Multi-Omics Studies towards Novel Modulators of Influenza A Virus–Host Interaction
Abstract
:1. Introduction
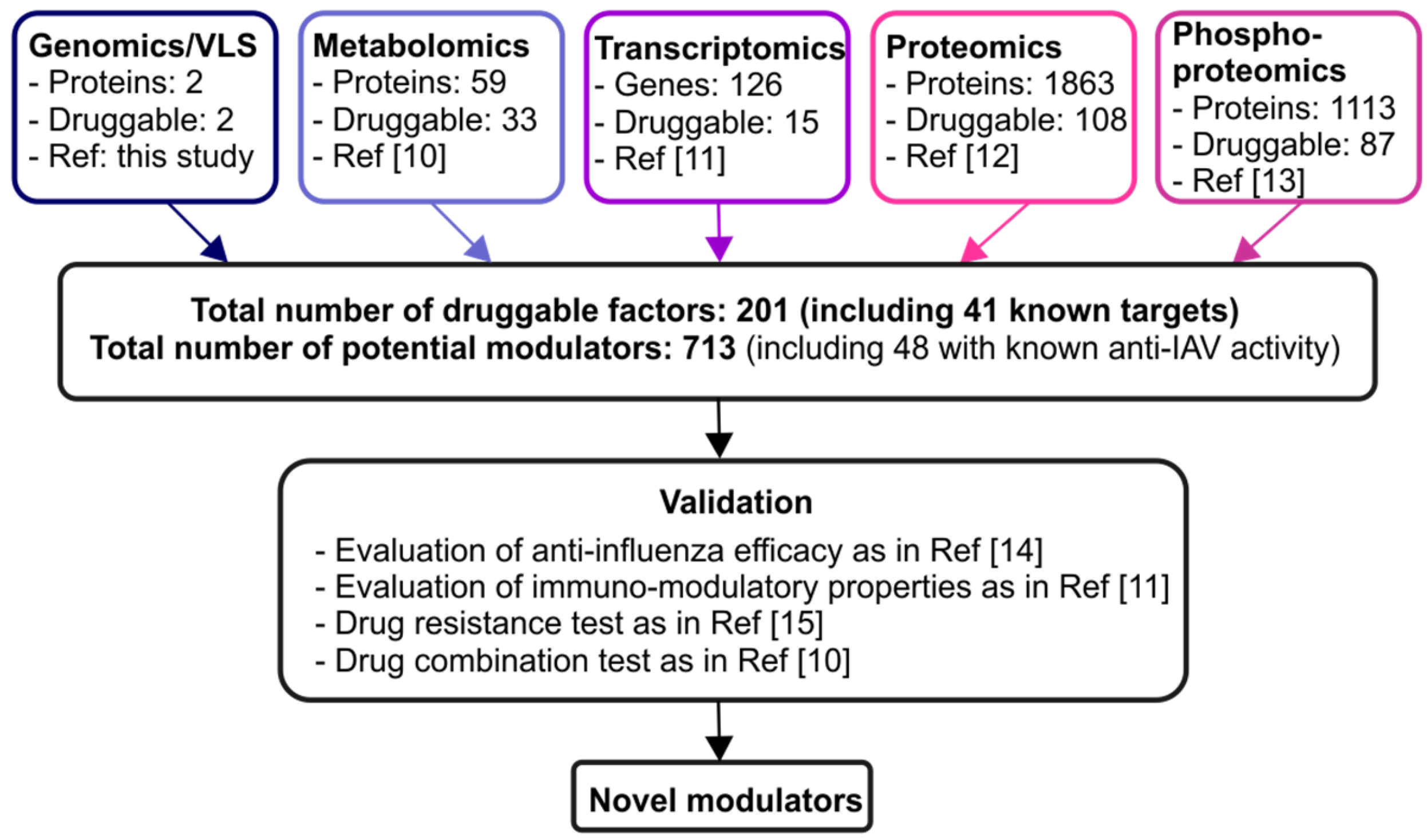
2. Combination of Various Omics Techniques Identifies Potential Novel Modulators of IAV–Host Interaction
3. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Belanov, S.S.; Bychkov, D.; Benner, C.; Ripatti, S.; Ojala, T.; Kankainen, M.; Kai Lee, H.; Wei-Tze Tang, J.; Kainov, D.E. Genome-wide analysis of evolutionary markers of human influenza A(H1N1)pdm09 and A(H3N2) viruses may guide selection of vaccine strain candidates. Genome Biol. Evol. 2015, 7, 3472–3483. [Google Scholar] [CrossRef] [PubMed]
- Rewar, S.; Mirdha, D.; Rewar, P. Treatment and prevention of pandemic H1N1 influenza. Ann. Glob. Health 2015, 81, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Kamps, B.S.; Hoffmann, C. Amantadine. Available online: www.influenzareport.com/ir/drugs/amanta.htm (accessed on 28 September 2016).
- Spanakis, N.; Pitiriga, V.; Gennimata, V.; Tsakris, A. A review of neuraminidase inhibitor susceptibility in influenza strains. Expert Rev. Anti Infect. Ther. 2014, 12, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Rao, M.; Wallis, R.S.; Kaufmann, S.H.; Rustomjee, R.; Mwaba, P.; Vilaplana, C.; Yeboah-Manu, D.; Chakaya, J.; Ippolito, G.; et al. Host-directed therapies for infectious diseases: Current status, recent progress, and future prospects. Lancet Infect. Dis. 2016, 16, e47–e63. [Google Scholar] [CrossRef]
- McKimm-Breschkin, J.L.; Fry, A.M. Meeting report: 4th ISIRV antiviral group conference: Novel antiviral therapies for influenza and other respiratory viruses. Antivir. Res. 2016, 129, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Loregian, A.; Mercorelli, B.; Nannetti, G.; Compagnin, C.; Palu, G. Antiviral strategies against influenza virus: Towards new therapeutic approaches. Cell. Mol. Life Sci. 2014, 71, 3659–3683. [Google Scholar] [CrossRef] [PubMed]
- Muller, K.H.; Kakkola, L.; Nagaraj, A.S.; Cheltsov, A.V.; Anastasina, M.; Kainov, D.E. Emerging cellular targets for influenza antiviral agents. Trends Pharmacol. Sci. 2012, 33, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Vanderlinden, E.; Naesens, L. Emerging antiviral strategies to interfere with influenza virus entry. Med. Res. Rev. 2014, 34, 301–339. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Gaelings, L.; Soderholm, S.; Belanov, S.; Nandania, J.; Nyman, T.A.; Matikainen, S.; Anders, S.; Velagapudi, V.; Kainov, D.E. JNJ872 inhibits influenza a virus replication without altering cellular antiviral responses. Antivir. Res. 2016, 133, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Soderholm, S.; Anastasina, M.; Islam, M.M.; Tynell, J.; Poranen, M.M.; Bamford, D.H.; Stenman, J.; Julkunen, I.; Sauliene, I.; de Brabander, J.K.; et al. Immuno-modulating properties of saliphenylhalamide, SNS-032, obatoclax, and gemcitabine. Antivir. Res. 2016, 126, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lietzen, N.; Ohman, T.; Rintahaka, J.; Julkunen, I.; Aittokallio, T.; Matikainen, S.; Nyman, T.A. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 2011, 7, e1001340. [Google Scholar] [CrossRef] [PubMed]
- Soderholm, S.; Kainov, D.; Ohman, T.; Denisova, O.; Schepens, B.; Kulesskiy, E.; Imanishi, S.Y.; Corthals, G.; Hintsanen, P.; Aittokallio, T.; et al. Phosphoproteomics to characterize host response during influenza A virus infection of human macrophages. Mol. Cell. Proteom. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Denisova, O.V.; Kakkola, L.; Feng, L.; Stenman, J.; Nagaraj, A.; Lampe, J.; Yadav, B.; Aittokallio, T.; Kaukinen, P.; Ahola, T.; et al. Obatoclax, saliphenylhalamide, and gemcitabine inhibit influenza A virus infection. J. Biol. Chem. 2012, 287, 35324–35332. [Google Scholar] [CrossRef] [PubMed]
- Denisova, O.V.; Soderholm, S.; Virtanen, S.; von Schantz, C.; Bychkov, D.; Vashchinkina, E.; Desloovere, J.; Tynell, J.; Ikonen, N.; Theisen, L.L.; et al. Akt inhibitor MK2206 prevents influenza pH1N1 virus infection in vitro. Antimicrob. Agents Chemother. 2014, 58, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Griffith, O.L.; Coffman, A.C.; Weible, J.V.; McMichael, J.F.; Spies, N.C.; Koval, J.; Das, I.; Callaway, M.B.; Eldred, J.M.; et al. DGidb: Mining the druggable genome. Nat. Methods 2013, 10, 1209–1210. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Acharya, B.; Thapa, K. Indoor staying during winter season makes people more susceptible to flu. J. Nepal Health Res. Counc. 2016, 14, 69–70. [Google Scholar] [PubMed]
- Yu, M.; Wang, Q.; Qi, W.; Zhang, K.; Liu, J.; Tao, P.; Ge, S.; Liao, M.; Ning, Z. Expression of inflammation-related genes in the lung of BALB/c mice response to H7N9 influenza A virus with different pathogenicity. Med. Microbiol. Immunol. 2016, 205, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Liu, X.; Zhang, P.; Li, D.; Xu, S.; Zhou, Q.; Guo, M.; Huai, W.; Chen, X.; Wang, Q.; et al. Rb selectively inhibits innate IFN-beta production by enhancing deacetylation of IFN-beta promoter through HDAC1 and HDAC8. J. Autoimmun. 2016, 73, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ou, R.; Rabelo de Souza, G.; Cunha, T.M.; Lemos, H.; Mohamed, E.; Li, L.; Pacholczyk, G.; Randall, J.; Munn, D.H.; et al. Virus infections incite pain hypersensitivity by inducing indoleamine 2,3 dioxygenase. PLoS Pathog. 2016, 12, e1005615. [Google Scholar] [CrossRef] [PubMed]
- Blank, T.; Detje, C.N.; Spiess, A.; Hagemeyer, N.; Brendecke, S.M.; Wolfart, J.; Staszewski, O.; Zoller, T.; Papageorgiou, I.; Schneider, J.; et al. Brain endothelial- and epithelial-specific interferon receptor chain 1 drives virus-induced sickness behavior and cognitive impairment. Immunity 2016, 44, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Marion, T.; Elbahesh, H.; Thomas, P.G.; DeVincenzo, J.P.; Webby, R.; Schughart, K. Respiratory mucosal proteome quantification in human influenza infections. PLoS ONE 2016, 11, e0153674. [Google Scholar] [CrossRef] [PubMed]
- Valkenburg, S.A.; Josephs, T.M.; Clemens, E.B.; Grant, E.J.; Nguyen, T.H.; Wang, G.C.; Price, D.A.; Miller, A.; Tong, S.Y.; Thomas, P.G.; et al. Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses. Proc. Natl. Acad. Sci. USA 2016, 113, 4440–4445. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Lim, W.; Bist, P.; Perumalsamy, R.; Lukman, H.M.; Li, F.; Welker, L.B.; Yan, B.; Sethi, G.; Tambyah, P.A.; et al. Influenza A virus enhances its propagation through the modulation of Annexin-A1 dependent endosomal trafficking and apoptosis. Cell Death Differ. 2016, 23, 1243–1256. [Google Scholar] [CrossRef] [PubMed]
- Nagesh, P.T.; Husain, M. Influenza A virus dysregulates host histone deacetylase 1 that inhibits viral infection in lung epithelial cells. J. Virol. 2016, 90, 4614–4625. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhou, Y.; Zhu, S.; Feng, J.; Chen, X.; Liu, S.; Peng, N.; Yang, X.; Xu, G.; Zhu, Y. Pyruvate carboxylase activates the RIG-I-like receptor-mediated antiviral immune response by targeting the MAVS signalosome. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.R.; Christensen, J.P.; Buus, S.; Rasmussen, M.; Korsholm, K.S.; Nielsen, M.; Claesson, M.H. Immunogenicity of HLA class I and II double restricted influenza A-derived peptides. PLoS ONE 2016, 11, e0145629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.H.; Park, E.B.; Lee, J.Y.; Min, J.Y. Identification of novel membrane-associated prostaglandin E synthase-1 (mPGES-1) inhibitors with anti-influenza activities in vitro. Biochem. Biophys. Res. Commun. 2016, 469, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.G.; Gamage, A.; Zyla, R.; Armstrong, S.M.; Advani, S.; Advani, A.; Wang, C.; Lee, W.L. Influenza virus infection induces platelet-endothelial adhesion which contributes to lung injury. J. Virol. 2016, 90, 1812–1823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tian, H.; Cui, J.; Xiao, J.; Wang, M.; Hu, Y. The C-Jun N-terminal kinase (JNK) is involved in H5N1 influenza A virus RNA and protein synthesis. Arch. Virol. 2016, 161, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Noti, J.D.; McMillen, C.M.; Beezhold, D.H. ICAM-1 regulates the survival of influenza virus in lung epithelial cells during the early stages of infection. Virology 2016, 487, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Aeffner, F.; Woods, P.S.; Davis, I.C. Ecto-5′-nucleotidase CD73 modulates the innate immune response to influenza infection but is not required for development of influenza-induced acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 309, L1313–L1322. [Google Scholar] [CrossRef] [PubMed]
- Checconi, P.; Salzano, S.; Bowler, L.; Mullen, L.; Mengozzi, M.; Hanschmann, E.M.; Lillig, C.H.; Sgarbanti, R.; Panella, S.; Nencioni, L.; et al. Redox proteomics of the inflammatory secretome identifies a common set of redoxins and other glutathionylated proteins released in inflammation, influenza virus infection and oxidative stress. PLoS ONE 2015, 10, e0127086. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Q.; Xie, J.; Xu, Y. Cigarette smoke inhibits BAFF expression and mucosal immunoglobulin a responses in the lung during influenza virus infection. Respir. Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.M.; Crabtree, J.M.; Sage, L.K.; Tompkins, S.M.; Tripp, R.A. Interferon lambda upregulates IDO1 expression in respiratory epithelial cells after influenza virus infection. J. Interferon Cytokine Res. 2015, 35, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, J.; Cheng, Z.; Yang, S.; Chu, H.; Fan, Y.; Li, C.; Wong, B.H.; Zheng, S.; Zhu, Y.; et al. Functional variants regulating LGALS1 (Galectin 1) expression affect human susceptibility to influenza A(H7N9). Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.R.; Nie, W.; Zang, Y.S.; Fang, Z.; Xiu, Q.Y.; Xu, X.X. Clinical aspects and cytokine response in adults with seasonal influenza infection. Int. J. Clin. Exp. Med. 2014, 7, 5593–5602. [Google Scholar] [PubMed]
- Nita-Lazar, M.; Banerjee, A.; Feng, C.; Amin, M.N.; Frieman, M.B.; Chen, W.H.; Cross, A.S.; Wang, L.X.; Vasta, G.R. Desialylation of airway epithelial cells during influenza virus infection enhances pneumococcal adhesion via galectin binding. Mol. Immunol. 2015, 65, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jolly, L.; Stavrou, A.; Vanderstoken, G.; Meliopoulos, V.A.; Habgood, A.; Tatler, A.L.; Porte, J.; Knox, A.; Weinreb, P.; Violette, S.; et al. Influenza promotes collagen deposition via alphaVbeta6 integrin-mediated transforming growth factor beta activation. J. Biol. Chem. 2014, 289, 35246–35263. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.Y.; Lewis, D.E.; Han, L.; Jaja, M.; Yao, S.; Li, F.; Robertson, M.J.; Zhou, B.; Sun, J.; Chang, H.C. Activation of dendritic cell function by soypeptide lunasin as a novel vaccine adjuvant. Vaccine 2014, 32, 5411–5419. [Google Scholar] [CrossRef] [PubMed]
- Oguin, T.H., 3rd; Sharma, S.; Stuart, A.D.; Duan, S.; Scott, S.A.; Jones, C.K.; Daniels, J.S.; Lindsley, C.W.; Thomas, P.G.; Brown, H.A. Phospholipase D facilitates efficient entry of influenza virus, allowing escape from innate immune inhibition. J. Biol. Chem. 2014, 289, 25405–25417. [Google Scholar] [CrossRef] [PubMed]
- Hrincius, E.R.; Liedmann, S.; Anhlan, D.; Wolff, T.; Ludwig, S.; Ehrhardt, C. Avian influenza viruses inhibit the major cellular signalling integrator c-Abl. Cell Microbiol. 2014, 16, 1854–1874. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Wu, Q.M.; Zhang, L.J.; Wang, Z.G.; Sun, E.Z.; Zhang, Z.L.; Pang, D.W. Three-dimensional tracking of Rab5- and Rab7-associated infection process of influenza virus. Small 2014, 10, 4746–4753. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, K.; Wang, T.; Wang, X.; Lu, X.; Peng, B.; Wu, W.; Zhang, R.; Chen, S.; Zhang, R.; et al. Primary study on the lesions and specific proteins in BEAS-2B cells induced with the 2009 A (H1N1) influenza virus. Appl. Microbiol. Biotechnol. 2014, 98, 9691–9701. [Google Scholar] [CrossRef] [PubMed]
- To, E.E.; Broughton, B.R.; Hendricks, K.S.; Vlahos, R.; Selemidis, S. Influenza A virus and TLR7 activation potentiate Nox2 oxidase-dependent Ros production in macrophages. Free Radic. Res. 2014, 48, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Chesarino, N.M.; McMichael, T.M.; Hach, J.C.; Yount, J.S. Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J. Biol. Chem. 2014, 289, 11986–11992. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Li, Y.; Pan, R.; Zou, X. Characterizing and controlling the inflammatory network during influenza A virus infection. Sci. Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Tsuda, M.; Nanbo, A.; Hattori, T.; Sasaki, J.; Sasaki, T.; Miyazaki, T.; Ohba, Y. A Ca2+-dependent signaling circuit regulates influenza A virus internalization and infection. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- McGuire, P.J.; Tarasenko, T.N.; Wang, T.; Levy, E.; Zerfas, P.M.; Moran, T.; Lee, H.S.; Bequette, B.J.; Diaz, G.A. Acute metabolic decompensation due to influenza in a mouse model of ornithine transcarbamylase deficiency. Dis. Model. Mech. 2014, 7, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.J.; Pritzl, C.J.; Vijayan, M.; Bomb, K.; McClain, M.E.; Alexander, S.; Hahm, B. Sphingosine kinase 1 serves as a pro-viral factor by regulating viral RNA synthesis and nuclear export of viral ribonucleoprotein complex upon influenza virus infection. PLoS ONE 2013, 8, e75005. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, T.; Asano, A.; Usui, T.; Takeuchi, T.; Watanabe, Y.; Yamano, Y. Antiviral and antiproliferative effects of canine interferon-λ1. Vet. Immunol. Immunopathol. 2013, 156, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Buggele, W.A.; Krause, K.E.; Horvath, C.M. Small RNA profiling of influenza A virus-infected cells identifies miR-449B as a regulator of histone deacetylase 1 and interferon β. PLoS ONE 2013, 8, e76560. [Google Scholar] [CrossRef] [PubMed]
- Naesens, L.; Guddat, L.W.; Keough, D.T.; van Kuilenburg, A.B.; Meijer, J.; Vande Voorde, J.; Balzarini, J. Role of human hypoxanthine guanine phosphoribosyltransferase in activation of the antiviral agent T-705 (favipiravir). Mol. Pharmacol. 2013, 84, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Chen, C.J.; Yen, C.T.; Yu, C.P.; Huang, P.N.; Kuo, R.L.; Lin, S.J.; Chang, C.K.; Shih, S.R. Caspase-1 deficient mice are more susceptible to influenza A virus infection with PA variation. J. Infect. Dis. 2013, 208, 1898–1905. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, L.; Klonowski, K.D.; Tompkins, S.M.; Tripp, R.A.; Mellor, A.L. Induction and role of indoleamine 2,3 dioxygenase in mouse models of influenza A virus infection. PLoS ONE 2013, 8, e66546. [Google Scholar] [CrossRef] [PubMed]
- Selemidis, S.; Seow, H.J.; Broughton, B.R.; Vinh, A.; Bozinovski, S.; Sobey, C.G.; Drummond, G.R.; Vlahos, R. Nox1 oxidase suppresses influenza A virus-induced lung inflammation and oxidative stress. PLoS ONE 2013, 8, e60792. [Google Scholar] [CrossRef] [PubMed]
- Skovgaard, K.; Cirera, S.; Vasby, D.; Podolska, A.; Breum, S.O.; Durrwald, R.; Schlegel, M.; Heegaard, P.M. Expression of innate immune genes, proteins and microRNAs in lung tissue of pigs infected experimentally with influenza virus (H1N2). Innate Immun. 2013, 19, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wang, Q.; Chen, S.; Gao, S.; Song, L.; Liu, P.; Huang, W. Influenza a virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein. J. Virol. 2013, 87, 3039–3052. [Google Scholar] [CrossRef] [PubMed]
- Bauer, R.N.; Brighton, L.E.; Mueller, L.; Xiang, Z.; Rager, J.E.; Fry, R.C.; Peden, D.B.; Jaspers, I. Influenza enhances caspase-1 in bronchial epithelial cells from asthmatic volunteers and is associated with pathogenesis. J. Allergy Clin. Immunol. 2012, 130, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Leung, H.S.; Li, O.T.; Chan, R.W.; Chan, M.C.; Nicholls, J.M.; Poon, L.L. Entry of influenza A virus with a alpha2,6-linked sialic acid binding preference requires host fibronectin. J. Virol. 2012, 86, 10704–10713. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, I.; McNally, B.; Willette, M.; Peeples, M.E.; Chaussabel, D.; Durbin, J.E.; Ramilo, O.; Mejias, A.; Flano, E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J. Virol. 2012, 86, 5422–5436. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Kien, F.; Maniere, M.; Zhang, Y.; Lagarde, N.; Tse, K.S.; Poon, L.L.; Nal, B. Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection. J. Virol. 2012, 86, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Jost, S.; Quillay, H.; Reardon, J.; Peterson, E.; Simmons, R.P.; Parry, B.A.; Bryant, N.N.; Binder, W.D.; Altfeld, M. Changes in cytokine levels and NK cell activation associated with influenza. PLoS ONE 2011, 6, e25060. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, J.; Torres, M.; Romo, J.; Torres, D.; Jimenez, L.; Ramirez, G.; Cruz, A.; Espinosa, E.; Herrera, T.; Buendia, I.; et al. Inflammatory profiles in severe pneumonia associated with the pandemic influenza A/H1N1 virus isolated in Mexico city. Autoimmunity 2011, 44, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.L.; Chen, Y.H.; Wang, S.W.; Huang, Y.J.; Leu, C.H.; Yeh, N.C.; Chu, C.Y.; Lin, C.C.; Shieh, G.S.; Chen, Y.L.; et al. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J. Virol. 2011, 85, 10010–10020. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Liu, Y.; Zheng, J.; Ng, I.H.; Xiang, Z.; Lam, K.T.; Mao, H.; Li, H.; Peiris, J.S.; Lau, Y.L.; Tu, W. Type 1 responses of human Vgamma9Vdelta2 T cells to influenza A viruses. J. Virol. 2011, 85, 10109–10116. [Google Scholar] [CrossRef] [PubMed]
- Carroll, T.D.; Matzinger, S.R.; Fritts, L.; McChesney, M.B.; Miller, C.J. Memory B cells and CD8+ lymphocytes do not control seasonal influenza A virus replication after homologous re-challenge of rhesus macaques. PLoS ONE 2011, 6, e21756. [Google Scholar] [CrossRef] [PubMed]
- Haidari, M.; Zhang, W.; Ganjehei, L.; Ali, M.; Chen, Z. Inhibition of MLC phosphorylation restricts replication of influenza virus—A mechanism of action for anti-influenza agents. PLoS ONE 2011, 6, e21444. [Google Scholar] [CrossRef] [PubMed]
- Allen, I.C.; Moore, C.B.; Schneider, M.; Lei, Y.; Davis, B.K.; Scull, M.A.; Gris, D.; Roney, K.E.; Zimmermann, A.G.; Bowzard, J.B.; et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity 2011, 34, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, R.; Maeda, N.; Shibata, K.; Yamada, H.; Kase, T.; Yoshikai, Y. Interleukin-15 is critical in the pathogenesis of influenza A virus-induced acute lung injury. J. Virol. 2010, 84, 5574–5582. [Google Scholar] [CrossRef] [PubMed]
- Rowe, T.; Leon, A.J.; Crevar, C.J.; Carter, D.M.; Xu, L.; Ran, L.; Fang, Y.; Cameron, C.M.; Cameron, M.J.; Banner, D.; et al. Modeling host responses in ferrets during A/California/07/2009 influenza infection. Virology 2010, 401, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Harmon, A.; Jin, J.; Francis, D.H.; Christopher-Hennings, J.; Nelson, E.; Montelaro, R.C.; Li, F. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J. Virol. 2010, 84, 4673–4681. [Google Scholar] [CrossRef] [PubMed]
- Wahl, A.; Schafer, F.; Bardet, W.; Hildebrand, W.H. HLA class I molecules reflect an altered host proteome after influenza virus infection. Hum. Immunol. 2010, 71, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Danishuddin, M.; Khan, S.N.; Khan, A.U. Molecular interactions between mitochondrial membrane proteins and the C-terminal domain of PB1-F2: An in silico approach. J. Mol. Model. 2010, 16, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.G.; Dash, P.; Aldridge, J.R., Jr.; Ellebedy, A.H.; Reynolds, C.; Funk, A.J.; Martin, W.J.; Lamkanfi, M.; Webby, R.J.; Boyd, K.L.; et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 2009, 30, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Melchjorsen, J.; Kristiansen, H.; Christiansen, R.; Rintahaka, J.; Matikainen, S.; Paludan, S.R.; Hartmann, R. Differential regulation of the OASL and OAS1 genes in response to viral infections. J. Interferon Cytokine Res. 2009, 29, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kakugawa, S.; Shimojima, M.; Goto, H.; Horimoto, T.; Oshimori, N.; Neumann, G.; Yamamoto, T.; Kawaoka, Y. Mitogen-activated protein kinase-activated kinase Rsk2 plays a role in innate immune responses to influenza virus infection. J. Virol. 2009, 83, 2510–2517. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhuang, X. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc. Natl. Acad. Sci. USA 2008, 105, 11790–11795. [Google Scholar] [CrossRef] [PubMed]
- LeBouder, F.; Morello, E.; Rimmelzwaan, G.F.; Bosse, F.; Pechoux, C.; Delmas, B.; Riteau, B. Annexin II incorporated into influenza virus particles supports virus replication by converting plasminogen into plasmin. J. Virol. 2008, 82, 6820–6828. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Song, W.; Wang, P.; Lee, K.; Chan, W.; Chen, H.; Cai, Z. Proteomics analysis of differential expression of cellular proteins in response to avian H9N2 virus infection in human cells. Proteomics 2008, 8, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Link, C.; Ebensen, T.; Standner, L.; Dejosez, M.; Reinhard, E.; Rharbaoui, F.; Guzman, C.A. An SopB-mediated immune escape mechanism of Salmonella enterica can be subverted to optimize the performance of live attenuated vaccine carrier strains. Microbes Infect. 2006, 8, 2262–2269. [Google Scholar] [CrossRef] [PubMed]
- Zamarin, D.; Garcia-Sastre, A.; Xiao, X.; Wang, R.; Palese, P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Olschlager, V.; Pleschka, S.; Fischer, T.; Rziha, H.J.; Wurzer, W.; Stitz, L.; Rapp, U.R.; Ludwig, S.; Planz, O. Lung-specific expression of active Raf kinase results in increased mortality of influenza A virus-infected mice. Oncogene 2004, 23, 6639–6646. [Google Scholar] [CrossRef] [PubMed]
- Sieczkarski, S.B.; Whittaker, G.R. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 2003, 4, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Norbury, C.C.; Cho, Y.; Yewdell, J.W.; Bennink, J.R. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8+ T cells at the levels of t cell repertoire and presentation of viral antigens. J. Exp. Med. 2001, 193, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Julkunen, I.; Sareneva, T.; Pirhonen, J.; Ronni, T.; Melen, K.; Matikainen, S. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev. 2001, 12, 171–180. [Google Scholar] [CrossRef]
- Swiergiel, A.H.; Smagin, G.N.; Johnson, L.J.; Dunn, A.J. The role of cytokines in the behavioral responses to endotoxin and influenza virus infection in mice: Effects of acute and chronic administration of the interleukin-1-receptor antagonist (IL-1ra). Brain Res. 1997, 776, 96–104. [Google Scholar] [CrossRef]
- Akaike, T.; Ando, M.; Oda, T.; Doi, T.; Ijiri, S.; Araki, S.; Maeda, H. Dependence on O2− generation by xanthine oxidase of pathogenesis of influenza virus infection in mice. J. Clin. Invest. 1990, 85, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Pierson, D.; Knight, V.; Hansard, P.; Chan, E. Hepatic carbamyl phosphate synthetase and ornithine transcarbamylase in mouse influenze A and influenze B infection. Proc. Soc. Exp. Biol. Med. 1976, 152, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Basaran, E.; Karakucuk-Iyidogan, A.; Schols, D.; Oruc-Emre, E.E. Synthesis of novel chiral sulfonamide-bearing 1,2,4-triazole-3-thione analogs derived from d- and l-phenylalanine esters as potential anti-influenza agents. Chirality 2016, 28, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Akpovwa, H. Chloroquine could be used for the treatment of filoviral infections and other viral infections that emerge or emerged from viruses requiring an acidic pH for infectivity. Cell Biochem. Funct. 2016, 34, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Hyuga, S.; Hyuga, M.; Oshima, N.; Maruyama, T.; Kamakura, H.; Yamashita, T.; Yoshimura, M.; Amakura, Y.; Hakamatsuka, T.; Odaguchi, H.; et al. Ephedrine alkaloids-free ephedra herb extract: A safer alternative to ephedra with comparable analgesic, anticancer, and anti-influenza activities. J. Nat. Med. 2016, 70, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Li, R.; Li, X.; He, J.; Jiang, S.; Liu, S.; Yang, J. Quercetin as an antiviral agent inhibits influenza A virus (IAV) entry. Viruses 2016. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.X.; Zhou, H.X.; Qi, W.B.; Ning, Z.Y.; Ma, Y.J.; Li, Y.L.; Wang, G.C.; Chen, J.X. Antiviral effects of the combination of glycyrrhizin and ribavirin against influenza a H1N1 virus infection in vivo. Yao Xue Xue Bao 2015, 50, 966–972. [Google Scholar] [PubMed]
- Lu, Y.; Hardes, K.; Dahms, S.O.; Bottcher-Friebertshauser, E.; Steinmetzer, T.; Than, M.E.; Klenk, H.D.; Garten, W. Peptidomimetic furin inhibitor MI-701 in combination with oseltamivir and ribavirin efficiently blocks propagation of highly pathogenic avian influenza viruses and delays high level oseltamivir resistance in MDCK cells. Antivir. Res. 2015, 120, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Dai, J.; Zhao, X.; Chen, Y.; Wang, G.; Li, K. Chloroquine enhances replication of influenza a virus A/WSN/33 (H1N1) in dose-, time-, and MOI-dependent manners in human lung epithelial cells A549. J. Med. Virol. 2015, 87, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, Y.F.; Tang, L.P.; Tsoi, B.; Chen, M.; Chen, H.; Chen, X.M.; Tan, R.R.; Kurihara, H.; He, R.R. A new mechanism of vitamin C effects on A/FM/1/47(H1N1) virus-induced pneumonia in restraint-stressed mice. BioMed Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Ammer, E.; Nietzsche, S.; Rien, C.; Kuhnl, A.; Mader, T.; Heller, R.; Sauerbrei, A.; Henke, A. The anti-obesity drug orlistat reveals anti-viral activity. Med. Microbiol. Immunol. 2015, 204, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, B.; Ghavami, S.; Kroeker, A.L.; Mahood, T.H.; Stelmack, G.L.; Klonisch, T.; Coombs, K.M.; Halayko, A.J. Suppression of influenza A virus replication in human lung epithelial cells by noncytotoxic concentrations bafilomycin A1. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L270–L286. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Shi, X.; Ju, D.; Huang, H.; Wei, W.; Dong, X. Anti-inflammatory effect of thalidomide on H1N1 influenza virus-induced pulmonary injury in mice. Inflammation 2014, 37, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Mehrbod, P.; Hair-Bejo, M.; Tengku Ibrahim, T.A.; Omar, A.R.; El Zowalaty, M.; Ajdari, Z.; Ideris, A. Simvastatin modulates cellular components in influenza A virus-infected cells. Int. J. Mol. Med. 2014, 34, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, F.; Jaworska, J.; Verway, M.; Tzelepis, F.; Massoud, A.; Gillard, J.; Wong, G.; Kobinger, G.; Xing, Z.; Couture, C.; et al. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity 2014, 40, 554–568. [Google Scholar] [CrossRef] [PubMed]
- Shahiduzzaman, M.; Ezatti, P.; Xin, G.; Coombs, K.M. Proteasomal serine hydrolases are up-regulated by and required for influenza virus infection. J. Proteome Res. 2014, 13, 2223–2238. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhou, W.; Huang, H.; Zhu, H.; Zhou, P.; Zhu, H.; Ju, D. Inhibition of the inflammatory cytokine tumor necrosis factor-α with etanercept provides protection against lethal H1N1 influenza infection in mice. Crit. Care 2013. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Zhang, C.; Qiu, X.; Zheng, W.; Ruan, J.; Shao, Y. Exploring the molecular mechanism of action between drug and RNA polymerase based on partially-resolved spatial structures. Curr. Comput. Aided Drug Des. 2013, 9, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Alsuwaidi, A.R.; Almarzooqi, S.; Albawardi, A.; Benedict, S.; Kochiyil, J.; Mustafa, F.; Hartwig, S.M.; Varga, S.M.; Souid, A.K. Cellular bioenergetics, caspase activity and glutathione in murine lungs infected with influenza A virus. Virology 2013, 446, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, J.I.; Lee, I.; Lee, S.; Hwang, M.W.; Bae, J.Y.; Heo, J.; Kim, D.; Han, S.Z.; Park, M.S. Aronia melanocarpa and its components demonstrate antiviral activity against influenza viruses. Biochem. Biophys. Res. Commun. 2013, 440, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Gluck, B.; Schmidtke, M.; Walther, M.; Meerbach, A.; Wutzler, P. Simvastatin treatment showed no prophylactic effect in influenza virus-infected mice. J. Med. Virol. 2013, 85, 1978–1982. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, H.; Bae, S.; Choi, J.; Lim, S.Y.; Lee, N.; Kong, J.M.; Hwang, Y.I.; Kang, J.S.; Lee, W.J. Vitamin C is an essential factor on the anti-viral immune responses through the production of interferon-α/β at the initial stage of influenza A virus (H3N2) infection. Immune Netw. 2013, 13, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kim, S.H.; Huh, S.Y.; Kong, S.Y.; Choi, Y.J.; Cheong, H.J.; Kim, H.J. Reduced antibody formation after influenza vaccination in patients with neuromyelitis optica spectrum disorder treated with rituximab. Eur. J. Neurol. 2013, 20, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Belser, J.A.; Szretter, K.J.; Katz, J.M.; Tumpey, T.M. Simvastatin and oseltamivir combination therapy does not improve the effectiveness of oseltamivir alone following highly pathogenic avian H5N1 influenza virus infection in mice. Virology 2013, 439, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Checconi, P.; Sgarbanti, R.; Celestino, I.; Limongi, D.; Amatore, D.; Iuvara, A.; Alimonti, A.; Garaci, E.; Palamara, A.T.; Nencioni, L. The environmental pollutant cadmium promotes influenza virus replication in MDCK cells by altering their redox state. Int. J. Mol. Sci. 2013, 14, 4148–4162. [Google Scholar] [CrossRef] [PubMed]
- Kumaki, Y.; Morrey, J.D.; Barnard, D.L. Effect of statin treatments on highly pathogenic avian influenza H5N1, seasonal and H1N1pdm09 virus infections in BALB/c mice. Future Virol. 2012, 7, 801–818. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, J.; Ye, X. Protein kinase inhibitor flavopiridol inhibits the replication of influenza virus in vitro. Wei Sheng Wu Xue Bao 2012, 52, 1137–1142. [Google Scholar] [PubMed]
- Chen, Y.H.; Wu, K.L.; Chen, C.H. Methamphetamine reduces human influenza A virus replication. PLoS ONE 2012, 7, e48335. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Ibrahim, M.; Eladl, A.E.; Saif, Y.M.; Lee, C.W. Enhanced replication of swine influenza viruses in dexamethasone-treated juvenile and layer turkeys. Vet. Microbiol. 2013, 162, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, R.A.; Jawad, A.F.; Boyer, J.; Maurer, K.; McDonald, K.; Prak, E.T.; Sullivan, K.E. Rituximab-treated patients have a poor response to influenza vaccination. J. Clin. Immunol. 2013, 33, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Yatmaz, S.; Seow, H.J.; Gualano, R.C.; Wong, Z.X.; Stambas, J.; Selemidis, S.; Crack, P.J.; Bozinovski, S.; Anderson, G.P.; Vlahos, R. Glutathione peroxidase-1 reduces influenza A virus-induced lung inflammation. Am. J. Respir. Cell Mol. Biol. 2013, 48, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jin, Y.; Han, D.; Zhang, G.; Cao, S.; Xie, J.; Xue, J.; Li, Y.; Meng, D.; Fan, X.; et al. Mast cell-induced lung injury in mice infected with H5N1 influenza virus. J. Virol. 2012, 86, 3347–3356. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Chompoo, J.; Taira, N.; Fukuta, M.; Gima, S.; Tawata, S. Solid-phase synthesis of mimosine tetrapeptides and their inhibitory activities on neuraminidase and tyrosinase. J. Agric. Food Chem. 2011, 59, 12858–12863. [Google Scholar] [CrossRef] [PubMed]
- Garigliany, M.M.; Desmecht, D.J. N-acetylcysteine lacks universal inhibitory activity against influenza A viruses. J. Negat. Results Biomed. 2011. [Google Scholar] [CrossRef] [PubMed]
- Uchide, N.; Toyoda, H. Antioxidant therapy as a potential approach to severe influenza-associated complications. Molecules 2011, 16, 2032–2052. [Google Scholar] [CrossRef] [PubMed]
- Canestri, A.; Krivine, A.; Assoumou, L.; le Corre, M.; Rozenberg, F.; Marcelin, A.G.; Schneider, L.; Samri, A.; Carcelain, G.; Autran, B.; et al. Maraviroc does not affect humoral response to the pandemic influenza A-H1N1V 2009 adjuvanted vaccine in HIV-1-infected patients. AIDS 2010, 24, 2887–2889. [Google Scholar] [CrossRef] [PubMed]
- Dudek, S.E.; Luig, C.; Pauli, E.K.; Schubert, U.; Ludwig, S. The clinically approved proteasome inhibitor PS-341 efficiently blocks influenza A virus and vesicular stomatitis virus propagation by establishing an antiviral state. J. Virol. 2010, 84, 9439–9451. [Google Scholar] [CrossRef] [PubMed]
- Ferkolj, I. How to improve the safety of biologic therapy in Crohn’s disease. J. Physiol. Pharmacol. 2009, 60, S67–S70. [Google Scholar]
- Serrato, V.A.; Azevedo, V.F.; Sabatoski, V.; Goncalves, B.P.; Machado, D.M. Influenza H1N1 infection in a patient with psoriatic arthritis in treatment with Adalimumab: A case report. Clin. Rheumatol. 2013, 32, S21–S23. [Google Scholar] [CrossRef] [PubMed]
- Yamaya, M.; Shinya, K.; Hatachi, Y.; Kubo, H.; Asada, M.; Yasuda, H.; Nishimura, H.; Nagatomi, R. Clarithromycin inhibits type a seasonal influenza virus infection in human airway epithelial cells. J. Pharmacol. Exp. Ther. 2010, 333, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Chase, G.; Deng, T.; Fodor, E.; Leung, B.W.; Mayer, D.; Schwemmle, M.; Brownlee, G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 2008, 377, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Vigerust, D.J.; McCullers, J.A. Chloroquine is effective against influenza A virus in vitro but not in vivo. Influenza Other Respir. Viruses 2007, 1, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Garozzo, A.; Tempera, G.; Ungheri, D.; Timpanaro, R.; Castro, A. N-acetylcysteine synergizes with oseltamivir in protecting mice from lethal influenza infection. Int. J. Immunopathol. Pharmacol. 2007, 20, 349–354. [Google Scholar] [PubMed]
- Kido, H.; Okumura, Y.; Yamada, H.; Le, T.Q.; Yano, M. Proteases essential for human influenza virus entry into cells and their inhibitors as potential therapeutic agents. Curr. Pharm. Des. 2007, 13, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Maeda, N.; Beck, M.A. Vitamin C deficiency increases the lung pathology of influenza virus-infected gulo−/− mice. J. Nutr. 2006, 136, 2611–2616. [Google Scholar] [PubMed]
- Ooi, E.E.; Chew, J.S.; Loh, J.P.; Chua, R.C. In vitro inhibition of human influenza a virus replication by chloroquine. Virol. J. 2006. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Khanna, M.; Srivastava, V.; Tyagi, Y.K.; Raj, H.G.; Ravi, K. Effect of quercetin supplementation on lung antioxidants after experimental influenza virus infection. Exp. Lung Res. 2005, 31, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Yokozawa, T.; Sekiya, M.; Cho, E.J.; Kurokawa, M.; Shiraki, K. Effect of wen-pi-tang extract on lung damage by influenza virus infection. Phytomedicine 2004, 11, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sharma, S.; Khanna, M.; Raj, H.G. Effect of quercetin on lipid peroxidation and changes in lung morphology in experimental influenza virus infection. Int. J. Exp. Pathol. 2003, 84, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Ungheri, D.; Pisani, C.; Sanson, G.; Bertani, A.; Schioppacassi, G.; Delgado, R.; Sironi, M.; Ghezzi, P. Protective effect of N-acetylcysteine in a model of influenza infection in mice. Int. J. Immunopathol. Pharmacol. 2000, 13, 123–128. [Google Scholar] [PubMed]
- Sieczkarski, S.B.; Whittaker, G.R. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 2002, 76, 10455–10464. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yao, D.F.; Ohuchi, M.; Ide, M.; Yano, M.; Okumura, Y.; Kido, H. Ambroxol suppresses influenza-virus proliferation in the mouse airway by increasing antiviral factor levels. Eur. Respir. J. 2002, 19, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Jeon, W.K.; Ko, B.S. Flavanone glycosides from Citrus junos and their anti-influenza virus activity. Planta Med. 2001, 67, 548–549. [Google Scholar] [CrossRef] [PubMed]
- Kastner, S.B.; Haines, D.M.; Archer, J.; Townsend, H.G. Investigations on the ability of clenbuterol hydrochloride to reduce clinical signs and inflammation associated with equine influenza A infection. Equine Vet. J. 1999, 31, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.; Simons, K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J. Cell Biol. 1998, 140, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Dolganova, A.; Sharonov, B.P. Application of various antioxidants in the treatment of influenza. Braz. J. Med. Biol. Res. 1997, 30, 1333–1336. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Suga, M.; Akaike, T.; Fujii, S.; Muranaka, H.; Doi, T.; Maeda, H.; Ando, M. Therapeutic effect of erythromycin on influenza virus-induced lung injury in mice. Am. J. Respir. Crit. Care Med. 1998, 157, 853–857. [Google Scholar] [CrossRef] [PubMed]
- Moser, C.A.; Speaker, T.J.; Offit, P.A. Effect of microencapsulation on immunogenicity of a bovine herpes virus glycoprotein and inactivated influenza virus in mice. Vaccine 1997, 15, 1767–1772. [Google Scholar] [CrossRef]
- Yokochi, S.; Ishiwata, Y.; Saito, H.; Ebinuma, H.; Tsuchiya, M.; Ishii, H. Stimulation of antiviral activities of interferon by a liver extract preparation. Arzneimittelforschung 1997, 47, 968–974. [Google Scholar] [PubMed]
- Ochiai, H.; Sakai, S.; Hirabayashi, T.; Shimizu, Y.; Terasawa, K. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antivir. Res. 1995, 27, 425–430. [Google Scholar] [CrossRef]
- Guinea, R.; Carrasco, L. Requirement for vacuolar proton-ATPase activity during entry of influenza virus into cells. J. Virol. 1995, 69, 2306–2312. [Google Scholar] [PubMed]
- Conti, G.; Portincasa, P.; Chezzi, C. Cerulenin inhibits production of mature virion particles in chick embryo fibroblasts infected by influenza A viruses. Res. Virol. 1995, 146, 141–149. [Google Scholar] [CrossRef]
- Broni, B.; Julkunen, I.; Condra, J.H.; Davies, M.E.; Berry, M.J.; Krug, R.M. Parental influenza virion nucleocapsids are efficiently transported into the nuclei of murine cells expressing the nuclear interferon-induced mx protein. J. Virol. 1990, 64, 6335–6340. [Google Scholar] [PubMed]
- Hagiwara, S. Mechanisms of human platelet aggregation and release reaction induced by influenza virus. Nihon Ketsueki Gakkai Zasshi 1989, 52, 906–915. [Google Scholar] [PubMed]
- Gorse, G.J.; Kopp, W.C. Modulation by immunosuppressive agents of peripheral blood mononuclear cell responses to influenza A virus. J. Lab. Clin. Med. 1987, 110, 592–601. [Google Scholar] [PubMed]
- Kozhukharova, M.S.; Slepushkin, A.N.; Radeva Kh, T.; Lavrukhina, L.A.; Demidova, S.A. Evaluation of dipyridamole efficacy as an agent for preventing acute respiratory viral diseases. Vopr. Virusol. 1987, 32, 294–297. [Google Scholar] [PubMed]
- Versluis, D.J.; Beyer, W.E.; Masurel, N.; Wenting, G.J.; Weimar, W. Impairment of the immune response to influenza vaccination in renal transplant recipients by cyclosporine, but not azathioprine. Transplantation 1986, 42, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Kuzmov, K.; Galabov, A.S.; Radeva, K.; Kozhukharova, M.; Milanov, K. Epidemiological trial of the prophylactic effectiveness of the interferon inducer dipyridamole with respect to influenza and acute respiratory diseases. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 1985, 6, 26–30. [Google Scholar]
- Schiltknecht, E.; Ada, G.L. In vivo effects of cyclosporine on influenza A virus-infected mice. Cell. Immunol. 1985, 91, 227–239. [Google Scholar] [CrossRef]
- Bernasconi, P.; Massera, E. Evaluation of a new pharmaceutical form of nimesulide for the treatment of influenza. Drugs Exp. Clin. Res. 1985, 11, 739–743. [Google Scholar] [PubMed]
- Nugent, K.M.; Shanley, J.D. Verapamil inhibits influenza A virus replication. Arch. Virol. 1984, 81, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Semkow, R.; Loza-Tulimowska, M.; Wilczynski, J.; Krus, S. The dynamics of serum antibodies and metaplasia of the lung respiratory epithelium in influenza-virus-preimmunized mice subjected to immunosuppression. Acta Microbiol. Pol. 1984, 33, 51–55. [Google Scholar] [PubMed]
- Krizanova, O.; Ciampor, F.; Veber, P. Influence of chlorpromazine on the replication of influenza virus in chick embryo cells. Acta Virol. 1982, 26, 209–216. [Google Scholar] [PubMed]
- Tonew, E.; Indulen, M.K.; Dzeguze, D.R. Antiviral action of dipyridamole and its derivatives against influenza virus A. Acta Virol. 1982, 26, 125–129. [Google Scholar] [PubMed]
- Pan, Y.T.; Elbein, A.D. The formation of lipid-linked oligosaccharides in Madin-Darby canine kidney cells. Changes in oligosaccharide profiles induced by glucosamine. J. Biol. Chem. 1982, 257, 2795–2801. [Google Scholar] [PubMed]
- Hamzehei, M.; Ledinko, N. Inhibition of influenza a virus replication by rifampicin and selenocystamine. J. Med. Virol. 1980, 6, 169–174. [Google Scholar] [PubMed]
- Theisen, L.L.; Erdelmeier, C.A.; Spoden, G.A.; Boukhallouk, F.; Sausy, A.; Florin, L.; Muller, C.P. Tannins from hamamelis virginiana bark extract: Characterization and improvement of the antiviral efficacy against influenza a virus and human papillomavirus. PLoS ONE 2014, 9, e88062. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, M.; Minakuchi, M.; Kawaguchi, A.; Nagata, K.; Kamatari, Y.O.; Kuwata, K. Structure-based discovery of anti-influenza virus a compounds among medicines. Biochim. Biophys. Acta 2012, 1820, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Nobata, K.; Fujimura, M.; Ishiura, Y.; Myou, S.; Nakao, S. Ambroxol for the prevention of acute upper respiratory disease. Clin. Exp. Med. 2006, 6, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Perwitasari, O.; Yan, X.; O’Donnell, J.; Johnson, S.; Tripp, R.A. Repurposing kinase inhibitors as antiviral agents to control influenza A virus replication. Assay Drug Dev. Technol. 2015, 13, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Kakkola, L.; Denisova, O.V.; Tynell, J.; Viiliainen, J.; Ysenbaert, T.; Matos, R.C.; Nagaraj, A.; Ohman, T.; Kuivanen, S.; Paavilainen, H.; et al. Anticancer compound ABT-263 accelerates apoptosis in virus-infected cells and imbalances cytokine production and lowers survival rates of infected mice. Cell Death Dis. 2013. [Google Scholar] [CrossRef] [PubMed]
- Rialdi, A.; Campisi, L.; Zhao, N.; Lagda, A.C.; Pietzsch, C.; Ho, J.S.; Martinez-Gil, L.; Fenouil, R.; Chen, X.; Edwards, M.; et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science 2016. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Caldas, J.; Gehlenborg, N.; Faisal, A.; Brazma, A.; Kaski, S. Probabilistic retrieval and visualization of biologically relevant microarray experiments. Bioinformatics 2009, 25, i145–i153. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, C.N.; Penders, T.M.; Rouse, E.M. Recreational use of dextromethorphan, “Robotripping”—A brief review. Am. J. Addict. 2016, 25, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; MacIsaac, R.; Thompson, J.L.; Levin, B.; Buchsbaum, R.; Haley, E.C., Jr.; Levi, C.; Campbell, B.; Bladin, C.; Parsons, M.; et al. Tenecteplase versus alteplase in stroke thrombolysis: An individual patient data meta-analysis of randomized controlled trials. Int. J. Stroke 2016, 11, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Vitaliti, G.; Matin, N.; Tabatabaie, O.; di Traglia, M.; Pavone, P.; Lubrano, R.; Falsaperla, R. Natalizumab in multiple sclerosis: Discontinuation, progressive multifocal leukoencephalopathy and possible use in children. Expert Rev. Neurother. 2015, 15, 1321–1341. [Google Scholar] [CrossRef] [PubMed]
- Bain, G.; King, C.D.; Rewolinski, M.; Schaab, K.; Santini, A.M.; Shapiro, D.; Moran, M.; van de Wetering de Rooij, S.; Roffel, A.F.; Schuilenga-Hut, P.; et al. Pharmacodynamics and pharmacokinetics of AM103, a novel inhibitor of 5-lipoxygenase-activating protein (FLAP). Clin. Pharmacol. Ther. 2010, 87, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Beauparlant, P.; Bedard, D.; Bernier, C.; Chan, H.; Gilbert, K.; Goulet, D.; Gratton, M.O.; Lavoie, M.; Roulston, A.; Turcotte, E.; et al. Preclinical development of the nicotinamide phosphoribosyl transferase inhibitor prodrug GMX1777. Anticancer Drugs 2009, 20, 346–354. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Söderholm, S.; Fu, Y.; Gaelings, L.; Belanov, S.; Yetukuri, L.; Berlinkov, M.; Cheltsov, A.V.; Anders, S.; Aittokallio, T.; Nyman, T.A.; et al. Multi-Omics Studies towards Novel Modulators of Influenza A Virus–Host Interaction. Viruses 2016, 8, 269. https://doi.org/10.3390/v8100269
Söderholm S, Fu Y, Gaelings L, Belanov S, Yetukuri L, Berlinkov M, Cheltsov AV, Anders S, Aittokallio T, Nyman TA, et al. Multi-Omics Studies towards Novel Modulators of Influenza A Virus–Host Interaction. Viruses. 2016; 8(10):269. https://doi.org/10.3390/v8100269
Chicago/Turabian StyleSöderholm, Sandra, Yu Fu, Lana Gaelings, Sergey Belanov, Laxman Yetukuri, Mikhail Berlinkov, Anton V. Cheltsov, Simon Anders, Tero Aittokallio, Tuula A. Nyman, and et al. 2016. "Multi-Omics Studies towards Novel Modulators of Influenza A Virus–Host Interaction" Viruses 8, no. 10: 269. https://doi.org/10.3390/v8100269





