Begomovirus-Associated Satellite DNA Diversity Captured Through Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Whitefly Collection, Sample Processing, Metagenomic Sequencing and Data Analysis
| Crop/Plant (Dataset) 1 | Location | Collection Date | Whiteflies 2 |
|---|---|---|---|
| Squash | Imperial Valley, California, USA | June 2012 | B. tabaci (MEAM1) |
| Tomato | Salamá, Baja Verapaz, Guatemala | January 2012 | B. tabaci (NW), T. vaporariorum |
| Tomato (Guat_T2) | El Progreso, Jutiapa, Guatemala | January 2012 | B. tabaci (NW), T. vaporariorum |
| Squash | Teculután, Zacapa, Guatemala | January 2012 | B. tabaci (MEAM1) |
| Bean | Torrox, Málaga, Spain | September 2011 | B. tabaci (MED) |
| Squash (Spain_S) | Torrox, Málaga, Spain | September 2011 | B. tabaci (MED) |
| Tomato (Spain_T) | Torrox, Málaga, Spain | September 2011 | B. tabaci (MED) |
| European black nightshade | Algarrobo-Costa, Málaga, Spain | September 2011 | B. tabaci (MED) |
| European black nightshade | Algarrobo-Costa, Málaga, Spain | September 2011 | B. tabaci (MED) |
| Tomato | Bet Dagan, Israel | March 2011 | B. tabaci (MEAM1) |
| Squash (Israel_S) | Bet Dagan, Israel | March 2011 | B. tabaci (MEAM1) |
| Tomato (PR_T1) | Santa Isabel, Puerto Rico | November 2010 | B. tabaci (MEAM1) |
| Tomato (PR_T2) | Santa Isabel, Puerto Rico | November 2010 | B. tabaci (MEAM1) |
| Pumpkin (PR_P) | Santa Isabel, Puerto Rico | November 2010 | B. tabaci (MEAM1) |
| Eggplant (PR_E) | Santa Isabel, Puerto Rico | November 2010 | B. tabaci (MEAM1) |
2.2. Satellite DNA Molecule Completion
2.3. Satellite DNA Sequence Analysis
3. Results
3.1. Overview
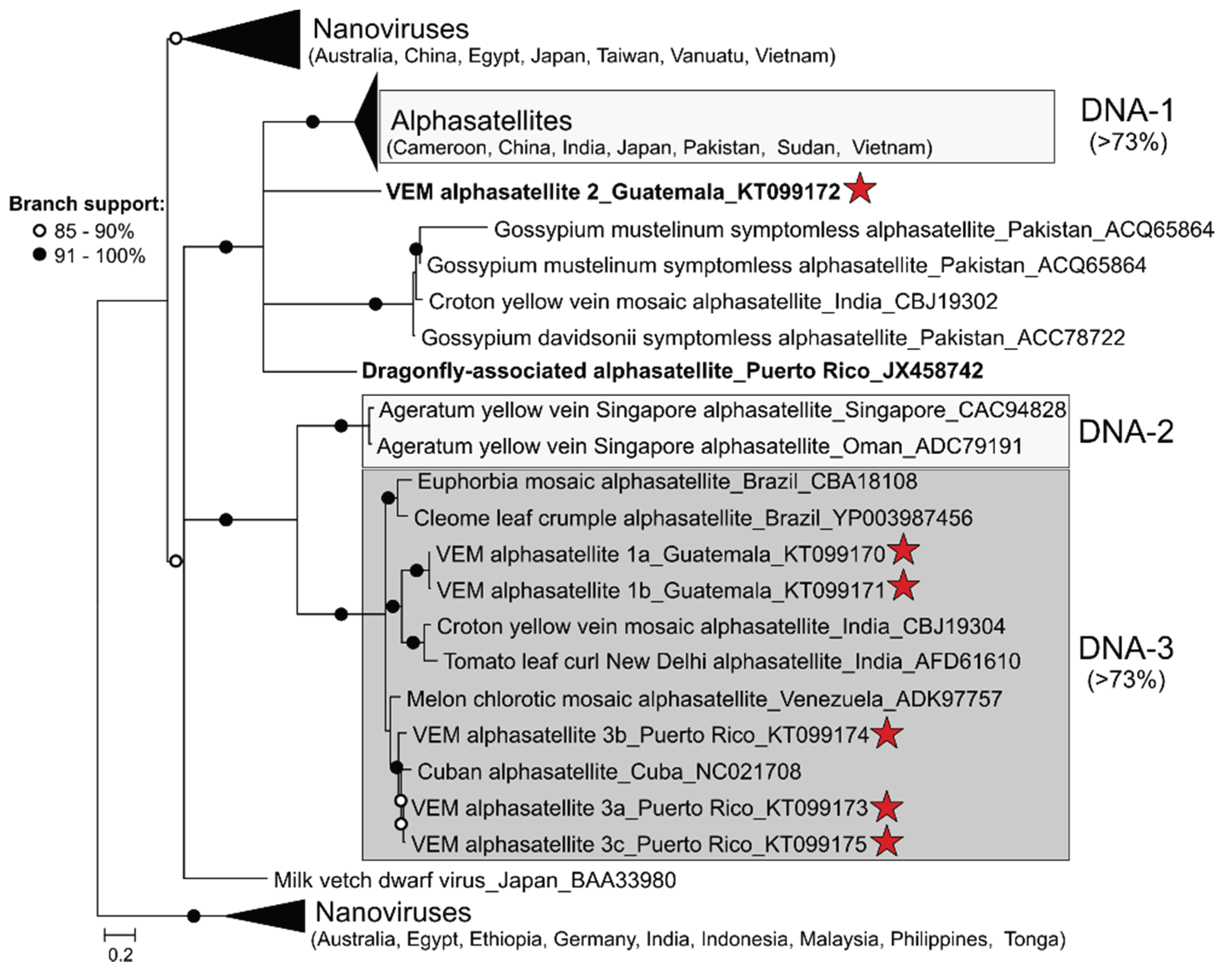
3.2. Detection of Protein-Encoding Satellite DNAs and Identification of a New Alphasatellite Clade
| Dataset | Genome a (Accession Number) | Best Match b (% Identity)/Accession Number | Satellite Size (nt) |
|---|---|---|---|
| Guat_T2 | VEM alphasatellite 1a (KT099170) | MeCMV-associated alphasatellite, Venezuela (76%)/KF670648 | 1370 |
| VEM alphasatellite 1b (KT099171) | MeCMV-associated alphasatellite, Venezuela (74%)/KF670648 | 1329 | |
| VEM alphasatellite 2 (KT099172) | Ageratum enation virus associated DNA 1, India (71%)/FN543100 | 1359 | |
| Israel_S | VEM Cotton leaf curl Gezira betasatellite * (KT099176) | Cotton leaf curl Gezira betasatellite, Egypt(95%)/AF397215 | 1324 |
| VEM Cotton leaf curl Gezira betasatellite * (KT099177) | Cotton leaf curl Gezira betasatellite, Egypt (99%)/AF397217 | 1348 | |
| VEM Cotton leaf curl Gezira betasatellite (KU095847) | Cotton leaf curl Gezira betasatellite, Egypt(98%)/AF397215 | 1310 | |
| VEM Cotton leaf curl Gezira virus alphasatellite * (KT099178) | Cotton leaf curl Gezira virus DNA 1, Burkina Faso (96%)/FN554581 | 1359 | |
| PR_T1 | VEM Ipomoea begomovirus satellite 1 (KT099179) | Ipomoea begomovirus satellite DNA beta isolate, Spain (70%)/FJ914403 | 731 |
| VEM Ipomoea begomovirus satellite 2 (KT099180) | Ipomoea begomovirus satellite DNA beta isolate, Spain (70%)/FJ914403 | 713 | |
| VEM Ipomoea begomovirus satellite 3 (KT099181) | SPLCLaV replication-associated protein, Spain (95%) **/EU839579 | 743 | |
| VEM alphasatellite 3a (KT099173) | Cuban alphasatellite 1, Cuba (86%)/HE806451 | 1300 | |
| PR_P | VEM alphasatellite 3b (KT099174) | Cuban alphasatellite 1, Cuba (85%)/HE806451 | 1307 |
| VEM alphasatellite 3c (KT099175) | Cuban alphasatellite 1, Cuba (86%)/HE806451 | 1303 | |
| PR_T1, T2, P, and E | VEM Ipomoea begomovirus satellite (Contigs, n = 49) * | Ipomoea begomovirus satellite DNA beta isolate, Spain (< 80%)/Various | |
| Spain T and S | VEM Ipomoea begomovirus satellite (Contigs, n = 10) * | Ipomoea begomovirus satellite DNA beta isolate, Spain (> 90%)/Various |
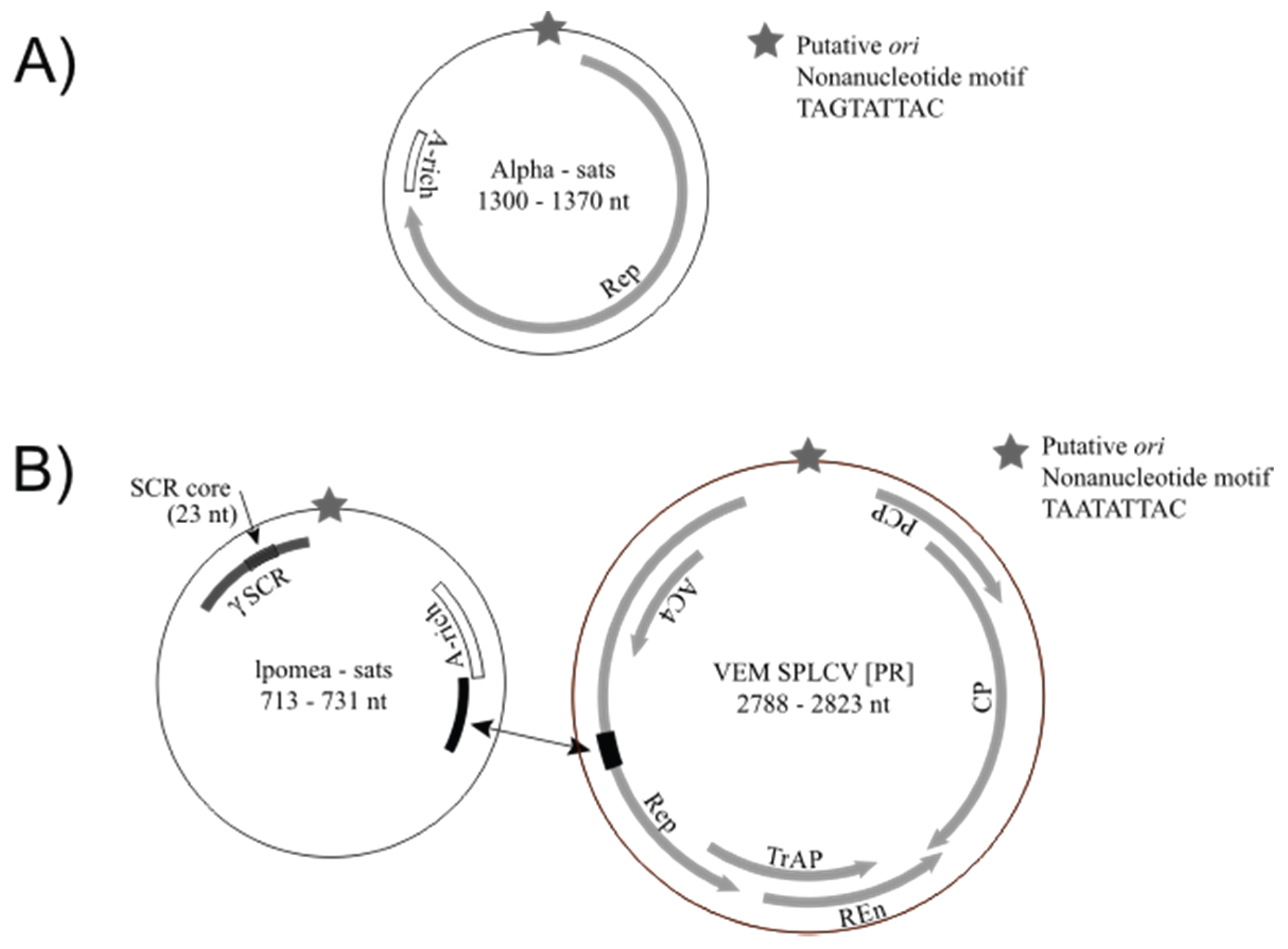
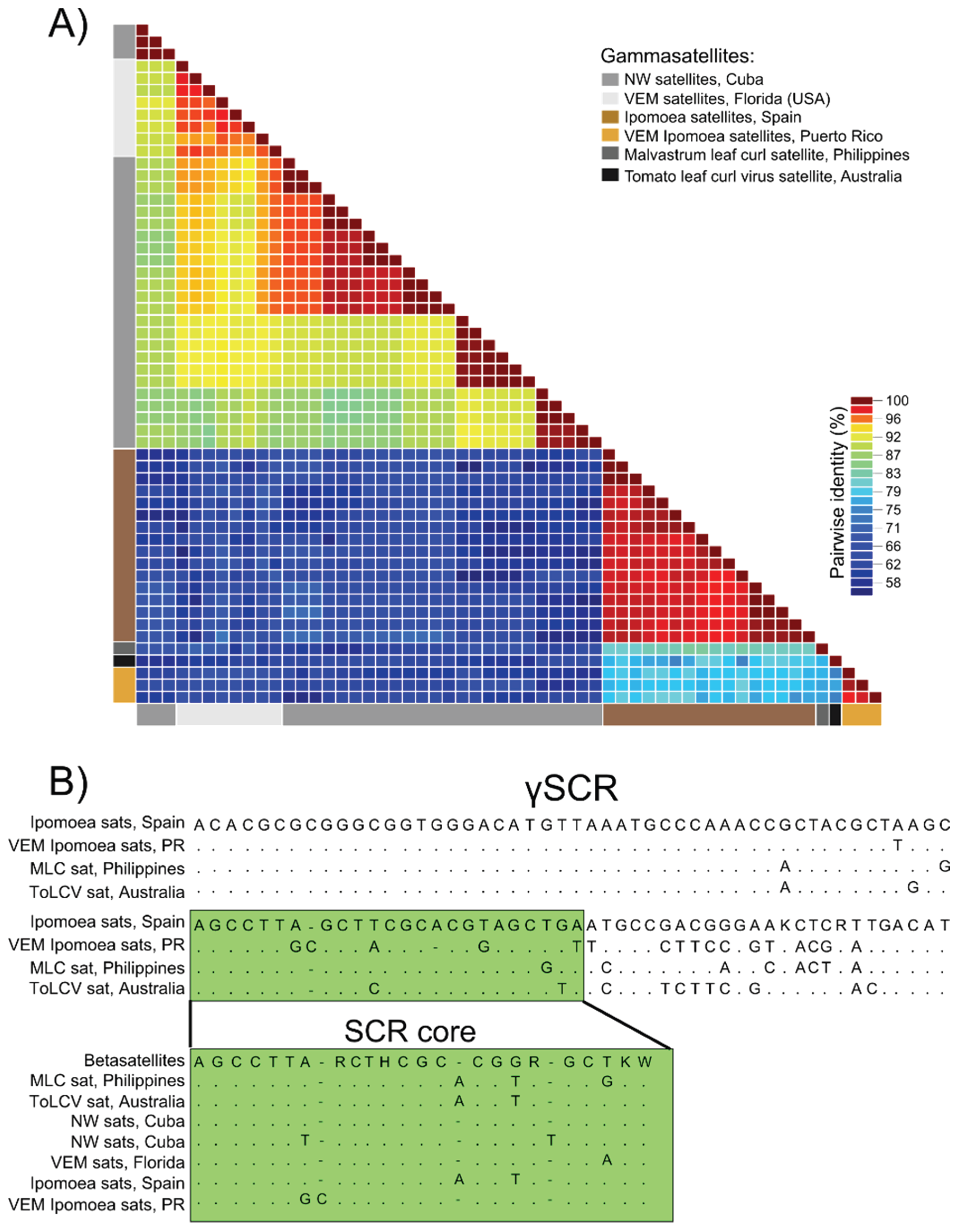
3.3. Detection of Non-Coding Small Satellite DNAs (Gammasatellites)
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Leke, W.; Mignouna, D.; Brown, J.; Kvarnheden, A. Begomovirus disease complex: Emerging threat to vegetable production systems of West and Central Africa. Agric. Food Secur. 2015, 4. [Google Scholar] [CrossRef]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Briddon, R.W.; Zafar, Y.; Stanley, J. Geminivirus disease complexes: An emerging threat. Trends Plant Sci. 2003, 8, 128–134. [Google Scholar] [CrossRef]
- Mansoor, S.; Zafar, Y.; Briddon, R.W. Geminivirus disease complexes: The threat is spreading. Trends Plant Sci. 2006, 11, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.; Coombs, S.; Revill, P.; Harding, R.; Vu, M.; Dale, J. Molecular characterization of begomoviruses and DNA satellites from Vietnam: Additional evidence that the New World geminiviruses were present in the Old World prior to continental separation. J. Gen. Virol. 2008, 89, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Evolution of geminiviruses and their satellites. FEBS Lett. 2009, 583, 1825–1832. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Mansoor, S.; Bedford, I.D.; Rishi, N.; Siwatch, S.S.; Zafar, Y.; Abdel-Salam, A.M.; Markham, P.G. Diversity of DNA 1: A satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology 2004, 324, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, P.; Liu, P.; Gong, H.; Zhou, X. Characterization of alphasatellites associated with monopartite begomovirus/betasatellite complexes in Yunnan, China. Virol. J. 2010, 7. [Google Scholar] [CrossRef]
- Romay, G.; Chirinos, D.; Geraud-Pouey, F.; Desbiez, C. Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch. Virol. 2010, 155, 1843–1847. [Google Scholar] [CrossRef] [PubMed]
- Paprotka, T.; Metzler, V.; Jeske, H. The first DNA 1-like α satellites in association with New World begomoviruses in natural infections. Virology 2010, 404, 148–157. [Google Scholar] [CrossRef]
- Briddon, R.W.; Stanley, J. Subviral agents associated with plant single-stranded DNA viruses. Virology 2006, 344, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.D.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M.; et al. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef]
- Rosario, K.; Duffy, S.; Breitbart, M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch. Virol. 2012, 157, 1851–1871. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Norman, A.; Gucciardo, S.; Stanley, J. The DNA β satellite component associated with ageratum yellow vein disease encodes an essential pathogenicity protein (βC1). Virology 2004, 324, 37–47. [Google Scholar] [CrossRef]
- Saeed, M.; Behjatnia, S.A.A.; Mansoor, S.; Zafar, Y.; Hasnain, S.; Rezaian, M.A. A single complementary-sense transcript of a geminiviral DNA β satellite is determinant of pathogenicity. Mol. Plant Microbe Interact. 2005, 18, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A begomovirus DNA β-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Dogra, S.C.; Dry, I.B.; Randles, J.W. Silencing suppressor activity of a begomovirus DNA β encoded protein and its effect on heterologous helper virus replication. Virus Res. 2012, 167, 97–101. [Google Scholar] [CrossRef]
- Gopal, P.; Pravin Kumar, P.; Sinilal, B.; Jose, J.; Kasin Yadunandam, A.; Usha, R. Differential roles of C4 and βC1 in mediating suppression of post-transcriptional gene silencing: Evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 2007, 123, 9–18. [Google Scholar] [CrossRef]
- Sharma, P.; Ikegami, M.; Kon, T. Identification of the virulence factors and suppressors of posttranscriptional gene silencing encoded by Ageratum yellow vein virus, a monopartite begomovirus. Virus Res. 2010, 149, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Fauquet, C.M. Differential interaction between cassava mosaic geminiviruses and geminivirus satellites. J. Gen. Virol. 2010, 91, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.M.; Shahid, M.S.; Briddon, R.W.; Khan, A.J.; Zhu, J.-K.; Brown, J.K. An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation. J. Gen. Virol. 2011, 92, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-Ul-Rehman, M.S.; Nahid, N.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology 2010, 405, 300–308. [Google Scholar] [CrossRef]
- Nawaz-ul-Rehman, M.S.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Maintenance of an Old World betasatellite by a New World helper begomovirus and possible rapid adaptation of the betasatellite. J. Virol. 2009, 83, 9347–9355. [Google Scholar] [CrossRef]
- Saeed, M.; Zafar, Y.; Randles, J.W.; Rezaian, M.A. A monopartite begomovirus-associated DNA β satellite substitutes for the DNA β of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 2007, 88, 2881–2889. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, Y.; Raja, P.; Li, S.; Wolf, J.N.; Shen, Q.; Bisaro, D.M.; Zhou, X. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011, 7, e1002329. [Google Scholar] [CrossRef]
- Zhou, X.; Xie, Y.; Tao, X.; Zhang, Z.; Li, Z.; Fauquet, C.M. Characterization of DNA β associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J. Gen. Virol. 2003, 84, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Bedford, I.D.; Stanley, J. Adaptation from whitefly to leafhopper transmission of an autonomously replicating nanovirus-like DNA component associated with ageratum yellow vein disease. J. Gen. Virol. 2002, 83, 907–913. [Google Scholar] [CrossRef]
- Saunders, K.; Stanley, J. A nanovirus-like DNA component associated with yellow vein disease of Ageratum conyzoides: Evidence for interfamilial recombination between plant DNA viruses. Virology 1999, 264, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Chattopadhyay, B.; Chakraborty, S. Biology and interactions of two distinct monopartite begomoviruses and betasatellites associated with radish leaf curl disease in India. Virol. J. 2012, 9. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Kumar, J.; Singh, S.P.; Tuli, R. Association of satellites with a mastrevirus in natural infection: Complexity of Wheat dwarf India virus disease. J. Virol. 2014, 88, 7093–7104. [Google Scholar] [CrossRef]
- Huang, C.; Xie, Y.; Zhao, L.; Ren, H.; Li, Z. A naturally occurring defective DNA satellite associated with a monopartite begomovirus: Evidence for recombination between alphasatellite and betasatellite. Viruses 2013, 5, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.E.; vandenBosch, F.; Jeger, M.J. Factors influencing begomovirus evolution and their increasing global significance: Implications for sustainable control. Crit. Rev. Plant Sci. 2006, 25, 23–46. [Google Scholar] [CrossRef]
- Harrison, B.D.; Robinson, D.J. Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (begomoviruses). Annu. Rev. Phytopathol. 1999, 37, 369–398. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Capobianco, H.; Ng, T.F.; Breitbart, M.; Polston, J.E. RNA viral metagenome of whiteflies leads to the discovery and characterization of a whitefly-transmitted carlavirus in North America. PLoS ONE 2014, 9, e86748. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.F.; Duffy, S.; Polston, J.E.; Bixby, E.; Vallad, G.E.; Breitbart, M. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE 2011, 6, e19050. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.F.; Willner, D.L.; Lim, Y.W.; Schmieder, R.; Chau, B.; Nilsson, C.; Anthony, S.; Ruan, Y.J.; Rohwer, F.; Breitbart, M. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE 2011, 6, e20579. [Google Scholar] [CrossRef]
- Rosario, K.; Seah, Y.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.; Duffy, S.; Breitbart, M. Vector-enabled metagenomic (VEM) surveys using whiteflies (Aleyrodidae) reveal novel begomovirus species in the New and Old Worlds. Viruses 2015, 7, 5553–5570. [Google Scholar] [CrossRef]
- Shatters, R.G.; Powell, C.A.; Boykin, L.; Liansheng, H.; McKenzie, C.L. Improved DNA barcoding method for Bemisia tabaci and related Aleyrodidae: Development of universal and Bemisia tabaci biotype-specific mitochonddrial cytochrome c oxidase I polymerase chain reaction primers. J. Econ. Entomol. 2009, 102, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Inoue-Nagata, A.K.; Albuquerque, L.C.; Rocha, W.B.; Nagata, T. A simple method for cloning the complete begomovirus genome using the bacteriophage φ29 DNA polymerase. J. Virol. Methods 2004, 116, 209–211. [Google Scholar] [CrossRef]
- Niu, B.; Fu, L.; Sun, S.; Li, W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Tournayre, J.; Mahul, A.; Debroas, D.; Enault, F. Metavir 2: New tools for viral metagenome comparison and assembled virome analysis. BMC Bioinform. 2014, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mubin, M.; Briddon, R.W.; Mansoor, S. Complete nucleotide sequence of chili leaf curl virus and its associated satellites naturally infecting potato in Pakistan. Arch. Virol. 2009, 154, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X.; Fauquet, C.M. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch. Virol. 2008, 153, 763–781. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Hodcroft, E. TreeCollapser CL4. Available online: http://emmahodcroft.com/TreeCollapseCL.html (accessed on 22 October 2015).
- Tiendrebeogo, F.; Lefeuvre, P.; Hoareau, M.; Villemot, J.; Konate, G.; Traore, A.; Barro, N.; Traore, V.; Reynaud, B.; Traore, O.; et al. Molecular diversity of Cotton leaf curl Gezira virus isolates and their satellite DNAs associated with okra leaf curl disease in Burkina Faso. Virol. J. 2010, 7. [Google Scholar] [CrossRef] [Green Version]
- Jeske, H.; Kober, S.; Schäfer, B.; Strohmeier, S. Circomics of Cuban geminiviruses reveals the first alpha-satellite DNA in the Caribbean. Virus Genes 2014, 49, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Padilla-Rodriguez, M.; Kraberger, S.; Stainton, D.; Martin, D.P.; Breitbart, M.; Varsani, A. Discovery of a novel mastrevirus and alphasatellite-like circular DNA in dragonflies (Epiprocta) from Puerto Rico. Virus Res. 2013, 171, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Zaffalon, V.; Mukherjee, S.; Reddy, V.; Thompson, J.; Tepfer, M. A survey of geminiviruses and associated satellite DNAs in the cotton-growing areas of northwestern India. Arch. Virol. 2012, 157, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Martínez-Zubiaur, Y.; Moriones, E.; Navas-Castillo, J. A novel class of DNA satellites associated with New World begomoviruses. Virology 2012, 426, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dry, I.B.; Krake, L.R.; Rigden, J.E.; Rezaian, M.A. A novel subviral agent associated with a geminivirus: The first report of a DNA satellite. Proc. Natl. Acad. Sci. USA 1997, 94, 7088–7093. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Khan, S.H.; Bashir, A.; Saeed, M.; Zafar, Y.; Malik, K.A.; Briddon, R.; Stanley, J.; Markham, P.G. Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology 1999, 259, 190–199. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosario, K.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Breitbart, M. Begomovirus-Associated Satellite DNA Diversity Captured Through Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae). Viruses 2016, 8, 36. https://doi.org/10.3390/v8020036
Rosario K, Marr C, Varsani A, Kraberger S, Stainton D, Moriones E, Polston JE, Breitbart M. Begomovirus-Associated Satellite DNA Diversity Captured Through Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae). Viruses. 2016; 8(2):36. https://doi.org/10.3390/v8020036
Chicago/Turabian StyleRosario, Karyna, Christian Marr, Arvind Varsani, Simona Kraberger, Daisy Stainton, Enrique Moriones, Jane E. Polston, and Mya Breitbart. 2016. "Begomovirus-Associated Satellite DNA Diversity Captured Through Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae)" Viruses 8, no. 2: 36. https://doi.org/10.3390/v8020036






