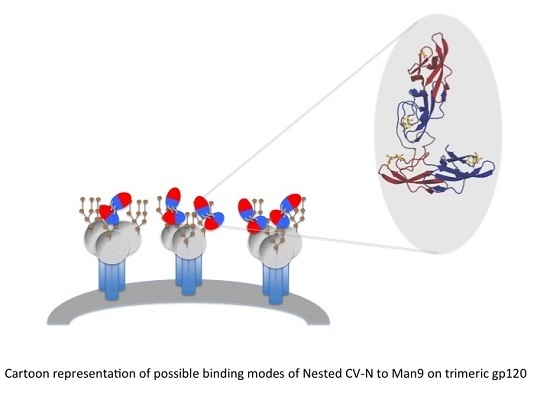
A Designed “Nested” Dimer of Cyanovirin-N Increases Antiviral Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
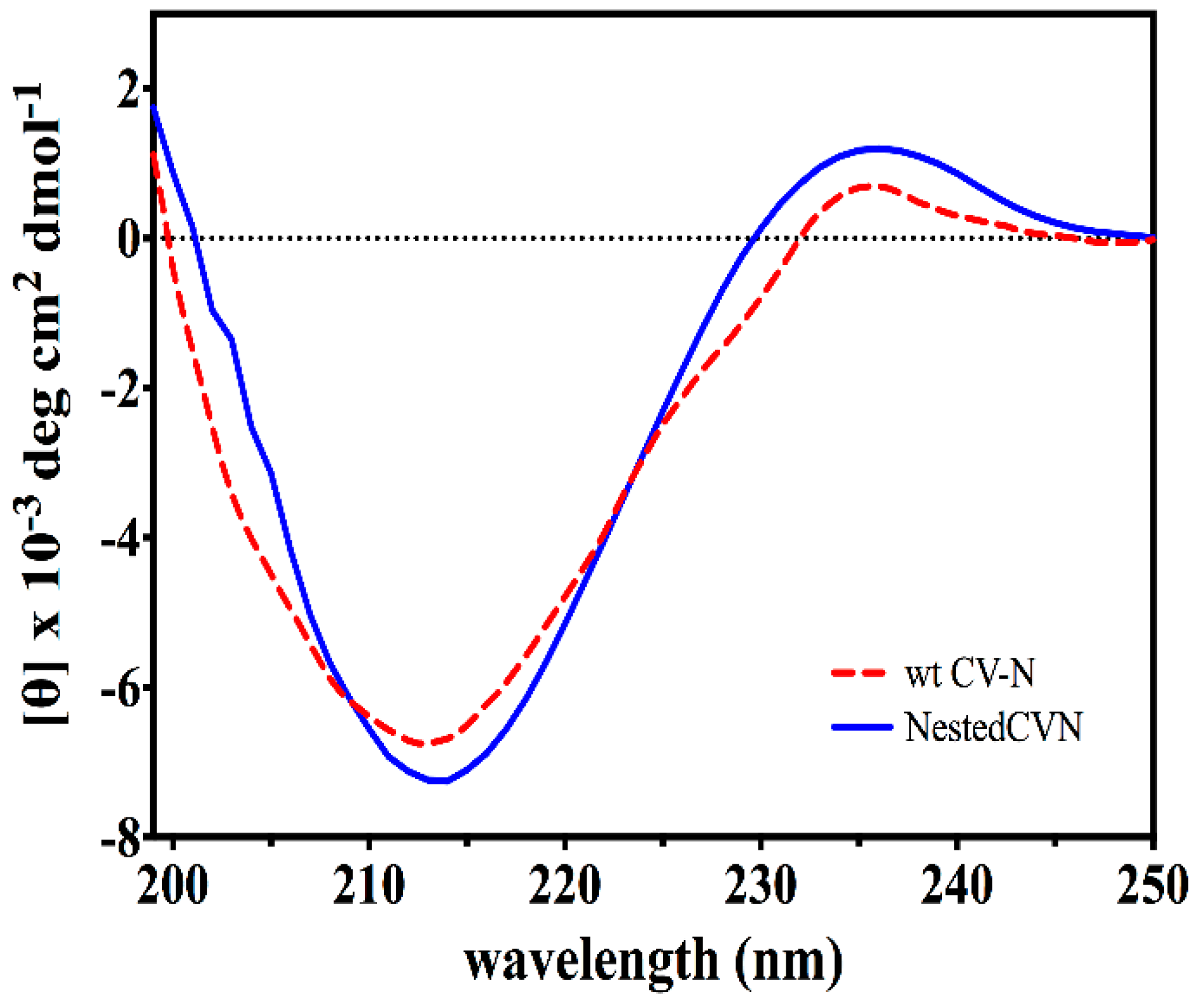
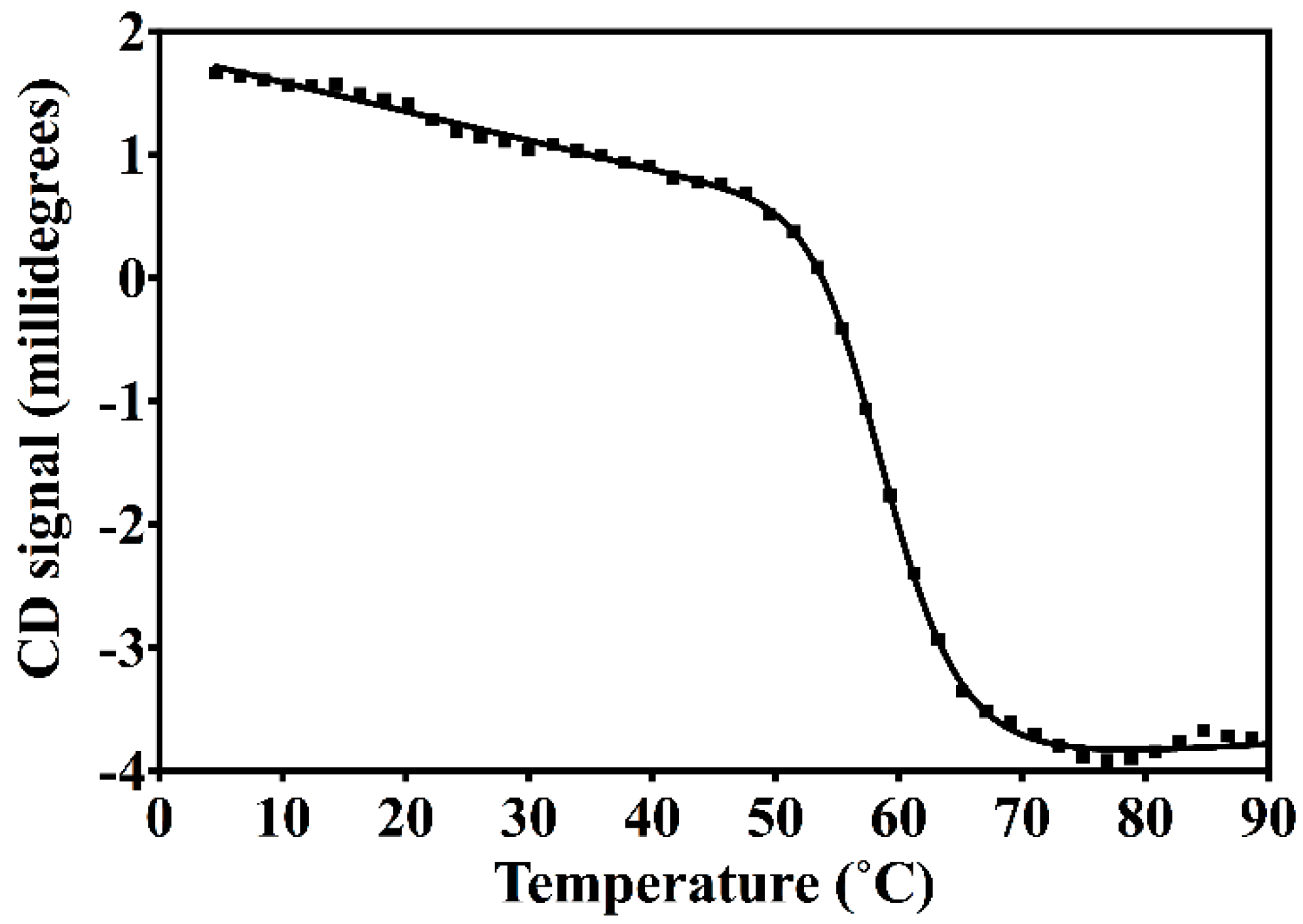
2.2. Circular Dichroism Spectroscopy (CD)
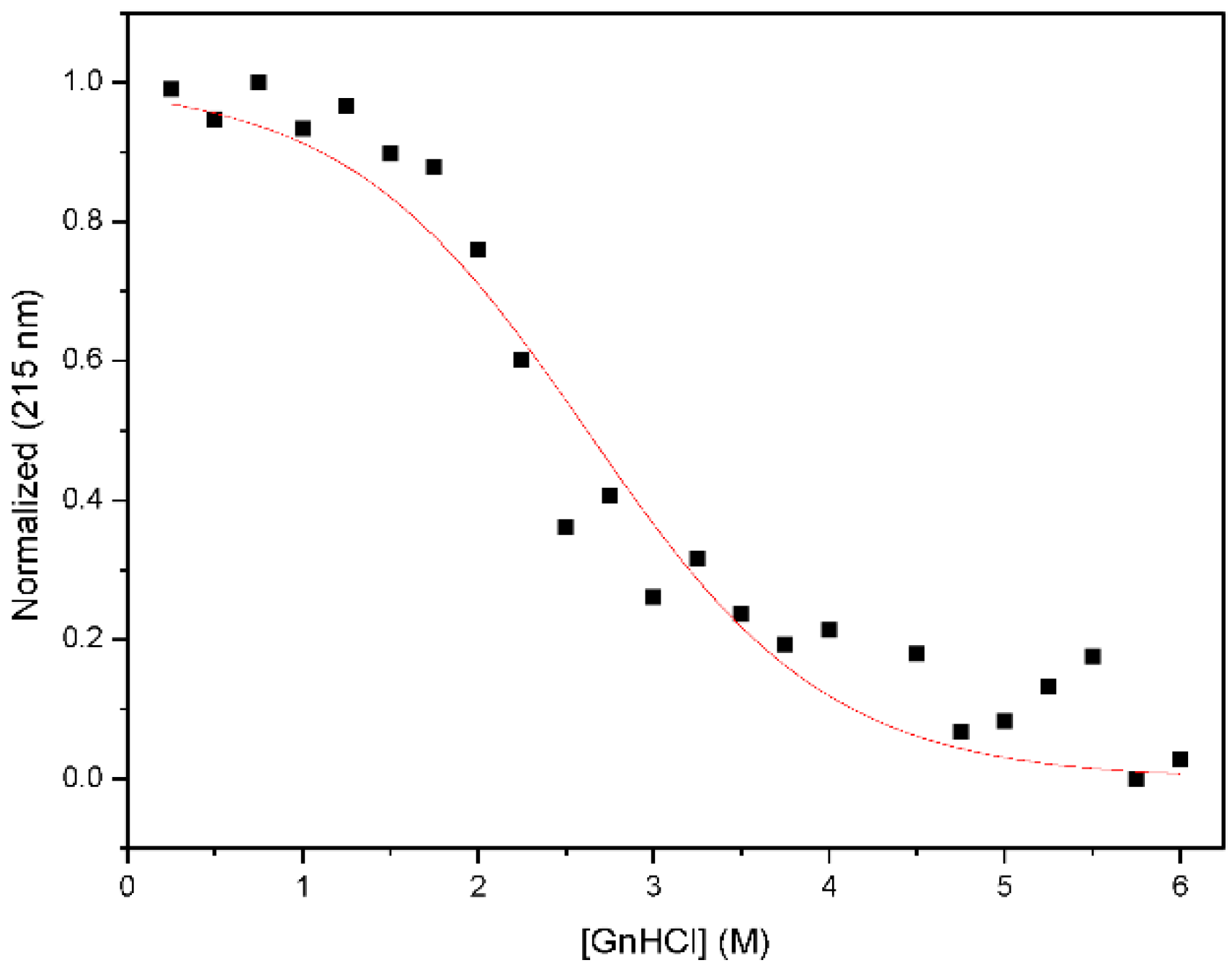
2.3. Chemical Denaturation Analysis
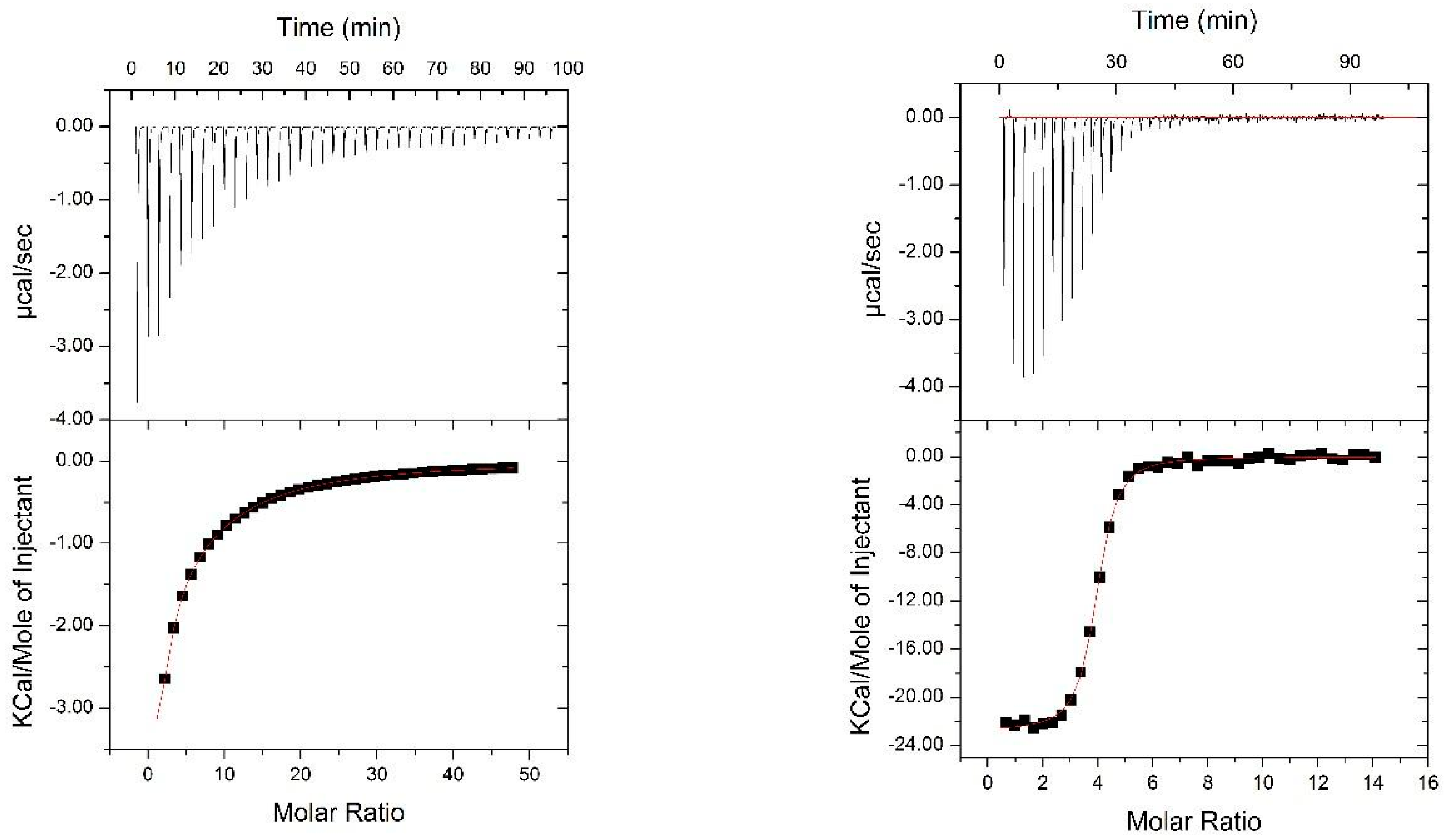
2.4. Isothermal Titration Calorimery
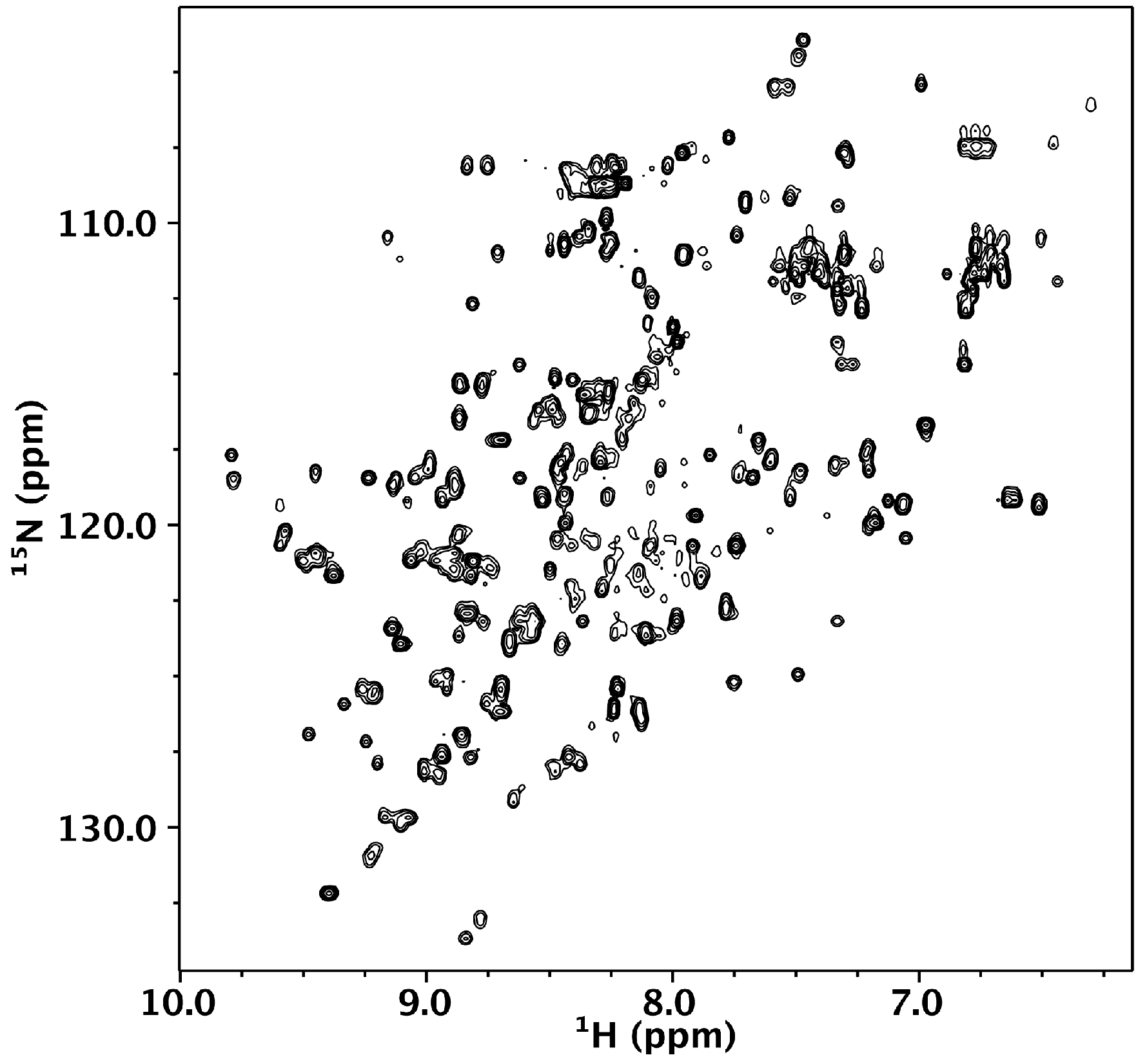
2.5. Nuclear Magnetic Resonance
2.6. Binding to gp120
2.7. Antiviral Assays
2.8. Cell Fusion Assay
3. Results
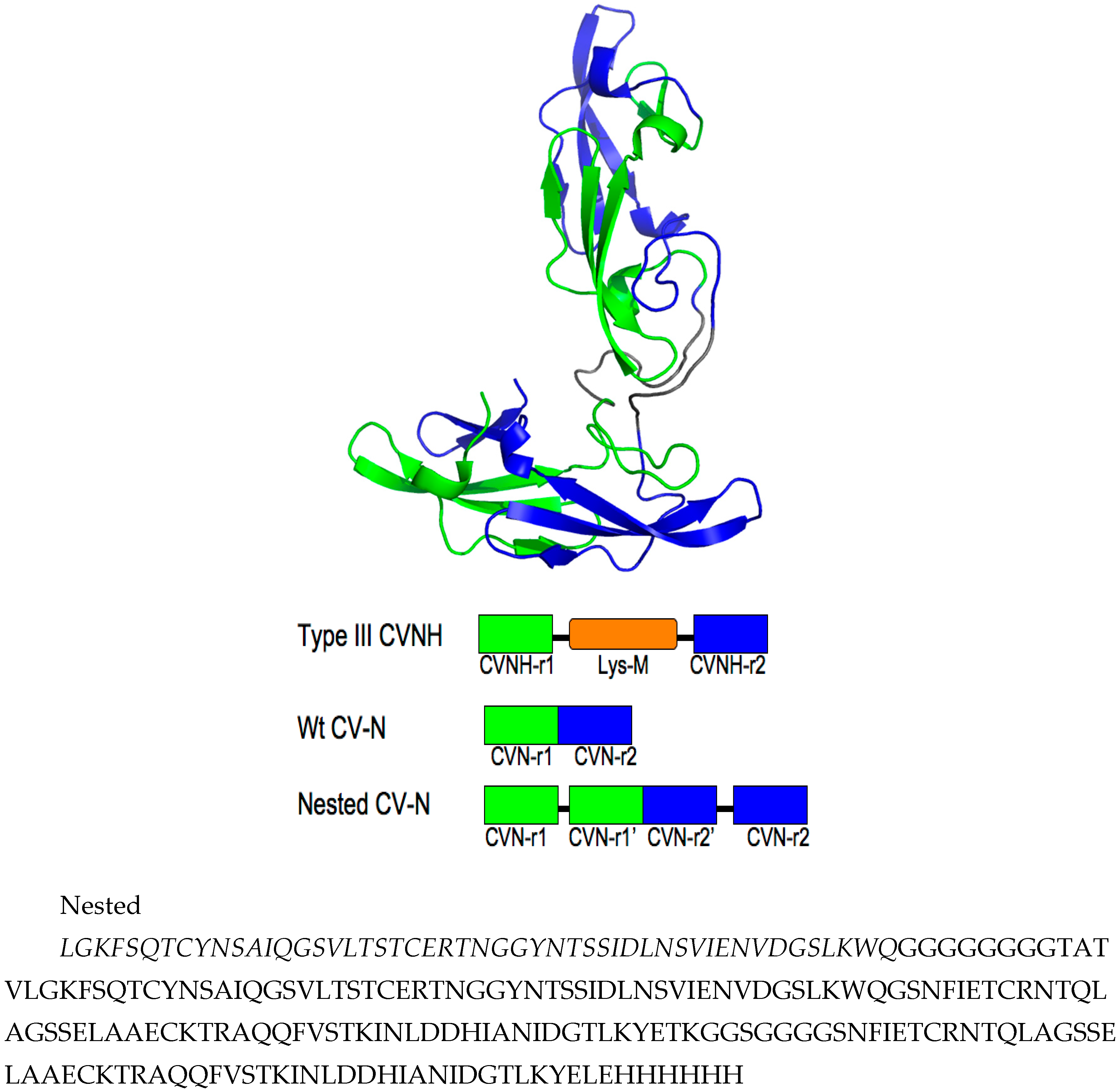
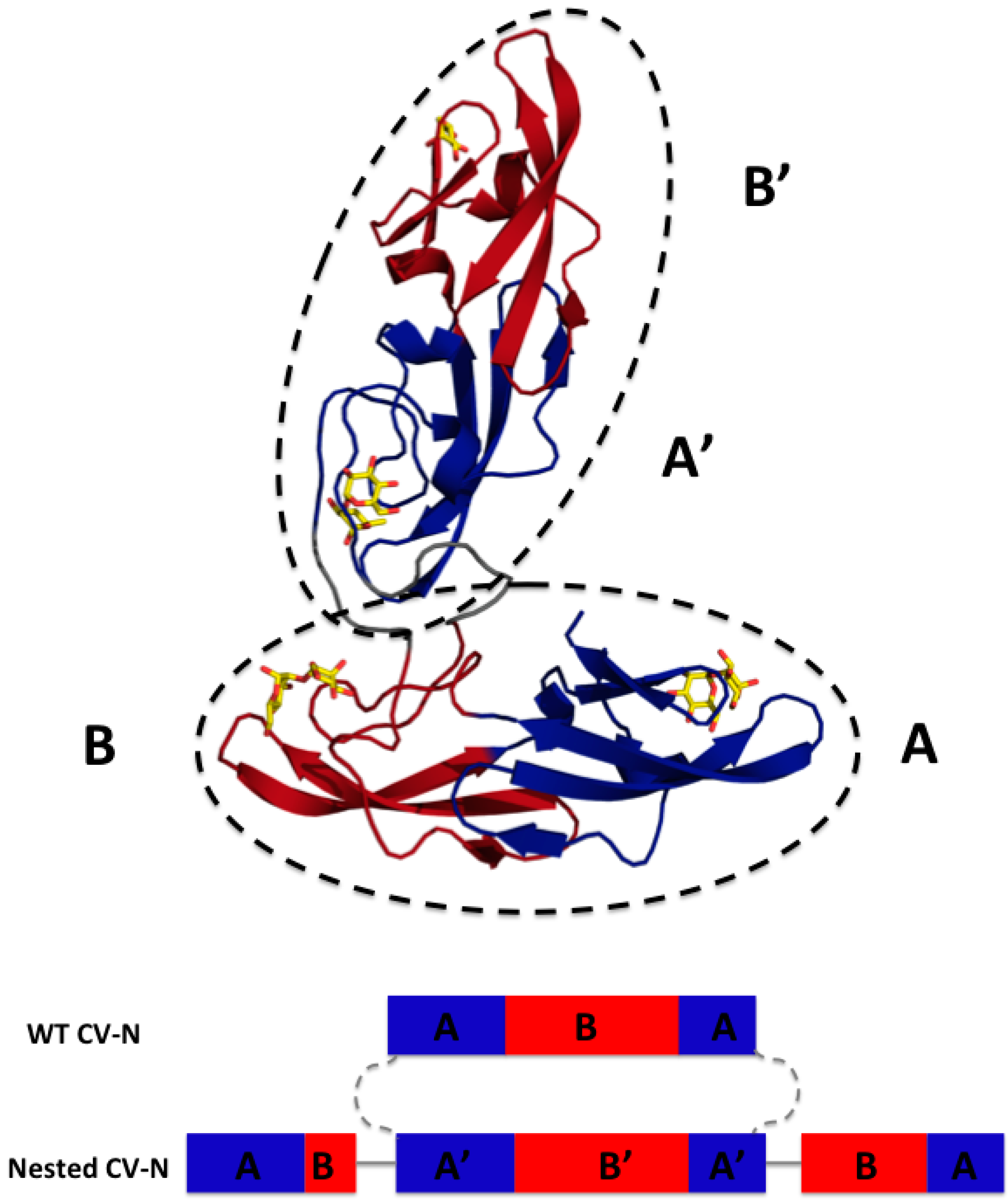
3.1. Design of Nested CV-N
3.2. Nested CV-N Adopts a Native-Like Fold
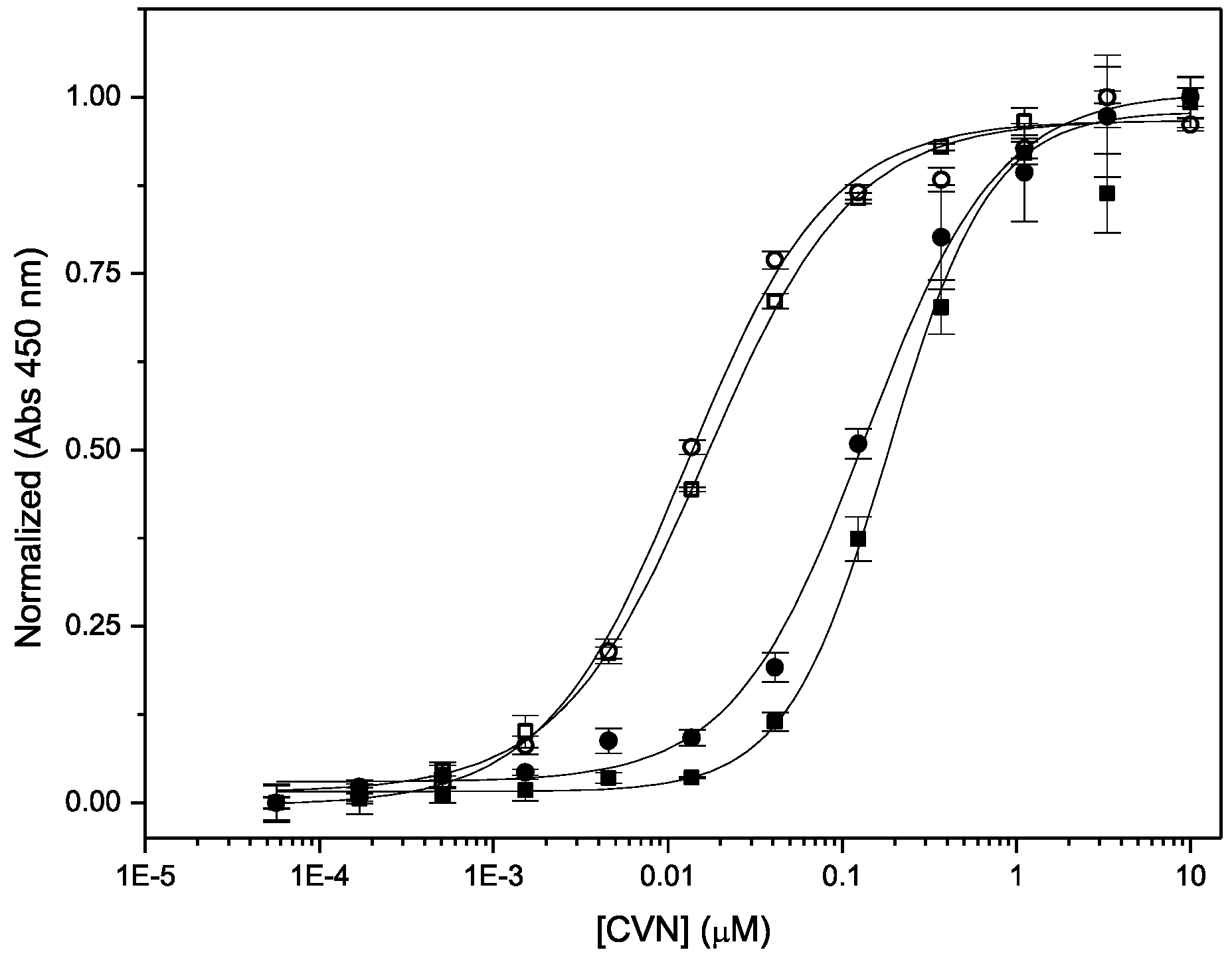
3.3. Binding to Glycans and Antiviral Activity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A

References
- O’Keefe, B.R.; Smee, D.F.; Turpin, J.A.; Saucedo, C.J.; Gustafson, K.R.; Mori, T.; Blakeslee, D.; Buckheit, R.; Boyd, M.R. Antimicrob. Agents Chemother. 2003, 47, 2518–2525.
- Helle, F.; Wychowski, C.; Vu-Dac, N.; Gustafson, K.R.; Voisset, C.; Dubuisson, J. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J. Biol. Chem. 2006, 281, 25177–25183. [Google Scholar] [CrossRef] [PubMed]
- Balzarini, J.; Van Laethem, K.; Hatse, S.; Froeyen, M.; Peumans, W.; Van Damme, E.; Schols, D. Carbohydrate-binding agents cause deletions of highly conserved glycosylation sites in HIV GP120: A new therapeutic concept to hit the achilles heel of HIV. J. Biol. Chem. 2005, 280, 41005–41014. [Google Scholar] [CrossRef] [PubMed]
- Balzarini, J. Carbohydrate-binding agents: A potential future cornerstone for the chemotherapy of enveloped viruses? Antivir. Chem. Chemother. 2007, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Koharudin, L.M.; Gronenborn, A.M. Antiviral lectins as potential HIV microbicides. Curr. Opin. Virol. 2014, 7, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Bewley, C.A.; Gustafson, K.R.; Boyd, M.R.; Covell, D.G.; Bax, A.; Clore, G.M.; Gronenborn, A.M. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat. Struct. Biol. 1998, 5, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Bewley, C.A. Solution structure of a cyanovirin-N:Man alpha 1-2Man alpha complex: Structural basis for high-affinity carbohydrate-mediated binding to gp120. Structure 2001, 9, 931–940. [Google Scholar] [CrossRef]
- Margulis, C.J. Computational study of the dynamics of mannose disaccharides free in solution and bound to the potent anti-HIV virucidal protein cyanovirin. J. Phys. Chem. B 2005, 109, 3639–3647. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.K.; Green, D.F. Carbohydrate recognition by the antiviral lectin cyanovirin-N. J. Am. Chem. Soc. 2012, 134, 19639–19651. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.G.; Matei, E.; Lasala, F.; Delgado, R.; Gronenborn, A.M. Dissecting carbohydrate-Cyanovirin-N binding by structure-guided mutagenesis: Functional implications for viral entry inhibition. Protein Eng. Des. Sel. 2006, 19, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Bolia, A.; Woodrum, B.W.; Cereda, A.; Ruben, M.A.; Wang, X.; Ozkan, S.B.; Ghirlanda, G. A flexible docking scheme efficiently captures the energetics of glycan-cyanovirin binding. Biophys. J. 2014, 106, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Vorontsov, I.I.; Miyashita, O. Solution and crystal molecular dynamics simulation study of m4-cyanovirin-N mutants complexed with di-mannose. Biophys. J. 2009, 97, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.K.; Terbush, R.N.; Patsalo, V.; Green, D.F. Computational models explain the oligosaccharide specificity of cyanovirin-N. Protein Sci. 2008, 17, 2008–2014. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.G.; Gronenborn, A.M. The domain-swapped dimer of cyanovirin-N contains two sets of oligosaccharide binding sites in solution. Biochem. Biophys. Res. Commun. 2002, 298, 598–602. [Google Scholar] [CrossRef]
- Yang, F.; Bewley, C.A.; Louis, J.M.; Gustafson, K.R.; Boyd, M.R.; Gronenborn, A.M.; Clore, G.M.; Wlodawer, A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J. Mol. Biol. 1999, 288, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Botos, I.; Mori, T.; Cartner, L.K.; Boyd, M.R.; Wlodawer, A. Domain-swapped structure of a mutant of cyanovirin-N. Biochem. Biophys. Res. Commun. 2002, 294, 184–190. [Google Scholar] [CrossRef]
- Ziolkowska, N.E.; O’Keefe, B.R.; Mori, T.; Zhu, C.; Giomarelli, B.; Vojdani, F.; Palmer, K.E.; McMahon, J.B.; Wlodawer, A. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure 2006, 14, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.G.; Lasala, F.; Delgado, R.; Sanchez, A.; Gronenborn, A.M. Flipping the switch from monomeric to dimeric CV-N has little effect on antiviral activity. Structure 2004, 12, 1799–1807. [Google Scholar] [CrossRef] [PubMed]
- Fromme, R.; Katiliene, Z.; Giomarelli, B.; Bogani, F.; Mahon, J.M.; Mori, T.; Fromme, P.; Ghirlanda, G. A Monovalent Mutant of Cyanovirin-N Provides Insight into the Role of Multiple Interactions with gp120 for Antiviral Activity. Biochemistry 2007, 46, 9199–9207. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S.R.; Barrientos, L.G.; Ratner, D.M.; O’Keefe, B.R.; Seeberger, P.H.; Gronenborn, A.M.; Boyd, M.R. Multisite and multivalent binding between cyanovirin-N and branched oligomannosides: Calorimetric and NMR characterization. Chem. Biol. 2002, 9, 1109–1118. [Google Scholar] [PubMed]
- Liu, Y.; Carroll, J.R.; Holt, L.A.; McMahon, J.; Giomarelli, B.; Ghirlanda, G. Multivalent interactions with gp120 are required for the anti-HIV activity of Cyanovirin. Biopolymers 2009, 92, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Keeffe, J.R.; Gnanapragasam, P.N.; Gillespie, S.K.; Yong, J.; Bjorkman, P.J.; Mayo, S.L. Designed oligomers of cyanovirin-N show enhanced HIV neutralization. Proc. Natl. Acad. Sci. USA 2011, 108, 14079–14084. [Google Scholar] [CrossRef] [PubMed]
- Koharudin, L.M.I.; Gronenborn, A.M. Sweet entanglements-protein: Glycan interactions in two HIV-inactivating lectin families. Biopolymers 2012, 9, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Matei, E.; Furey, W.; Gronenborn, A.M. Solution and crystal structures of a sugar binding site mutant of cyanovirin-N: No evidence of domain swapping. Structure 2008, 16, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Matei, E.; Zheng, A.; Furey, W.; Rose, J.; Aiken, C.; Gronenborn, A.M. Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization. J. Biol. Chem. 2010, 285, 13057–13065. [Google Scholar] [CrossRef] [PubMed]
- Woodrum, B.W.; Maxwell, J.D.; Bolia, A.; Ozkan, S.B.; Ghirlanda, G. The antiviral lectin cyanovirin-N: Probing multivalency and glycan recognition through experimental and computational approaches. Biochem. Soc. Trans. 2013, 41, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Fromme, R.; Katiliene, Z.; Fromme, P.; Ghirlanda, G. Conformational gating of dimannose binding to the antiviral protein cyanovirin revealed from the crystal structure at 1.35 A resolution. Protein Sci. 2008, 17, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Patsalo, V.; Raleigh, D.P.; Green, D.F. Rational and computational design of stabilized variants of cyanovirin-N that retain affinity and specificity for glycan ligands. Biochemistry 2011, 50, 10698–10712. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Blevins, R.A. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004, 278, 313–352. [Google Scholar] [PubMed]
- Boyd, M.R.; Gustafson, K.R.; McMahon, J.B.; Shoemaker, R.H.; O’Keefe, B.R.; Mori, T.; Gulakowski, R.J.; Wu, L.; Rivera, M.I.; Laurencot, C.M.; et al. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: Potential applications to microbicide development. Antimicrob. Agents Chemother. 1997, 41, 1521–1530. [Google Scholar] [PubMed]
- Shenoy, S.R.; O’Keefe, B.R.; Bolmstedt, A.J.; Cartner, L.K.; Boyd, M.R. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J. Pharmacol. Exp. Ther. 2001, 297, 704–710. [Google Scholar] [PubMed]
- Gulakowski, R.J.; McMahon, J.B.; Staley, P.G.; Moran, R.A.; Boyd, M.R. A semiautomated multiparameter approach for anti-HIV drug screening. J. Virol. Methods 1991, 33, 87–100. [Google Scholar] [CrossRef]
- Nussbaum, O.; Broder, C.C.; Berger, E.A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 1994, 68, 5411–5422. [Google Scholar] [PubMed]
- Alkhatib, G.; Broder, C.C.; Berger, E.A. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J. Virol. 1996, 70, 5487–5494. [Google Scholar] [PubMed]
- Fuerst, T.R.; Niles, E.G.; Studier, F.W.; Moss, B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 1986, 83, 8122–8126. [Google Scholar] [CrossRef] [PubMed]
- Broder, C.C.; Berger, E.A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc. Natl. Acad. Sci. USA 1995, 92, 9004–9008. [Google Scholar] [CrossRef] [PubMed]
- Koharudin, L.M.I.; Viscomi, A.R.; Montanini, B.; Kershaw, M.J.; Talbot, N.J.; Ottonello, S.; Gronenborn, A.M. Structure-Function Analysis of a CVNH-LysM Lectin Expressed during Plant Infection by the Rice Blast Fungus Magnaporthe oryzae. Structure 2011, 19, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Bewley, C.A.; Otero-Quintero, S. The Potent Anti-HIV Protein Cyanovirin-N Contains Two Novel Carbohydrate Binding Sites That Selectively Bind to Man 8D1D3 and Man 9with Nanomolar Affinity: Implications for Binding to the HIV Envelope Protein gp120. J. Am. Chem. Soc. 2001, 123, 3892–3902. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.G.; Louis, J.M.; Botos, I.; Mori, T.; Han, Z.; O’Keefe, B.R.; Boyd, M.R.; Wlodawer, A.; Gronenborn, A.M. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: Reconciliation of X-ray and NMR structures. Structure 2002, 10, 673–686. [Google Scholar] [CrossRef]
- Barrientos, L.G.; Louis, J.M.; Hung, J.; Smith, T.H.; O’Keefe, B.R.; Gardella, R.S.; Mori, T.; Boyd, M.R.; Gronenborn, A.M. Design and initial characterization of a circular permuted variant of the potent HIV-inactivating protein cyanovirin-N. Proteins 2002, 46, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, C.N.; Offer, J.; Zitzmann, N.; Dwek, R.A. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature 2007, 446, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Prien, J.M.; Ashline, D.J.; Lapadula, A.J.; Zhang, H. The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J. Am. Soc. Mass Spectrom. 2009, 20, 539–556. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, N.; Ohta, M.; Hyuga, S.; Hashimoto, O.; Hayakawa, T. Analysis of carbohydrate heterogeneity in a glycoprotein using liquid chromatography/mass spectrometry and liquid chromatography with tandem mass spectrometry. Anal. Biochem. 1999, 269, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Sandstrom, C.; Berteau, O.; Gemma, E.; Oscarson, S.; Kenne, L.; Gronenborn, A.M. Atomic mapping of the interactions between the antiviral agent cyanovirin-N and oligomannosides by saturation-transfer difference NMR. Biochemistry 2004, 43, 13926–13931. [Google Scholar] [CrossRef] [PubMed]
- Koharudin, L.M.I.; Furey, W.; Gronenborn, A.M. A designed chimeric cyanovirin-N homolog lectin: Structure and molecular basis of sucrose binding. Proteins 2009, 77, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Percudani, R.; Montanini, B.; Ottonello, S. The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes. Proteins 2005, 60, 670–678. [Google Scholar] [CrossRef] [PubMed]
| Protein | Binding to gp120 EC50 (nM) | Antiviral Activity EC50 (nM) | Fusion Inhibition IC50 (nM) |
|---|---|---|---|
| Nested CV-N | 1.29 ± 0.11 | 0.1 ± 0.02 | 7.5 ± 0.6 |
| WT CV-N | 1.66 ± 0.35 | 0.2 ± 0.04 | 18 ± 0.4 |
| WT CV-N (MTL) a | N/A | 0.5 ± 0.08 | |
| P51G-CVN | 4 b | 0.8 ± 0.04 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woodrum, B.W.; Maxwell, J.; Allen, D.M.; Wilson, J.; Krumpe, L.R.H.; Bobkov, A.A.; Hill, R.B.; Kibler, K.V.; O’Keefe, B.R.; Ghirlanda, G. A Designed “Nested” Dimer of Cyanovirin-N Increases Antiviral Activity. Viruses 2016, 8, 158. https://doi.org/10.3390/v8060158
Woodrum BW, Maxwell J, Allen DM, Wilson J, Krumpe LRH, Bobkov AA, Hill RB, Kibler KV, O’Keefe BR, Ghirlanda G. A Designed “Nested” Dimer of Cyanovirin-N Increases Antiviral Activity. Viruses. 2016; 8(6):158. https://doi.org/10.3390/v8060158
Chicago/Turabian StyleWoodrum, Brian W., Jason Maxwell, Denysia M. Allen, Jennifer Wilson, Lauren R.H. Krumpe, Andrey A. Bobkov, R. Blake Hill, Karen V. Kibler, Barry R. O’Keefe, and Giovanna Ghirlanda. 2016. "A Designed “Nested” Dimer of Cyanovirin-N Increases Antiviral Activity" Viruses 8, no. 6: 158. https://doi.org/10.3390/v8060158