J. Infect. Dis. 2013, 207, 198–199. [Google Scholar] [CrossRef] [PubMed]
- Lehners, N.; Schnitzler, P.; Geis, S.; Puthenparambil, J.; Benz, M.A.; Alber, B.; Luft, T.; Dreger, P.; Eisenbach, C.; Kunz, C.; et al. Risk factors and containment of respiratory syncytial virus outbreak in a hematology and transplant unit. Bone Marrow Transplant. 2013, 48, 1548–1553. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.Y.; Renaud, C.; Ficken, E.; Thomson, B.; Kuypers, J.; Englund, J.A. Respiratory tract infections due to human metapneumovirus in immunocompromised children. J. Pediatr. Infect. Dis. Soc. 2014, 3, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Hellermann, G.R.; Patton, G.; Kumar, M.; Behera, A.; Randall, T.S.; Zhang, J.; Lockey, R.F.; Mohapatra, S.S. An immunocompromised BALB/c mouse model for respiratory syncytial virus infection. Virol. J. 2005, 2, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boukhvalova, M.; Blanco, J.C.; Falsey, A.R.; Mond, J. Treatment with novel RSV Ig RI-002 controls viral replication and reduces pulmonary damage in immunocompromised sigmodon hispidus. Bone Marrow Transplant. 2016, 51, 119–126. [Google Scholar] [CrossRef] [PubMed]
| Exp # | Group # | # Ferrets | Route | Dose | IC | PZ | Necropsy |
| 1 | 1 | 3 | IT | 105 TCID50 | No | No | 4 DPI |
| 1 | 2 | 3 | IT | 105 TCID50 | No | No | 21 DPI |
| 1 | 3 | 3 | IT | 105 TCID50 | Yes | No | 4 DPI |
| 1 | 4 | 3 | IT | 105 TCID50 | Yes | No | 21 DPI |
| 1 | 5 | 3 | IT | 105 TCID50 | Yes | Yes | 21 DPI |
| 2 | 6 | 3 | IT | 105 TCID50 | No | No | 7 DPI |
| 2 | 7 | 3 | IN | 105 TCID50 | No | No | 21 DPI |
| 2 | 8 | 3 | IT | 105 TCID50 | Yes | No | 7 DPI |
| 2 | 9 | 3 | IN | 105 TCID50 | Yes | No | 21 DPI |
| Exp # | Group # | # Cotton rats | Route | Dose | IC | PZ | Necropsy |
| 1 | 10 | 3 | IN * | 105 TCID50 | No | No | 4 DPI |
| 1 | 11 | 3 | IN * | 105 TCID50 | No | No | 21 DPI |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stittelaar, K.J.; De Waal, L.; Van Amerongen, G.; Veldhuis Kroeze, E.J.B.; Fraaij, P.L.A.; Van Baalen, C.A.; Van Kampen, J.J.A.; Van der Vries, E.; Osterhaus, A.D.M.E.; De Swart, R.L. Ferrets as a Novel Animal Model for Studying Human Respiratory Syncytial Virus Infections in Immunocompetent and Immunocompromised Hosts. Viruses 2016, 8, 168. https://doi.org/10.3390/v8060168
Stittelaar KJ, De Waal L, Van Amerongen G, Veldhuis Kroeze EJB, Fraaij PLA, Van Baalen CA, Van Kampen JJA, Van der Vries E, Osterhaus ADME, De Swart RL. Ferrets as a Novel Animal Model for Studying Human Respiratory Syncytial Virus Infections in Immunocompetent and Immunocompromised Hosts. Viruses. 2016; 8(6):168. https://doi.org/10.3390/v8060168
Chicago/Turabian StyleStittelaar, Koert J., Leon De Waal, Geert Van Amerongen, Edwin J.B. Veldhuis Kroeze, Pieter L.A. Fraaij, Carel A. Van Baalen, Jeroen J.A. Van Kampen, Erhard Van der Vries, Albert D.M.E. Osterhaus, and Rik L. De Swart. 2016. "Ferrets as a Novel Animal Model for Studying Human Respiratory Syncytial Virus Infections in Immunocompetent and Immunocompromised Hosts" Viruses 8, no. 6: 168. https://doi.org/10.3390/v8060168