Big Data Offers Novel Insights for Oncolytic Virus Immunotherapy
Abstract
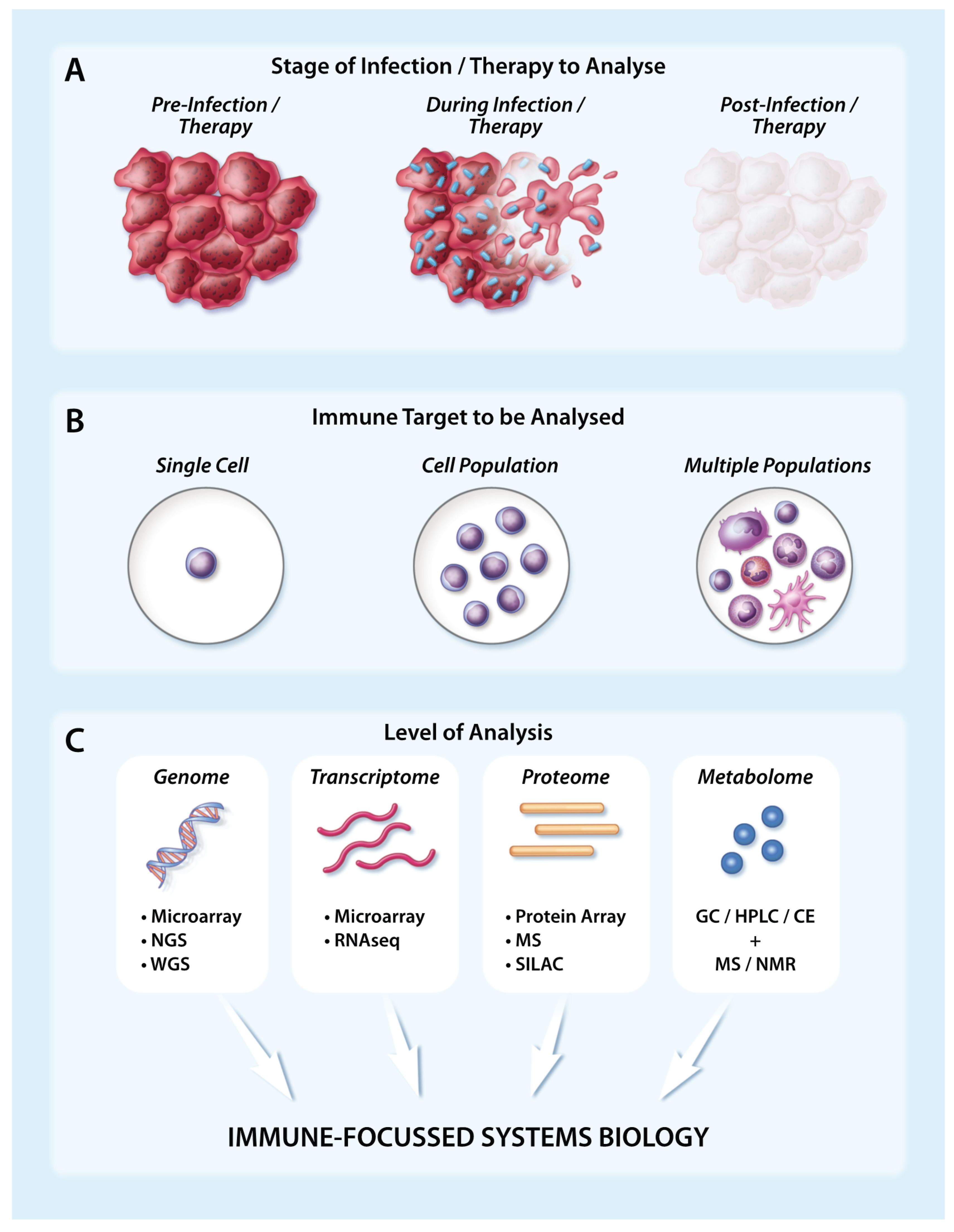
:1. Introduction
2. Viruses vs. Host Immunity
3. Using Large-Scale Approaches to Capture Cellular Immune Responses
4. Using Large-Scale Approaches to Track Global Immune Changes
5. Using Large-Scale Approaches to Enhance Immune Engagement during Oncolytic Virotherapy
6. Using Large-Scale Approaches to Analyse Tumour Adaptations to Immunotherapy and Signatures of Success
7. Future Directions
8. Final Thoughts
Acknowledgments
Conflicts of Interest
References
- Sinkovics, J.G.; Horvath, J.C. Natural and genetically engineered viral agents for oncolysis and gene therapy of human cancers. Arch. Immunol. Ther. Exp. 2008, 56, 3s–59s. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.W.; Senzer, N.; Cerullo, V.; Templeton, N.S.; Hemminki, A.; Nemunaitis, J. Oncolytic viruses for induction of anti-tumor immunity. Curr. Pharm. Biotechnol. 2012, 13, 1750–1760. [Google Scholar] [CrossRef] [PubMed]
- Vaha-Koskela, M.J.; Heikkila, J.E.; Hinkkanen, A.E. Oncolytic viruses in cancer therapy. Cancer Lett. 2007, 254, 178–216. [Google Scholar] [CrossRef] [PubMed]
- Breitbach, C.J.; Arulanandam, R.; de Silva, N.; Thorne, S.H.; Patt, R.; Daneshmand, M.; Moon, A.; Ilkow, C.; Burke, J.; Hwang, T.H.; et al. Oncolytic vaccinia virus disrupts tumor-associated vasculature in humans. Cancer Res. 2013, 73, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Breitbach, C.J.; Paterson, J.M.; Lemay, C.G.; Falls, T.J.; McGuire, A.; Parato, K.A.; Stojdl, D.F.; Daneshmand, M.; Speth, K.; Kirn, D.; et al. Targeted inflammation during oncolytic virus therapy severely compromises tumor blood flow. Mol. Ther. 2007, 15, 1686–1693. [Google Scholar] [CrossRef] [PubMed]
- Bridle, B.W.; Chen, L.; Lemay, C.G.; Diallo, J.S.; Pol, J.; Nguyen, A.; Capretta, A.; He, R.; Bramson, J.L.; Bell, J.C.; et al. HDAC inhibition suppresses primary immune responses, enhances secondary immune responses, and abrogates autoimmunity during tumor immunotherapy. Mol. Ther. 2013, 21, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Bridle, B.W.; Hanson, S.; Lichty, B.D. Combining oncolytic virotherapy and tumour vaccination. Cytokine Growth Factor Rev. 2010, 21, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Bridle, B.W.; Stephenson, K.B.; Boudreau, J.E.; Koshy, S.; Kazdhan, N.; Pullenayegum, E.; Brunelliere, J.; Bramson, J.L.; Lichty, B.D.; Wan, Y. Potentiating cancer immunotherapy using an oncolytic virus. Mol. Ther. 2010, 18, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Granot, T.; Yamanashi, Y.; Meruelo, D. Sindbis viral vectors transiently deliver tumor-associated antigens to lymph nodes and elicit diversified antitumor CD8+ T-cell immunity. Mol. Ther. 2014, 22, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Grekova, S.P.; Raykov, Z.; Zawatzky, R.; Rommelaere, J.; Koch, U. Activation of a glioma-specific immune response by oncolytic parvovirus minute virus of mice infection. Cancer Gene Ther. 2012, 19, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Kleijn, A.; Kloezeman, J.; Treffers-Westerlaken, E.; Fulci, G.; Leenstra, S.; Dirven, C.; Debets, R.; Lamfers, M. The in vivo therapeutic efficacy of the oncolytic adenovirus Delta24-RGD is mediated by tumor-specific immunity. PLoS ONE 2014, 9, e97495. [Google Scholar]
- Parviainen, S.; Ahonen, M.; Diaconu, I.; Kipar, A.; Siurala, M.; Vaha-Koskela, M.; Kanerva, A.; Cerullo, V.; Hemminki, A. GMCSF-armed vaccinia virus induces an antitumor immune response. Int. J. Cancer 2015, 136, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Todo, T.; Rabkin, S.D.; Sundaresan, P.; Wu, A.; Meehan, K.R.; Herscowitz, H.B.; Martuza, R.L. Systemic antitumor immunity in experimental brain tumor therapy using a multimutated, replication-competent herpes simplex virus. Hum. Gene Ther. 1999, 10, 2741–2755. [Google Scholar] [CrossRef] [PubMed]
- Zamarin, D.; Holmgaard, R.B.; Subudhi, S.K.; Park, J.S.; Mansour, M.; Palese, P.; Merghoub, T.; Wolchok, J.D.; Allison, J.P. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Diaz, R.M.; Galivo, F.; Kottke, T.; Wongthida, P.; Qiao, J.; Thompson, J.; Valdes, M.; Barber, G.; Vile, R.G. Oncolytic immunovirotherapy for melanoma using vesicular stomatitis virus. Cancer Res. 2007, 67, 2840–2848. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, M.; Fu, X.; Rousseau, R.; Chen, S.Y.; Zhang, X. Destruction of nonimmunogenic mammary tumor cells by a fusogenic oncolytic herpes simplex virus induces potent antitumor immunity. Mol. Ther. 2004, 9, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Wimmer, E.; Cello, J. Oncolytic poliovirus therapy and immunization with poliovirus-infected cell lysate induces potent antitumor immunity against neuroblastoma in vivo. Int. J. Oncol. 2011, 38, 81–87. [Google Scholar] [PubMed]
- Sharma, D.; Priyadarshini, P.; Vrati, S. Unraveling the web of viroinformatics: Computational tools and databases in virus research. J. Virol. 2015, 89, 1489–1501. [Google Scholar] [CrossRef] [PubMed]
- Heng, T.S.; Painter, M.W. The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 2008, 9, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.C.; Lanier, L.L. Beyond the transcriptome: Completion of act one of the immunological genome project. Curr. Opin. Immunol. 2013, 25, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Fensterl, V.; Wetzel, J.L.; Ramachandran, S.; Ogino, T.; Stohlman, S.A.; Bergmann, C.C.; Diamond, M.S.; Virgin, H.W.; Sen, G.C. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012, 8, e1002712. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Katze, M.G.; Fornek, J.L.; Palermo, R.E.; Walters, K.A.; Korth, M.J. Innate immune modulation by RNA viruses: Emerging insights from functional genomics. Nat. Rev. Immunol. 2008, 8, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Sadler, A.J.; Williams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sharma, A. Neutrophils: Cinderella of innate immune system. Int. Immunopharmacol. 2010, 10, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Ilkow, C.S.; Swift, S.L.; Bell, J.C.; Diallo, J.S. From scourge to cure: Tumour-selective viral pathogenesis as a new strategy against cancer. PLoS Pathog. 2014, 10, e1003836. [Google Scholar] [CrossRef] [PubMed]
- Stojdl, D.F.; Lichty, B.D.; tenOever, B.R.; Paterson, J.M.; Power, A.T.; Knowles, S.; Marius, R.; Reynard, J.; Poliquin, L.; Atkins, H.; et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 2003, 4, 263–275. [Google Scholar] [CrossRef]
- Ruotsalainen, J.J.; Kaikkonen, M.U.; Niittykoski, M.; Martikainen, M.W.; Lemay, C.G.; Cox, J.; de Silva, N.S.; Kus, A.; Falls, T.J.; Diallo, J.S.; et al. Clonal variation in interferon response determines the outcome of oncolytic virotherapy in mouse CT26 colon carcinoma model. Gene Ther. 2015, 22, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Brandes, M.; Klauschen, F.; Kuchen, S.; Germain, R.N. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell 2013, 154, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Kash, J.C.; Tumpey, T.M.; Proll, S.C.; Carter, V.; Perwitasari, O.; Thomas, M.J.; Basler, C.F.; Palese, P.; Taubenberger, J.K.; Garcia-Sastre, A.; et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 2006, 443, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Schwab, L.; Goroncy, L.; Palaniyandi, S.; Gautam, S.; Triantafyllopoulou, A.; Mocsai, A.; Reichardt, W.; Karlsson, F.J.; Radhakrishnan, S.V.; Hanke, K.; et al. Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance graft-versus-host disease via tissue damage. Nat. Med. 2014, 20, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Herz, J.; Johnson, K.R.; McGavern, D.B. Therapeutic antiviral T cells noncytopathically clear persistently infected microglia after conversion into antigen-presenting cells. J. Exp. Med. 2015, 212, 1153–1169. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, R.; Baldwin, N.; Cepika, A.M.; Athale, S.; Xue, Y.; Yu, C.I.; Metang, P.; Cheruku, A.; Berthier, I.; Gayet, I.; et al. Transcriptional specialization of human dendritic cell subsets in response to microbial vaccines. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kmiecik, J.; Poli, A.; Brons, N.H.; Waha, A.; Eide, G.E.; Enger, P.O.; Zimmer, J.; Chekenya, M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J. Neuroimmunol. 2013, 264, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Slifka, M.K.; Rodriguez, F.; Whitton, J.L. Rapid on/off cycling of cytokine production by virus-specific CD8+ T cells. Nature 1999, 401, 76–79. [Google Scholar] [CrossRef] [PubMed]
- McGray, A.J.; Hallett, R.; Bernard, D.; Swift, S.L.; Zhu, Z.; Teoderascu, F.; Vanseggelen, H.; Hassell, J.A.; Hurwitz, A.A.; Wan, Y.; et al. Immunotherapy-induced CD8+ T cells instigate immune suppression in the tumor. Mol. Ther. 2014, 22, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Best, J.A.; Blair, D.A.; Knell, J.; Yang, E.; Mayya, V.; Doedens, A.; Dustin, M.L.; Goldrath, A.W. Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat. Immunol. 2013, 14, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.D.; Kim, M.T.; Shan, Q.; Sompallae, R.; Xue, H.H.; Harty, J.T.; Badovinac, V.P. Phenotypic and functional alterations in circulating memory CD8 T cells with time after primary infection. PLoS Pathog. 2015, 11, e1005219. [Google Scholar] [CrossRef] [PubMed]
- Bridle, B.W.; Clouthier, D.; Zhang, L.; Pol, J.; Chen, L.; Lichty, B.D.; Bramson, J.L.; Wan, Y. Oncolytic vesicular stomatitis virus quantitatively and qualitatively improves primary CD8 T-cell responses to anticancer vaccines. Oncoimmunology 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Pol, J.G.; Zhang, L.; Bridle, B.W.; Stephenson, K.B.; Resseguier, J.; Hanson, S.; Chen, L.; Kazdhan, N.; Bramson, J.L.; Stojdl, D.F.; et al. Maraba virus as a potent oncolytic vaccine vector. Mol. Ther. 2014, 22, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Shaffer, D.R.; Alvarez Arias, D.A.; Nakazaki, Y.; Pos, W.; Torres, A.J.; Cremasco, V.; Dougan, S.K.; Cowley, G.S.; Elpek, K.; et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 2014, 506, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Wongthida, P.; Diaz, R.M.; Galivo, F.; Kottke, T.; Thompson, J.; Melcher, A.; Vile, R. VSV oncolytic virotherapy in the B16 model depends upon intact MyD88 signaling. Mol. Ther. 2011, 19, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Rommelfanger, D.M.; Wongthida, P.; Diaz, R.M.; Kaluza, K.M.; Thompson, J.M.; Kottke, T.J.; Vile, R.G. Systemic combination virotherapy for melanoma with tumor antigen-expressing vesicular stomatitis virus and adoptive T-cell transfer. Cancer Res. 2012, 72, 4753–4764. [Google Scholar] [CrossRef] [PubMed]
- Tahtinen, S.; Gronberg-Vaha-Koskela, S.; Lumen, D.; Merisalo-Soikkeli, M.; Siurala, M.; Airaksinen, A.J.; Vaha-Koskela, M.; Hemminki, A. Adenovirus improves the efficacy of adoptive T-cell therapy by recruiting immune cells to and promoting their activity at the tumor. Cancer Immunol. Res. 2015, 3, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Mahalingam, J.; Chiang, J.M.; Su, P.J.; Chu, Y.Y.; Lai, H.Y.; Fang, J.H.; Huang, C.T.; Chiu, C.T.; Lin, C.Y. Activated but not resting regulatory T cells accumulated in tumor microenvironment and correlated with tumor progression in patients with colorectal cancer. Int. J. Cancer 2013, 132, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Albert, M.H.; Mannert, J.; Fleischmann, K.K.; Schiemann, M.; Pagel, P.; Schmid, I.; Magg, T. Mirnome and transcriptome aided pathway analysis in human regulatory T cells. Genes Immun. 2014, 15, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.B.; Lu, M.H.; Fan, Y.H.; Cao, Y.L.; Zhang, Z.R.; Yang, S.M. Macrophages in tumor microenvironments and the progression of tumors. Clin. Dev. Immunol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Huang, S.C.; Sergushichev, A.; Lampropoulou, V.; Ivanova, Y.; Loginicheva, E.; Chmielewski, K.; Stewart, K.M.; Ashall, J.; Everts, B.; et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Chaussabel, D.; Quinn, C.; Shen, J.; Patel, P.; Glaser, C.; Baldwin, N.; Stichweh, D.; Blankenship, D.; Li, L.; Munagala, I.; et al. A modular analysis framework for blood genomics studies: Application to systemic lupus erythematosus. Immunity 2008, 29, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Pommerenke, C.; Wilk, E.; Srivastava, B.; Schulze, A.; Novoselova, N.; Geffers, R.; Schughart, K. Global transcriptome analysis in influenza-infected mouse lungs reveals the kinetics of innate and adaptive host immune responses. PLoS ONE 2012, 7, e41169. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulou, K.; Dimitrakopoulos, G.N.; Wilk, E.; Tsimpouris, C.; Sgarbas, K.N.; Schughart, K.; Bezerianos, A. Influenza A immunomics and public health omics: The dynamic pathway interplay in host response to H1N1 infection. OMICS 2014, 18, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Vanwolleghem, T.; Hou, J.; van Oord, G.; Andeweg, A.C.; Osterhaus, A.D.; Pas, S.D.; Janssen, H.L.; Boonstra, A. Re-evaluation of hepatitis B virus clinical phases by systems biology identifies unappreciated roles for the innate immune response and B cells. Hepatology 2015, 62, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, D.J.; Lefebvre, C.; Allan, K.; Brun, J.; Sanaei, C.A.; Baird, S.; Pearce, N.; Gronberg, S.; Wilson, B.; Prakesh, M.; et al. Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell 2011, 20, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Al-Yahya, S.; Mahmoud, L.; Al-Zoghaibi, F.; Al-Tuhami, A.; Amer, H.; Almajhdi, F.N.; Polyak, S.J.; Khabar, K.S. Human cytokinome analysis for interferon response. J. Virol. 2015, 89, 7108–7119. [Google Scholar] [CrossRef] [PubMed]
- Varble, A.; Benitez, A.A.; Schmid, S.; Sachs, D.; Shim, J.V.; Rodriguez-Barrueco, R.; Panis, M.; Crumiller, M.; Silva, J.M.; Sachidanandam, R.; et al. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe 2013, 14, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, E.; Weibel, S.; Szalay, A.A. Combination treatment with oncolytic vaccinia virus and cyclophosphamide results in synergistic antitumor effects in human lung adenocarcinoma bearing mice. J. Transl. Med. 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Ottolino-Perry, K.; Acuna, S.A.; Angarita, F.A.; Sellers, C.; Zerhouni, S.; Tang, N.; McCart, J.A. Oncolytic vaccinia virus synergizes with irinotecan in colorectal cancer. Mol. Oncol. 2015, 9, 1539–1552. [Google Scholar] [CrossRef] [PubMed]
- Siurala, M.; Bramante, S.; Vassilev, L.; Hirvinen, M.; Parviainen, S.; Tahtinen, S.; Guse, K.; Cerullo, V.; Kanerva, A.; Kipar, A.; et al. Oncolytic adenovirus and doxorubicin-based chemotherapy results in synergistic antitumor activity against soft-tissue sarcoma. Int. J. Cancer 2015, 136, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Pandha, H.S.; Heinemann, L.; Simpson, G.R.; Melcher, A.; Prestwich, R.; Errington, F.; Coffey, M.; Harrington, K.J.; Morgan, R. Synergistic effects of oncolytic reovirus and cisplatin chemotherapy in murine malignant melanoma. Clin. Cancer Res. 2009, 15, 6158–6166. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic cell death in cancer therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.S.; Liu, Z.; Bartlett, D.L. Oncolytic immunotherapy: Dying the right way is a key to eliciting potent antitumor immunity. Front. Oncol. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Workenhe, S.T.; Mossman, K.L. Oncolytic virotherapy and immunogenic cancer cell death: Sharpening the sword for improved cancer treatment strategies. Mol. Ther. 2014, 22, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Bezu, L.; Gomes-de-Silva, L.C.; Dewitte, H.; Breckpot, K.; Fucikova, J.; Spisek, R.; Galluzzi, L.; Kepp, O.; Kroemer, G. Combinatorial strategies for the induction of immunogenic cell death. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kepp, O.; Senovilla, L.; Vitale, I.; Vacchelli, E.; Adjemian, S.; Agostinis, P.; Apetoh, L.; Aranda, F.; Barnaba, V.; Bloy, N.; et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Lesterhuis, W.J.; Rinaldi, C.; Jones, A.; Rozali, E.N.; Dick, I.M.; Khong, A.; Boon, L.; Robinson, B.W.; Nowak, A.K.; Bosco, A.; et al. Network analysis of immunotherapy-induced regressing tumours identifies novel synergistic drug combinations. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, N.; Breinig, M.; Speck, T.; Michels, T.; Kreutzer, C.; Sorrentino, A.; Sharma, A.K.; Umansky, L.; Conrad, H.; Poschke, I.; et al. A high-throughput RNAi screen for detection of immune-checkpoint molecules that mediate tumor resistance to cytotoxic T lymphocytes. EMBO Mol. Med. 2015, 7, 450–463. [Google Scholar] [CrossRef] [PubMed]
- Zloza, A.; Kim, D.W.; Kim-Schulze, S.; Jagoda, M.C.; Monsurro, V.; Marincola, F.M.; Kaufman, H.L. Immunoglobulin-like transcript 2 (ILT2) is a biomarker of therapeutic response to oncolytic immunotherapy with vaccinia viruses. J. Immunother. Cancer 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Liikanen, I.; Monsurro, V.; Ahtiainen, L.; Raki, M.; Hakkarainen, T.; Diaconu, I.; Escutenaire, S.; Hemminki, O.; Dias, J.D.; Cerullo, V.; et al. Induction of interferon pathways mediates in vivo resistance to oncolytic adenovirus. Mol. Ther. 2011, 19, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Diallo, J.S.; le Boeuf, F.; Lai, F.; Cox, J.; Vaha-Koskela, M.; Abdelbary, H.; MacTavish, H.; Waite, K.; Falls, T.; Wang, J.; et al. A high-throughput pharmacoviral approach identifies novel oncolytic virus sensitizers. Mol. Ther. 2010, 18, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Moerdyk-Schauwecker, M.; Shah, N.R.; Murphy, A.M.; Hastie, E.; Mukherjee, P.; Grdzelishvili, V.Z. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: Role of type I interferon signaling. Virology 2013, 436, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Song, T.J.; Haddad, D.; Adusumilli, P.; Kim, T.; Stiles, B.; Hezel, M.; Socci, N.D.; Gonen, M.; Fong, Y. Molecular network pathways and functional analysis of tumor signatures associated with development of resistance to viral gene therapy. Cancer Gene Ther. 2012, 19, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Blackham, A.U.; Northrup, S.A.; Willingham, M.; Sirintrapun, J.; Russell, G.B.; Lyles, D.S.; Stewart, J.H.T. Molecular determinants of susceptibility to oncolytic vesicular stomatitis virus in pancreatic adenocarcinoma. J. Surg. Res. 2014, 187, 412–426. [Google Scholar] [CrossRef] [PubMed]
- Samuel, S.; Beljanski, V.; van Grevenynghe, J.; Richards, S.; Ben Yebdri, F.; He, Z.; Nichols, C.; Belgnaoui, S.M.; Steel, C.; Goulet, M.L.; et al. BCL-2 inhibitors sensitize therapy-resistant chronic lymphocytic leukemia cells to VSV oncolysis. Mol. Ther. 2013, 21, 1413–1423. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.C.; Tian, T.F.; Tseng, Y.J. 3Omics: A web-based systems biology tool for analysis, integration and visualization of human transcriptomic, proteomic and metabolomic data. BMC Syst. Biol. 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J., 3rd; Kaufmann, S.H.; Maertzdorf, J. High-throughput data analysis and data integration for vaccine trials. Vaccine 2015, 33, 5249–5255. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xue, Q.; Eisele, M.R.; Sulistijo, E.S.; Brower, K.; Han, L.; el Amir, A.D.; Pe’er, D.; Miller-Jensen, K.; Fan, R. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc. Natl. Acad. Sci. USA 2015, 112, E607–E615. [Google Scholar] [CrossRef] [PubMed]
- Becher, B.; Schlitzer, A.; Chen, J.; Mair, F.; Sumatoh, H.R.; Teng, K.W.; Low, D.; Ruedl, C.; Riccardi-Castagnoli, P.; Poidinger, M.; et al. High-dimensional analysis of the murine myeloid cell system. Nat. Immunol. 2014, 15, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.R.; Jung, S.; Amit, I. Making the case for chromatin profiling: A new tool to investigate the immune-regulatory landscape. Nat. Rev. Immunol. 2015, 15, 585–594. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swift, S.L.; Stojdl, D.F. Big Data Offers Novel Insights for Oncolytic Virus Immunotherapy. Viruses 2016, 8, 45. https://doi.org/10.3390/v8020045
Swift SL, Stojdl DF. Big Data Offers Novel Insights for Oncolytic Virus Immunotherapy. Viruses. 2016; 8(2):45. https://doi.org/10.3390/v8020045
Chicago/Turabian StyleSwift, Stephanie L., and David F. Stojdl. 2016. "Big Data Offers Novel Insights for Oncolytic Virus Immunotherapy" Viruses 8, no. 2: 45. https://doi.org/10.3390/v8020045






