RNA-Sequencing Reveals the Progression of Phage-Host Interactions between φR1-37 and Yersinia enterocolitica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Phage Propagation, and Growth Conditions
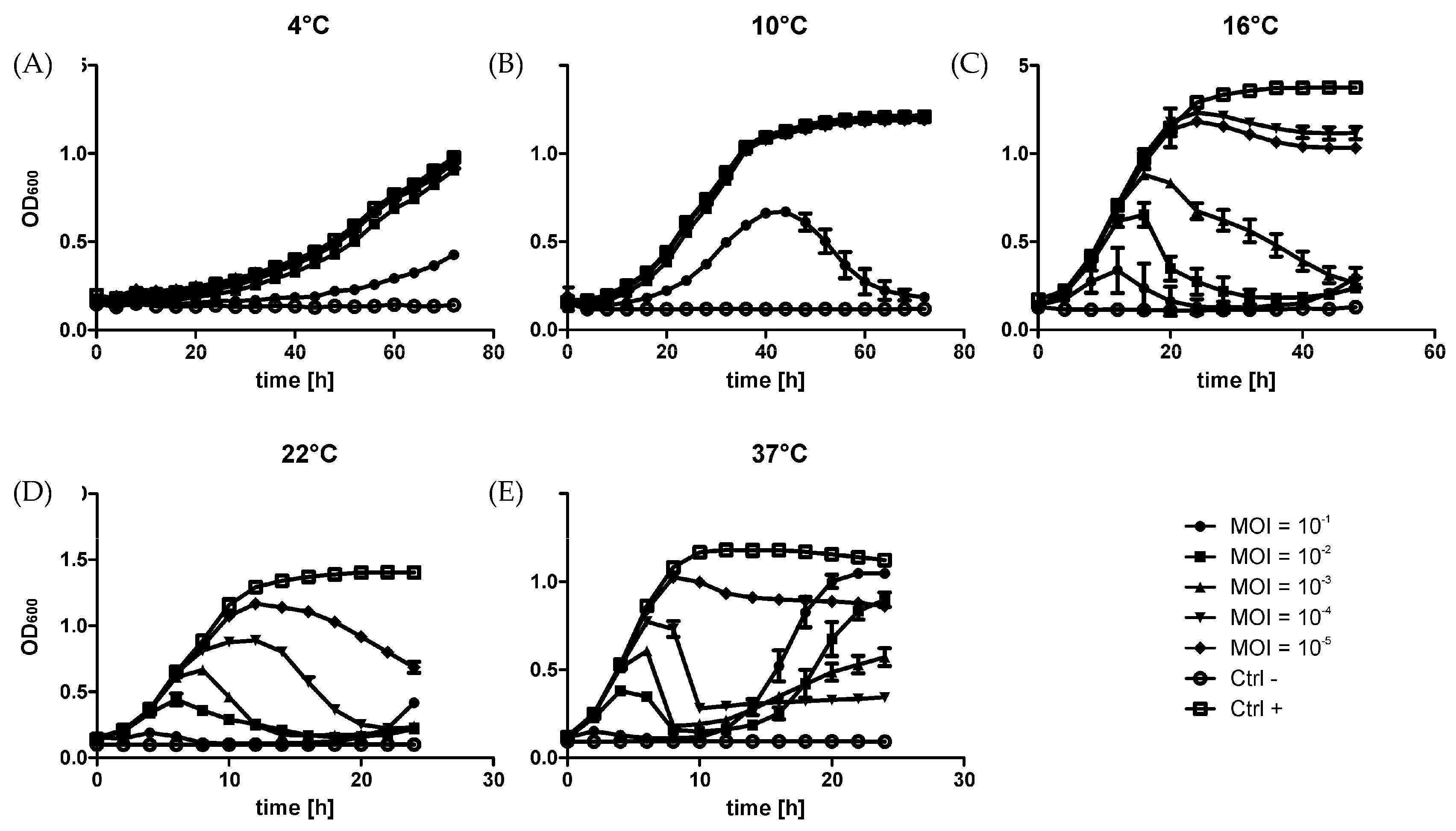
2.2. Growth Curves
2.3. Total RNA Extraction
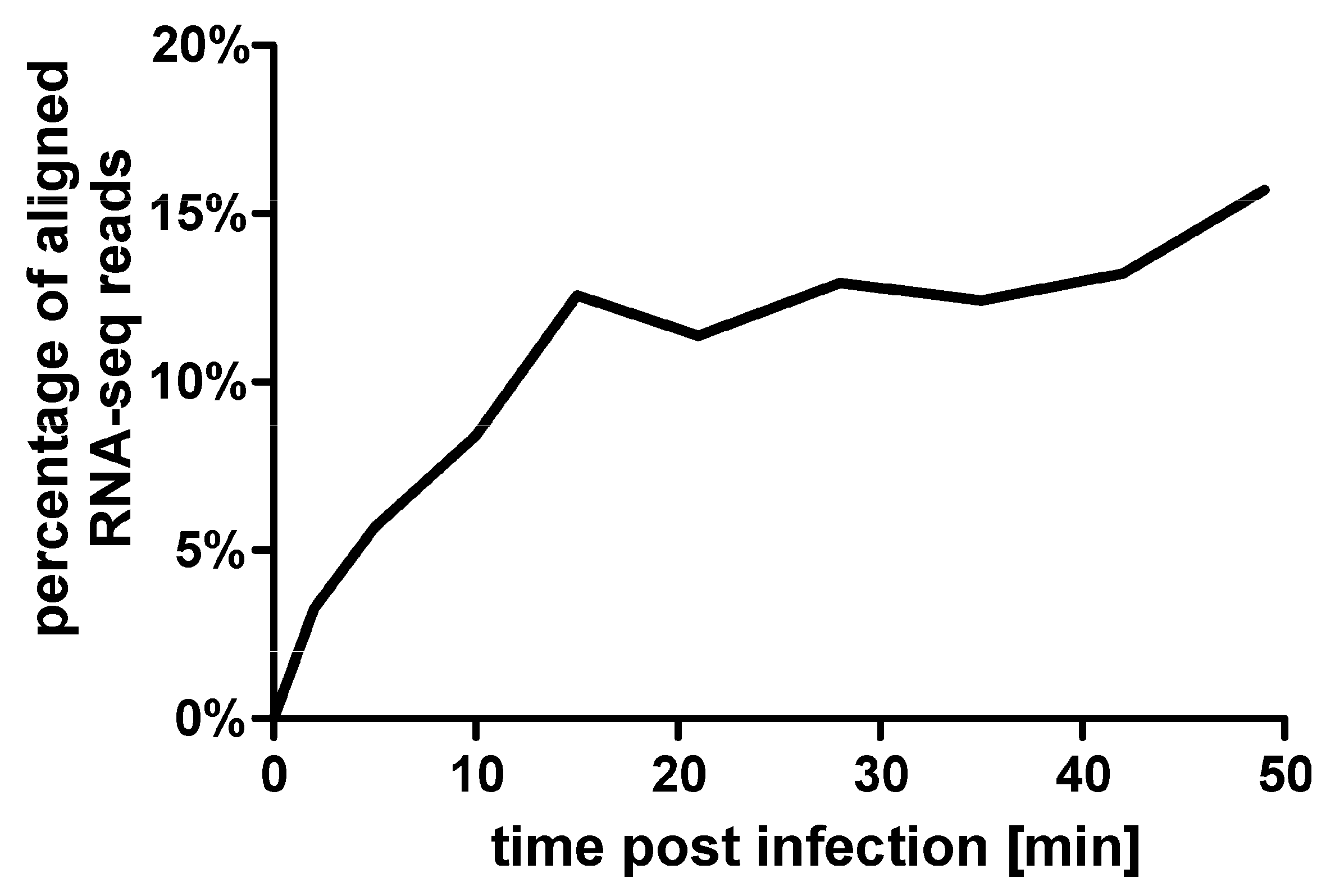
2.4. RNA Sequencing
2.5. Computational and Statistical Analyses
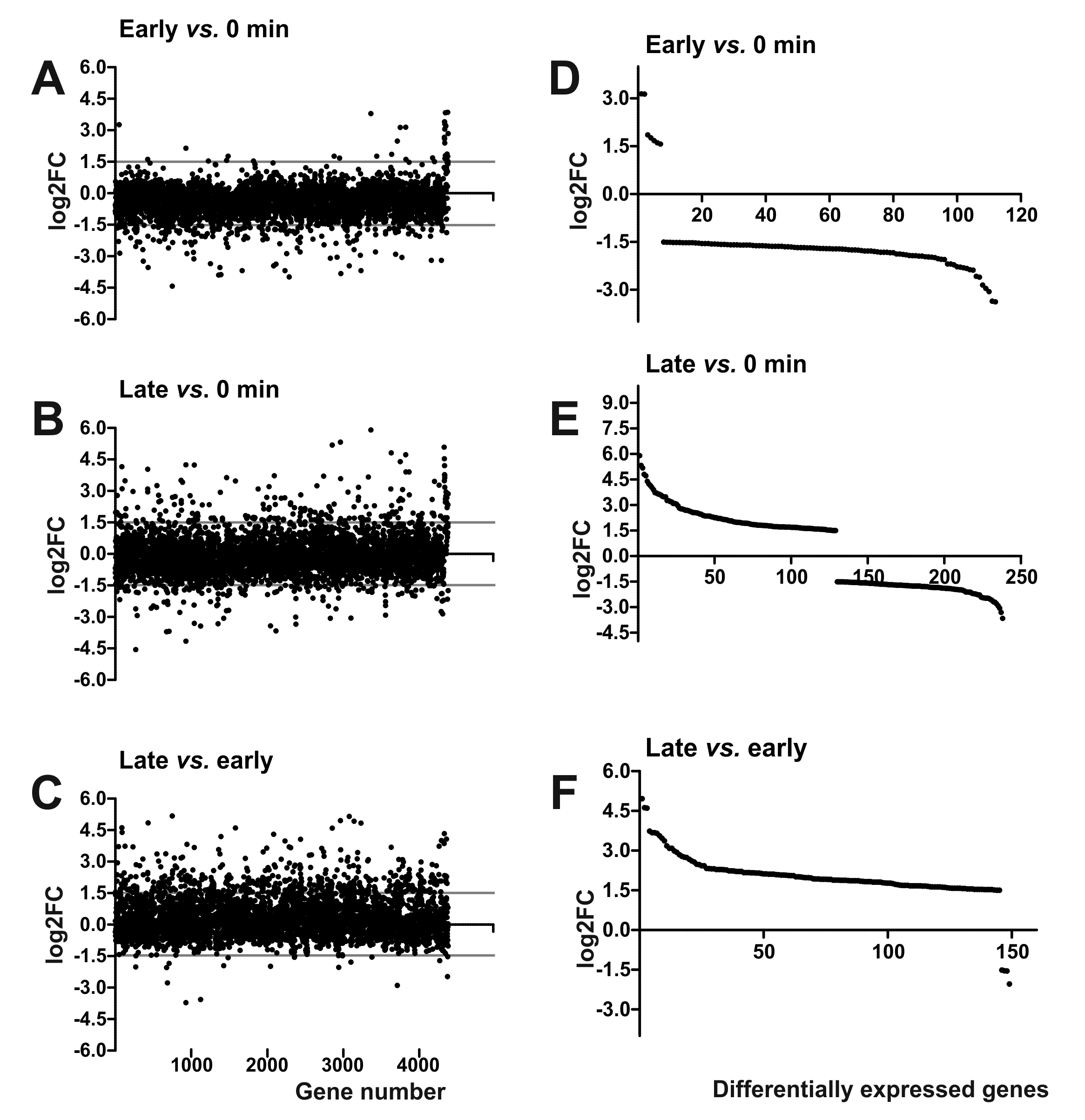
3. Results and Discussion
3.1. Microbiological Parameters of the φR1-37 Infection Process
3.2. General Features of the Transcription Analysis
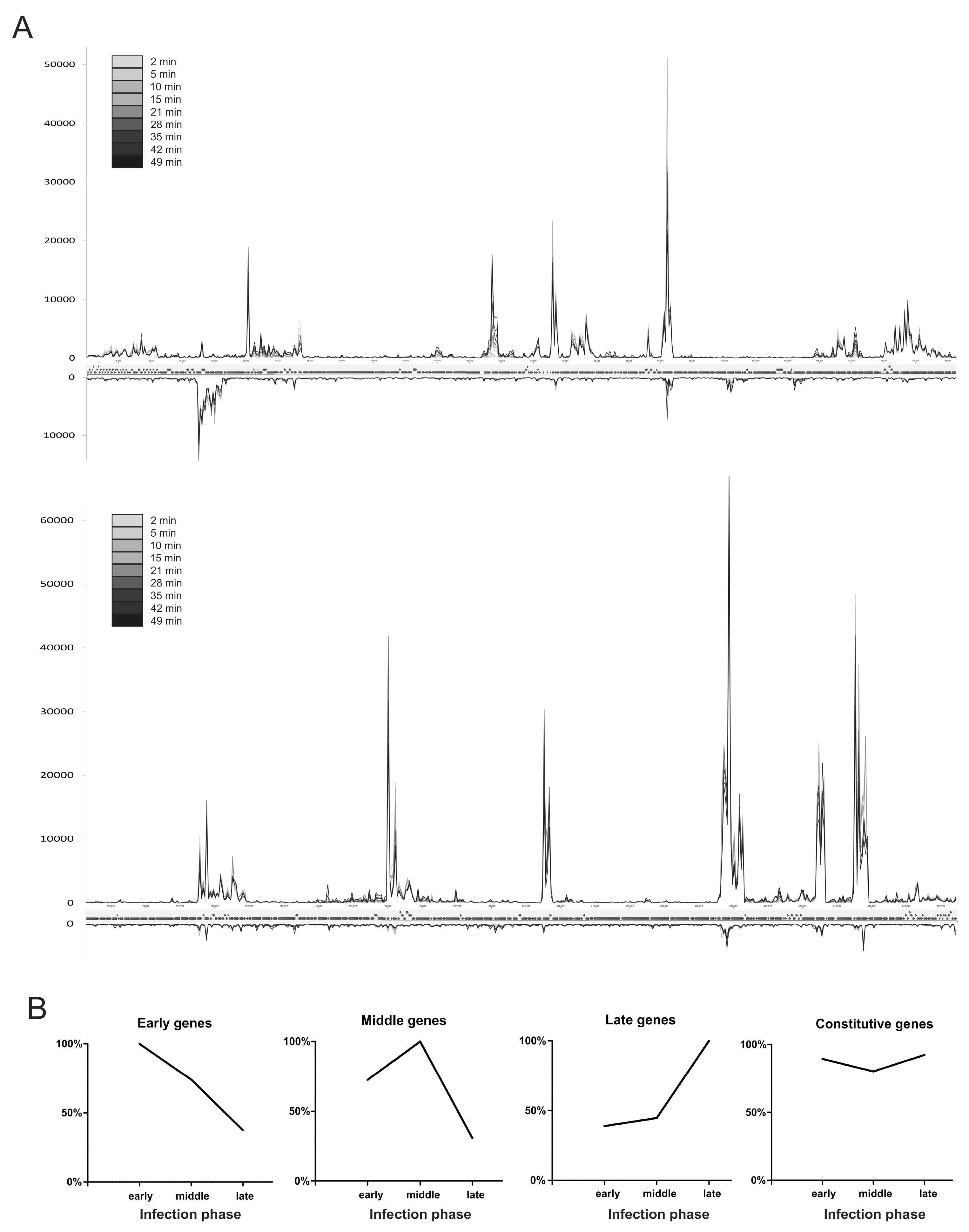
3.3. Temporal Regulation of φR1-37 Transcription
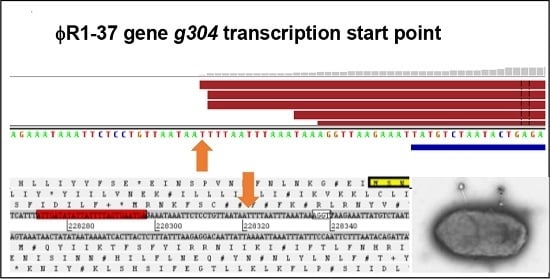
3.4. Validation of Phage Regulatory Elements
3.5. Novel RNA Species in φR1-37 Genome
3.6. Transcriptional Response of YeO3-R1 to φR1-37 Infection
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| asRNA | antisense RNA |
| LB | lysogeny broth |
| log2FC | log2 value of the fold change |
| LPS | lipopolysaccharide |
| MOI | multiplicity of infection |
| ncRNA | non-coding RNA |
| OC | outer core |
| PFU | plaque forming units |
| RNAP | RNA polymerase |
| sRNA | small RNA |
| TC | Total Count |
| TGR | Total Gene Reads |
References
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Guttman, B.; Raya, R.; Kutter, E. Basic Phage Biology. In Bacteriophages: Biology and Applications; Kutter, E., Sulakvelidze, A., Eds.; CRC Press: New York, NY, USA, 2004. [Google Scholar]
- Ceyssens, P.J.; Minakhin, L.; Van den Bossche, A.; Yakunina, M.; Klimuk, E.; Blasdel, B.; De Smet, J.; Noben, J.P.; Bläsi, U.; Severinov, K.; et al. Development of giant bacteriophage varphiKZ is independent of the host transcription apparatus. J. Virol. 2014, 88, 10501–10510. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Lecoutere, E.; Wagemans, J.; Cenens, W.; Aertsen, A.; Schoofs, L.; Landuyt, B.; Paeshuyse, J.; Scheer, M.; Schobert, M.; et al. A multifaceted study of Pseudomonas aeruginosa shutdown by virulent podovirus LUZ19. MBio 2013, 4, e00061-13. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.M.; Farmer, J.J., 3rd. New bacteriophage typing system for Yersinia enterocolitica, Yersinia kristensenii, Yersinia frederiksenii, and Yersinia intermedia: Correlation with serotyping, biotyping, and antibiotic susceptibility. J. Clin. Microbiol. 1982, 15, 491–502. [Google Scholar] [PubMed]
- Popp, A.; Hertwig, S.; Lurz, R.; Appel, B. Comparative study of temperate bacteriophages isolated from Yersinia. Syst. Appl. Microbiol. 2000, 23, 469–478. [Google Scholar] [CrossRef]
- Salem, M.; Virtanen, S.; Korkeala, H.; Skurnik, M. Isolation and characterization of Yersinia-specific bacteriophages from pig stools in Finland. J. Appl. Microbiol. 2015, 118, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, R.M.; Airdrie, D.W. Isolation of Yersinia ruckeri bacteriophages. Appl. Environ. Microbiol. 1984, 47, 1201–1205. [Google Scholar] [PubMed]
- Chan, B.K.; Abedon, S.T. Phage therapy pharmacology phage cocktails. Adv. Appl. Microbiol. 2012, 78, 1–23. [Google Scholar] [PubMed]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, M.; Strauch, E. Phage therapy: Facts and fiction. Int. J. Med. Microbiol. 2006, 296, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Citorik, R.J.; Mimee, M.; Lu, T.K. Bacteriophage-based synthetic biology for the study of infectious diseases. Curr. Opin. Microbiol. 2014, 19, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Majkowska-Skrobek, G.; Maciejewska, B.; Delattre, A.S.; Lavigne, R. Learning from bacteriophages-advantages and limitations of phage and phage-encoded protein applications. Curr. Protein Pept. Sci. 2012, 13, 699–722. [Google Scholar] [CrossRef] [PubMed]
- Hermoso, J.A.; Garcia, J.L.; Garcia, P. Taking aim on bacterial pathogens: From phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007, 10, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, M.; Venho, R.; Toivanen, P.; al-Hendy, A. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol. Microbiol. 1995, 17, 575–594. [Google Scholar] [CrossRef] [PubMed]
- Kiljunen, S.; Hakala, K.; Pinta, E.; Huttunen, S.; Pluta, P.; Gador, A.; Lonnberg, H.; Skurnik, M. Yersiniophage phiR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology 2005, 151, 4093–4102. [Google Scholar] [CrossRef] [PubMed]
- Pinta, E.; Duda, K.A.; Hanuszkiewicz, A.; Kaczynski, Z.; Lindner, B.; Miller, W.L.; Hyytiäinen, H.; Vogel, C.; Borowski, S.; Kasperkiewicz, K.; et al. Identification and role of a 6-deoxy-4-keto-hexosamine in the lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3. Chemistry 2009, 15, 9747–9754. [Google Scholar] [CrossRef] [PubMed]
- Pinta, E.; Duda, K.A.; Hanuszkiewicz, A.; Salminen, T.A.; Bengoechea, J.A.; Hyytiainen, H.; Lindner, B.; Radziejewska-Lebrecht, J.; Holst, O.; Skurnik, M. Characterization of the six glycosyltransferases involved in the biosynthesis of Yersinia enterocolitica serotype O:3 lipopolysaccharide outer core. J. Biol. Chem. 2010, 285, 28333–28342. [Google Scholar] [CrossRef] [PubMed]
- Beczala, A.; Duda, K.A.; Skurnik, M.; Holst, O. The structure of the O-specific polysaccharide of the lipopolysaccharide from Yersinia enterocolitica serotype O:50 strain 3229. Carbohydr. Res. 2012, 359, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Beczala, A.; Ovchinnikova, O.G.; Datta, N.; Mattinen, L.; Knapska, K.; Radziejewska-Lebrecht, J.; Holst, O.; Skurnik, M. Structure and genetic basis of Yersinia similis serotype O:9 O-specific polysaccharide. Innate Immun. 2015, 21, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, M.; Hyytiainen, H.J.; Happonen, L.J.; Kiljunen, S.; Datta, N.; Mattinen, L.; Williamson, K.; Kristo, P.; Szeliga, M.; Kalin-Manttari, L.; et al. Characterization of the genome, proteome, and structure of yersiniophage phiR1-37. J. Virol. 2012, 86, 12625–12642. [Google Scholar] [CrossRef] [PubMed]
- Mesyanzhinov, V.V.; Robben, J.; Grymonprez, B.; Kostyuchenko, V.A.; Bourkaltseva, M.V.; Sykilinda, N.N.; Krylov, V.N.; Volckaert, G. The genome of bacteriophage phi KZ of Pseudomonas aeruginosa. J. Mol. Biol. 2002, 317, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Dillies, M.A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, M.; Toivanen, P. Intervening sequences (IVSs) in the 23s ribosomal-RNA genes of pathogenic Yersinia enterocolitica strains - the IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol. Microbiol. 1991, 5, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Yonesaki, T. Phage-induced change in the stability of mRNAs. Virology 2004, 329, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Berdygulova, Z.; Westblade, L.F.; Florens, L.; Koonin, E.V.; Chait, B.T.; Ramanculov, E.; Washburn, M.P.; Darst, S.A.; Severinov, K.; Minakhin, L. Temporal regulation of gene expression of the Thermus thermophilus bacteriophage P23–45. J. Mol. Biol. 2011, 405, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Bolle, A.; Epstein, R.H.; Salser, W.; Geiduschek, E.P. Transcription during bacteriophage T4 development: Synthesis and relative stability of early and late RNA. J. Mol. Biol. 1968, 31, 325–348. [Google Scholar] [CrossRef]
- Bolle, A.; Epstein, R.H.; Salser, W.; Geiduschek, E.P. Transcription during bacteriophage T4 development: Requirements for late messenger synthesis. J. Mol. Biol. 1968, 33, 339–362. [Google Scholar] [CrossRef]
- Pavlova, O.; Lavysh, D.; Klimuk, E.; Djordjevic, M.; Ravcheev, D.A.; Gelfand, M.S.; Severinov, K.; Akulenko, N. Temporal regulation of gene expression of the Escherichia coli bacteriophage phiEco32. J. Mol. Biol. 2012, 416, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [PubMed]
- Abedon, S.T. Lysis of lysis-inhibited bacteriophage T4-infected cells. J. Bacteriol. 1992, 174, 8073–8080. [Google Scholar] [PubMed]
- Yakunina, M.; Artamonova, T.; Borukhov, S.; Makarova, K.S.; Severinov, K.; Minakhin, L. A non-canonical multisubunit RNA polymerase encoded by a giant bacteriophage. Nucl. Acids Res. 2015, 43, 10411–10420. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.A.; Rolando, M.R.; Carroll, C.A.; Shen, P.S.; Belnap, D.M.; Weintraub, S.T.; Serwer, P.; Hardies, S.C. Characterization of Pseudomonas chlororaphis myovirus 201varphi2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology 2008, 376, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucl. Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Sun, W.D.; Volckaert, G. PHIRE, a deterministic approach to reveal regulatory elements in bacteriophage genomes. Bioinformatics 2004, 20, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Leskinen, K.; Varjosalo, M.; Li, Z.; Li, C.M.; Skurnik, M. Expression of the Yersinia enterocolitica O:3 LPS O-antigen and outer core gene clusters is RfaH-dependent. Microbiology 2015, 161, 1282–1294. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009, 459, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Aravind, L.; Storz, G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007, 64, 738–754. [Google Scholar] [CrossRef] [PubMed]
- Stazic, D.; Lindell, D.; Steglich, C. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucl. Acids Res. 2011, 39, 4890–4899. [Google Scholar] [CrossRef] [PubMed]
- Kery, M.B.; Feldman, M.; Livny, J.; Tjaden, B. TargetRNA2: Identifying targets of small regulatory RNAs in bacteria. Nucl. Acids Res. 2014, 42, W124–W129. [Google Scholar] [CrossRef] [PubMed]
- Lindell, D.; Jaffe, J.D.; Coleman, M.L.; Futschik, M.E.; Axmann, I.M.; Rector, T.; Kettler, G.; Sullivan, M.B.; Steen, R.; Hess, W.R.; et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 2007, 449, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Ruger, W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003, 67, 86–156. [Google Scholar] [CrossRef] [PubMed]
- De Smet, J.; Zimmermann, M.; Kogadeeva, M.; Ceyssens, P.J.; Vermaelen, W.; Blasdel, B.G.; Bin Jang, H.; Sauer, U.; Lavigne, R. High coverage metabolomics analysis reveals phage-specific alterations of Pseudomonas aeruginosa physiology during infection. ISME J. 2016, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Zhan, X.; Gao, J.; Qiu, J.; Feng, Y.; Meganathan, R.; Cohen, S.N.; Georgiou, G. RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell 2003, 114, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Seto, D.; Bhatnagar, S.K.; Bessman, M.J. The purification and properties of deoxyguanosine triphosphate triphosphohydrolase from Escherichia coli. J. Biol. Chem. 1988, 263, 1494–1499. [Google Scholar] [PubMed]
- Loh, K.D.; Gyaneshwar, P.; Markenscoff Papadimitriou, E.; Fong, R.; Kim, K.S.; Parales, R.; Zhou, Z.; Inwood, W.; Kustu, S. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 5114–5119. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.S.; Rosenberg, M. Characterization of a 3rd CII-dependent, coordinately activated promoter on phage-lambda involved in lysogenic development. J. Biol. Chem. 1985, 260, 1838–1844. [Google Scholar]
- Stephenson, F.H. A CII-responsive promoter within the Q-gene of bacteriophage-lambda. Gene 1985, 35, 313–320. [Google Scholar] [CrossRef]
- Nejman-Falenczyk, B.; Bloch, S.; Licznerska, K.; Felczykowska, A.; Dydecka, A.; Wegrzyn, A.; Wegrzyn, G. Small regulatory RNAs in lambdoid bacteriophages and phage-derived plasmids: Not only antisense. Plasmid 2015, 78, 71–78. [Google Scholar] [CrossRef] [PubMed]
| Name | Location in the Genome | Strand | Location with Reference to Other Genes |
|---|---|---|---|
| misc_1 | 14,254–14,383 | − | antisense to 3′ end of g048 |
| misc_2 | 17,805–18,199 | + | antisense to 5′ end of g055 and 3′ end of g056 |
| misc_3 | 32,500–32,820 | − | antisense to 5′ end of g077 |
| misc_4 | 61,640–61,936 | + | intragenic region between g099 and g100 |
| misc_5 | 91,090–91,192 | − | antisense to middle part of g144 |
| misc_6 | 103,380–103,720 | + | antisense to g157 and 5′ end of g156 |
| misc_7 | 148,610–148,960 | + | antisense to 5′ end of g207 |
| misc_8 | 160,970–161,280 | + | antisense to middle part of g230 |
| misc_9 | 217,379–217,553 | + | antisense to middle part of g295 |
| misc_10 | 242,633–242,957 | − | antisense to middle part of g326 |
| Gene | Protein Names | Log2FC |
|---|---|---|
| Membrane Proteins | ||
| Y11_28711 | P pilus assembly/Cpx signaling pathway, periplasmic inhibitor | 5.18 |
| Y11_27571 | Putative inner membrane protein | 3.71 |
| Y11_38011 | UPF0391 membrane protein Y11_38011 | 2.56 |
| Y11_12421 | Putative inner membrane protein | 2.34 |
| Y11_36571 | Membrane protein | 2.30 |
| Y11_25791 | Integral membrane protein TerC | 2.07 |
| Y11_00031 | Putative permease PerM (=YfgO) | 1.98 |
| Y11_17401 | Outer membrane protein X | 1.70 |
| Y11_23901 | Conserved putative membrane protein | −2.12 |
| Y11_12601 | Putative membrane protein YPO2012 | −2.27 |
| Transporters | ||
| Y11_29791 | Phosphate ABC transporter, periplasmic phosphate-binding protein PstS | 5.33 |
| Y11_38431 | Na(+)/H(+) antiporter NhaA (Sodium/proton antiporter NhaA) | 4.72 |
| Y11_14731 | Mg(2+) transport ATPase protein C | 3.63 |
| Y11_29801 | Phosphate transport system permease protein PstC | 3.59 |
| Y11_07561 | Putative ABC sugar transporter | 2.70 |
| Y11_26421 | Uncharacterized ABC transporter, permease component YrbE | 1.99 |
| Y11_43381 | Phosphate transport system permease protein PstC | 1.70 |
| Y11_29811 | Phosphate transport system permease protein PstA | 1.68 |
| Y11_00591 | Sialic acid transporter (Permease) NanT | −2.18 |
| Stress response Proteins | ||
| Y11_08711 | Phage shock protein A | 3.51 |
| Y11_08721 | Phage shock protein B | 2.55 |
| Y11_08731 | Phage shock protein C | 2.41 |
| Y11_08741 | Phage shock protein D | 2.09 |
| Y11_04271 | Cold shock protein | 3.10 |
| Y11_04291 | Cold shock protein CspB | 2.96 |
| Y11_27291 | Cold shock protein CspG | 1.97 |
| Y11_27301 | Cold shock protein CspG | 1.87 |
| Y11_10461 | Osmotically inducible lipoprotein B | 4.24 |
| Y11_38001 | Osmotically inducible protein OsmY | 3.06 |
| Transcriptional Regulators | ||
| Y11_38441 | Transcriptional activator NhaR | 3.90 |
| Y11_21031 | Phosphate regulon transcriptional regulatory protein PhoB (SphR) | 3.73 |
| Y11_22431 | Transcriptional regulator, ArsR family | 2.67 |
| Y11_10431 | Transcriptional regulatory protein YciT | −2.44 |
| Y11_21251 | Transcriptional regulator, GntR family | −3.66 |
| Other | ||
| Y11_34281 | RNA polymerase sigma factor | 2.30 |
| Y11_28241 | Regulator of ribonuclease activity A | 2.26 |
| Y11_21021 | Phosphate regulon sensor protein PhoR (SphS) (EC 2.7.13.3) | 1.69 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leskinen, K.; Blasdel, B.G.; Lavigne, R.; Skurnik, M. RNA-Sequencing Reveals the Progression of Phage-Host Interactions between φR1-37 and Yersinia enterocolitica. Viruses 2016, 8, 111. https://doi.org/10.3390/v8040111
Leskinen K, Blasdel BG, Lavigne R, Skurnik M. RNA-Sequencing Reveals the Progression of Phage-Host Interactions between φR1-37 and Yersinia enterocolitica. Viruses. 2016; 8(4):111. https://doi.org/10.3390/v8040111
Chicago/Turabian StyleLeskinen, Katarzyna, Bob G. Blasdel, Rob Lavigne, and Mikael Skurnik. 2016. "RNA-Sequencing Reveals the Progression of Phage-Host Interactions between φR1-37 and Yersinia enterocolitica" Viruses 8, no. 4: 111. https://doi.org/10.3390/v8040111