Determinants of the Bovine Leukemia Virus Envelope Glycoproteins Involved in Infectivity, Replication and Pathogenesis
Abstract
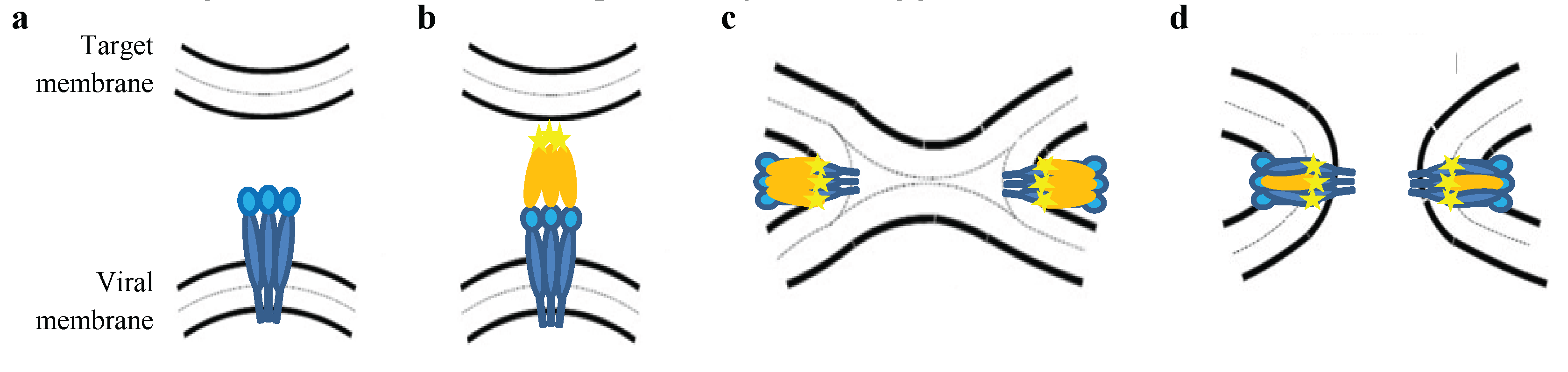
:1. Introduction
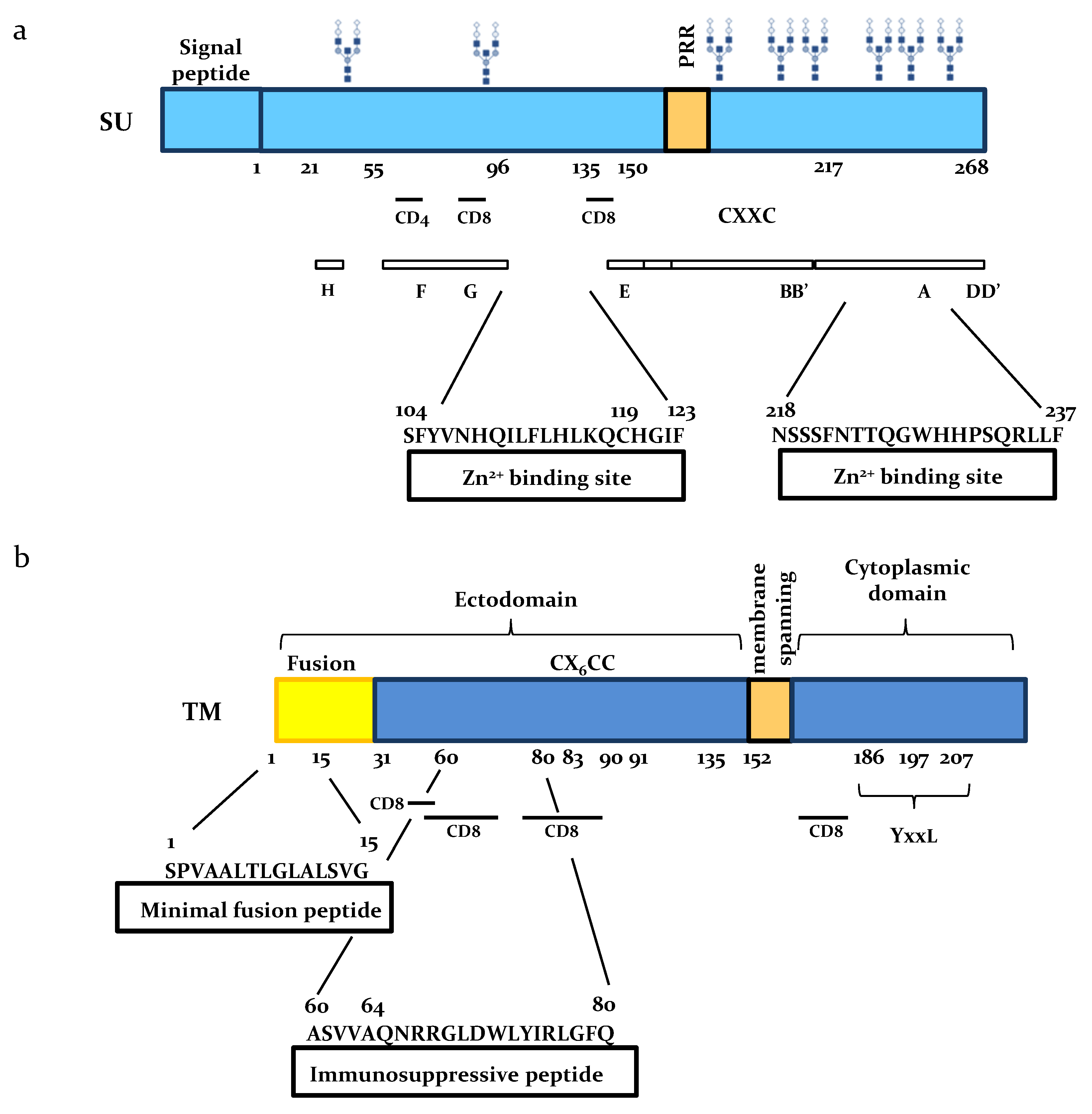
2. The SU Glycoprotein
2.1. SU Interacts with Zn
2.2. BLV SU is Immunogenic
2.3. Role of SU N-Linked Glycosylation
3. Functional Domains of the TM Subunit
3.1. The Ectodomain
3.2. The Cytoplasmic Tail
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wallin, M.; Ekström, M.; Garoff, H. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2004, 23, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Wallin, M.; Ekström, M.; Garoff, H. Receptor-triggered but alkylation-arrested env of murine leukemia virus reveals the transmembrane subunit in a prehairpin conformation. J. Virol. 2006, 80, 9921–9925. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, R.T.; Sodroski, J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science 1998, 280, 1884–1888. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, S.; Kronqvist, M.; Ekstrom, M.; Wallin, M.; Garoff, H. The conserved His8 of the Moloney murine leukemia virus Env SU subunit directs the activity of the SU-TM disulphide bond isomerase. Virology 2007, 361, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, G.; Rodríguez, S.M.; De Brogniez, A.; Gillet, N.; Golime, R.; Burny, A.; Jaworski, J.P.; Alvarez, I.; Vagnoni, L.; Trono, K.; et al. Vaccination against δ-retroviruses: The bovine leukemia virus paradigm. Viruses 2014, 6, 2416–2417. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.M.; Florins, A.; Gillet, N.; de Brogniez, A.; Sanchez-Alcaraz, M.T.; Boxus, M.; Boulanger, F.; Gutierrez, G.; Trono, K.; Alvarez, I.; et al. Preventive and therapeutic strategies for bovine leukemia virus: Lessons for HTLV. Viruses 2011, 3, 1210–1248. [Google Scholar] [CrossRef] [PubMed]
- Barez, P.Y.; de Brogniez, A.; Carpentier, A.; Gazon, H.; Gillet, N.; Gutiérrez, G.; Hamaidia, M.; Jacques, J.R.; Perike, S.; Neelature Sriramareddy, S.; et al. Recent Advances in BLV Research. Viruses 2015, 7, 6080–6088. [Google Scholar] [CrossRef] [PubMed]
- Florins, A.; Gillet, N.; Boxus, M.; Kerkhofs, P.; Kettmann, R.; Willems, L. Even attenuated bovine leukemia virus proviruses can be pathogenic in sheep. J. Virol. 2007, 81, 10195–10200. [Google Scholar] [CrossRef] [PubMed]
- Willems, L.; Kettmann, R.; Dequiedt, F.; Portetelle, D.; Vonèche, V.; Cornil, I.; Kerkhofs, P.; Burny, A.; Mammerickx, M. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 1993, 67, 4078–4085. [Google Scholar] [PubMed]
- Florins, A.; Gillet, N.; Asquith, B.; Boxus, M.; Burteau, C.; Twizere, J.C.; Urbain, P.; Vandermeers, F.; Debacq, C.; Sanchez-Alcaraz, M.T.; et al. Cell dynamics and immune response to BLV infection: A unifying model. Front. Biosci. 2007, 12, 1520–1531. [Google Scholar] [CrossRef] [PubMed]
- Willems, L.; Grimonpont, C.; Heremans, H.; Rebeyrotte, N.; Chen, G.; Portetelle, D.; Burny, A.; Kettmann, R. Mutations in the bovine leukemia virus Tax protein can abrogate the long terminal repeat-directed transactivating activity without concomitant loss of transforming potential. Proc. Natl. Acad. Sci. USA 1992, 89, 3957–3961. [Google Scholar] [CrossRef] [PubMed]
- Twizere, J.C.; Kruys, V.; Lefebvre, L.; Vanderplasschen, A.; Collete, D.; Debacq, C.; Lai, W.S.; Jauniaux, J.C.; Bernstein, L.R.; Semmes, O.J.; et al. Interaction of retroviral Tax oncoproteins with tristetraprolin and regulation of tumor necrosis factor-alpha expression. J. Natl. Cancer Inst. 2003, 95, 1846–1859. [Google Scholar] [CrossRef] [PubMed]
- Willems, L.; Kettmann, R.; Chen, G.; Portetelle, D.; Burny, A.; Derse, D. A cyclic AMP-responsive DNA-binding protein (CREB2) is a cellular transactivator of the bovine leukemia virus long terminal repeat. J. Virol. 1992, 66, 766–772. [Google Scholar] [PubMed]
- Kettmann, R.; Burny, A. Bovine Leukemia Virus. In The Retroviridae; Levy, J.A., Ed.; Plenum Press: New York, NY, USA, 1994; Volume 3, pp. 39–81. [Google Scholar]
- Gatot, J.S.; Callebaut, I.; Van Lint, C.; Demonté, D.; Kerkhofs, P.; Portetelle, D.; Burny, A.; Willems, L.; Kettmann, R. Bovine leukemia virus SU protein interacts with zinc, and mutations within two interacting regions differently affect viral fusion and infectivity in vivo. J. Virol. 2002, 76, 7956–7967. [Google Scholar] [CrossRef] [PubMed]
- Zarkik, S.; Defrise-Quertain, F.; Portetelle, D.; Burny, A.; Ruysschaert, J.M. Fusion of bovine leukemia virus with target cells monitored by R18 fluorescence and PCR assays. J. Virol. 1997, 71, 738–740. [Google Scholar] [PubMed]
- Bruck, C.; Mathot, S.; Portetelle, D.; Berte, C.; Franssen, J.D.; Herion, P.; Burny, A. Monoclonal antibodies define eight independent antigenic regions on the bovine leukemia virus (BLV) envelope glycoprotein gp51. Virology 1982, 122, 342–352. [Google Scholar] [CrossRef]
- Bai, L.; Takeshima, S.; Isogai, E.; Kohara, J.; Aida, Y. Novel CD8+ cytotoxic T cell epitopes in bovine leukemia virus with cattle. Vaccine 2015, 33, 7194–7202. [Google Scholar] [CrossRef] [PubMed]
- Mamoun, R.Z.; Morisson, M.; Rebeyrotte, N.; Busetta, B.; Couez, D.; Kettmann, R.; Hospital, M.; Guillemain, B. Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J. Virol. 1990, 64, 4180–4188. [Google Scholar] [PubMed]
- Pikora, C.A. Glycosylation of the ENV spike of primate immunodeficiency viruses and antibody neutralization. Curr. HIV Res. 2004, 2, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, H.C.; Matreyek, K.A.; Filone, C.M.; Hashimi, S.T.; Levroney, E.L.; Negrete, O.A.; Bertolotti-Ciarlet, A.; Choi, D.Y.; McHardy, I.; Fulcher, J.A.; et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 2006, 80, 4878–4889. [Google Scholar] [CrossRef] [PubMed]
- Von V, M.; Cattaneo, R. N-linked glycans with similar location in the fusion protein head modulate paramyxovirus fusion. J. Virol. 2003, 77, 10202–10212. [Google Scholar]
- Wei, X.; Decker, J.M.; Wang, S.; Hui, H.; Kappes, J.C.; Wu, X.; Salazar-Gonzalez, J.F.; Salazar, M.G.; Kilby, J.M.; Saag, M.S.; et al. Antibody neutralization and escape by HIV-1. Nature 2003, 422, 307–312. [Google Scholar] [CrossRef] [PubMed]
- De Brogniez, A.; Bouzar, A.B.; Jacques, J.R.; Cosse, J.P.; Gillet, N.; Callebaut, I.; Reichert, M.; Willems, L. Mutation of a Single Envelope N-Linked Glycosylation Site Enhances the Pathogenicity of Bovine Leukemia Virus. J. Virol. 2015, 89, 8945–8956. [Google Scholar] [CrossRef] [PubMed]
- Vonèche, V.; Portetelle, D.; Kettmann, R.; Willems, L.; Limbach, K.; Paoletti, E.; Ruysschaert, J.M.; Burny, A.; Brasseur, R. Fusogenic segments of bovine leukemia virus and simian immunodeficiency virus are interchangeable and mediate fusion by means of oblique insertion in the lipid bilayer of their target cells. Proc. Natl. Acad. Sci. USA 1992, 89, 3810–3814. [Google Scholar] [CrossRef] [PubMed]
- Gatot, J.S.; Callebaut, I.; Mornon, J.P.; Portetelle, D.; Burny, A.; Kerkhofs, P.; Kettmann, R.; Willems, L. Conservative mutations in the immunosuppressive region of the bovine leukemia virus transmembrane protein affect fusion but not infectivity in vivo. J. Biol. Chem. 1998, 273, 12870–12880. [Google Scholar] [CrossRef] [PubMed]
- Lorin, A.; Lins, L.; Stroobant, V.; Brasseur, R.; Charloteaux, B. Determination of the minimal fusion peptide of bovine leukemia virus gp30. Biochem. Biophys. Res. Commun. 2007, 355, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Beaufils, P.; Choquet, D.; Mamoun, R.Z.; Malissen, B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993, 12, 5105–5112. [Google Scholar] [PubMed]
- Willems, L.; Gatot, J.S.; Mammerickx, M.; Portetelle, D.; Burny, A.; Kerkhofs, P.; Kettmann, R. The YXXL signalling motifs of the bovine leukemia virus transmembrane protein are required for in vivo infection and maintenance of high viral loads. J. Virol. 1995, 69, 4137–4141. [Google Scholar] [PubMed]
- Inabe, K.; Nishizawa, M.; Tajima, S.; Ikuta, K.; Aida, Y. The YXXL sequences of a transmembrane protein of bovine leukemia virus are required for viral entry and incorporation of viral envelope protein into virions. J. Virol. 1999, 73, 1293–1301. [Google Scholar] [PubMed]
- Sagata, N.; Yasunaga, T.; Tsuzuku-Kawamura, J.; Ohishi, K.; Ogawa, Y.; Ikawa, Y. Complete nucleotide sequence of the genome of bovine leukemia virus: Its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 1985, 82, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Willems, L.; Thienpont, E.; Kerkhofs, P.; Burny, A.; Mammerickx, M.; Kettmann, R. Bovine leukemia virus, an animal model for the study of intrastrain variability. J. Virol. 1993, 67, 1086–1089. [Google Scholar] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Brogniez, A.; Mast, J.; Willems, L. Determinants of the Bovine Leukemia Virus Envelope Glycoproteins Involved in Infectivity, Replication and Pathogenesis. Viruses 2016, 8, 88. https://doi.org/10.3390/v8040088
De Brogniez A, Mast J, Willems L. Determinants of the Bovine Leukemia Virus Envelope Glycoproteins Involved in Infectivity, Replication and Pathogenesis. Viruses. 2016; 8(4):88. https://doi.org/10.3390/v8040088
Chicago/Turabian StyleDe Brogniez, Alix, Jan Mast, and Luc Willems. 2016. "Determinants of the Bovine Leukemia Virus Envelope Glycoproteins Involved in Infectivity, Replication and Pathogenesis" Viruses 8, no. 4: 88. https://doi.org/10.3390/v8040088




