Insect-Specific Virus Discovery: Significance for the Arbovirus Community
Abstract
:1. Introduction
2. Detection of Insect-Specific Viruses
| Taxa | Virus | Isolation | Sequence | Reference |
|---|---|---|---|---|
| Birnaviridae | Culex Y virus Espirito Santo virus | Yes Yes | Full genome Full genome | [21] [22] |
| Mosquito X virus | No | Full genome | [23] | |
| Bunyaviridae | Cumuto virus | Yes | Full genome | [24] |
| Gouléako | Yes | Full genome | [25] | |
| Herbert virus | Yes | Full genome | [26] | |
| Kibale virus | Yes | Full genome | [26] | |
| Phasi Charoen virus | Yes | Partial | [27] | |
| Tai virus | Yes | Full genome | [26] | |
| Flaviviridae | Aedes cinereus flavivirus | No | Partial | [28] |
| Aedes galloisi flavivirus | Yes | Partial | [29] | |
| Aedes flavivirus | Yes | Full genome | [4] | |
| Aedes vexans flavivirus | No | Partial | [28] | |
| Barkedji virus | Yes | Partial | [30] | |
| Calbertado virus | Yes | Partial | [16] | |
| Cell fusing agent virus | Yes | Full genome | [1,2] | |
| Chaoyang virus | Yes | Full genome | [31] | |
| Culex flavivirus | Yes | Full genome | [12] | |
| Culex theileri flavivirus | Yes | Full genome | [32] | |
| Czech Aedes vexans flavivirus | No | Partial | [28] | |
| Donggang virus | Yes | Full genome | Genbank accession #JQ086551 (2012) | |
| Hanko virus | Yes | Full genome | [33] | |
| Kamiti River virus | Yes | Full genome | [8,9] | |
| Taxa | Virus | Isolation | Sequence | Reference |
| Flaviviridae | Lammi virus | Yes | Partial | [34] |
| Mercadeo virus | Yes | Full genome | [35] | |
| Marisma mosquito virus | Yes | Partial | [36] | |
| Nakiwogo virus | Yes | Partial | [14] | |
| Nanay virus | Yes | Partial | [37] | |
| Nhumirim virus | Yes | Full genome | [38] | |
| Nounane virus | Yes | Full genome | [39] | |
| Ochlerotatus flavivirus | No | Partial | [28] | |
| Ochlerotatus caspius flavivirus | Yes | Full genome | [40] | |
| Palm Creek virus | Yes | Partial | [41] | |
| Quang Binh virus | Yes | Full genome | [42] | |
| Spanish Culex flavivirus | Yes | Partial | [36] | |
| Spanish Ochlerotatus flavivirus | Yes | Partial | [36] | |
| Yunan Culex flavivirus | Yes | Full genome | [43] | |
| Mesoniviridae | Bontag Baru virus | Yes | Full genome | [44] |
| Casuarina virus | Yes | Full genome | [45] | |
| Cavally virus | Yes | Full genome | [46] | |
| Dak Nong virus | Yes | Full genome | [47] | |
| Hana virus | Yes | Full genome | [48] | |
| Kamphaeng Phet virus | Yes | Full genome | [44] | |
| Méno virus | Yes | Full genome | [48] | |
| Moumo virus | Yes | Full genome | [48] | |
| Nam Dinh virus | Yes | Full genome | [49] | |
| Nsé virus | Yes | Full genome | [48] | |
| Negeviruses | Dezidougou virus | Yes | Full genome | [50] |
| Goutanap virus | Yes | Full genome | [51] | |
| Loreto virus | Yes | Full genome | [50] | |
| Negev virus | Yes | Full genome | [50] | |
| Ngewotan virus | Yes | Full genome | [50] | |
| Piura virus | Yes | Full genome | [50] | |
| Santana virus | Yes | Full genome | [50] | |
| Tanay virus | Yes | Full genome | [52] | |
| Wallerfield virus | Yes | Full genome | [23] | |
| Nodaviridae | Mosinovirus | Yes | Full genome | [53] |
| Reoviridae | Aedes pseudoscutellaris reovirus | Yes | Full genome | [54] |
| Cimodo virus | Yes | Full genome | [55] | |
| Fako virus | Yes | Full genome | [56] | |
| Rhabdoviridae | Arboretum virus | Yes | Full genome | [57] |
| Culex tritaeniorhynchus rhabdovirus | Yes | Full genome | [58] | |
| Moussa virus | Yes | Full genome | [59] | |
| Puerto Almendras virus | Yes | Full genome | [57] | |
| Togaviridae | Eilat virus | Yes | Full genome | [60] |
| Tymoviridae | Culex tymovirus-like virus | Yes | Full genome | [61] |
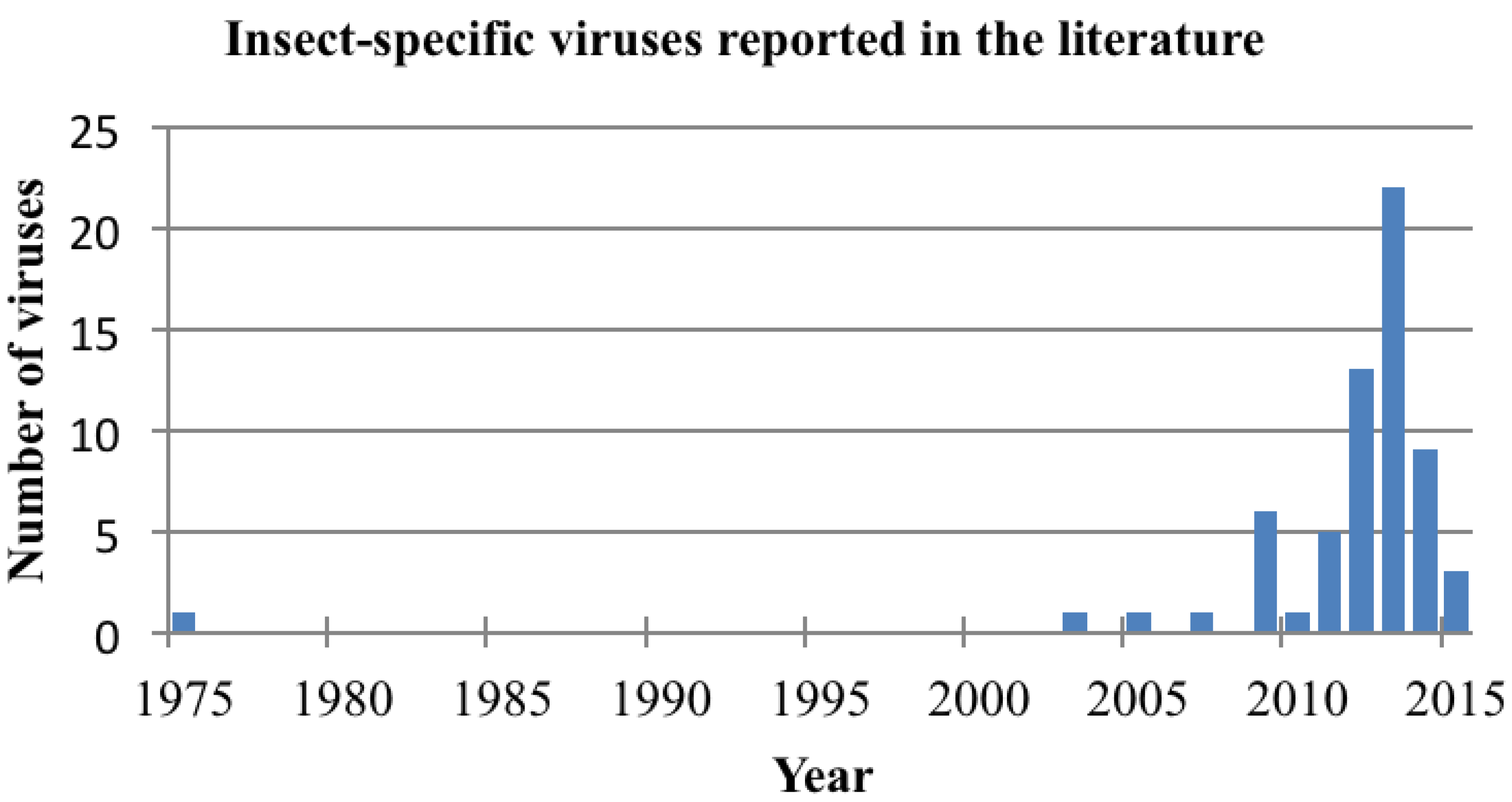
3. Viral Maintenance in Nature
4. Viral Diversity and Evolution

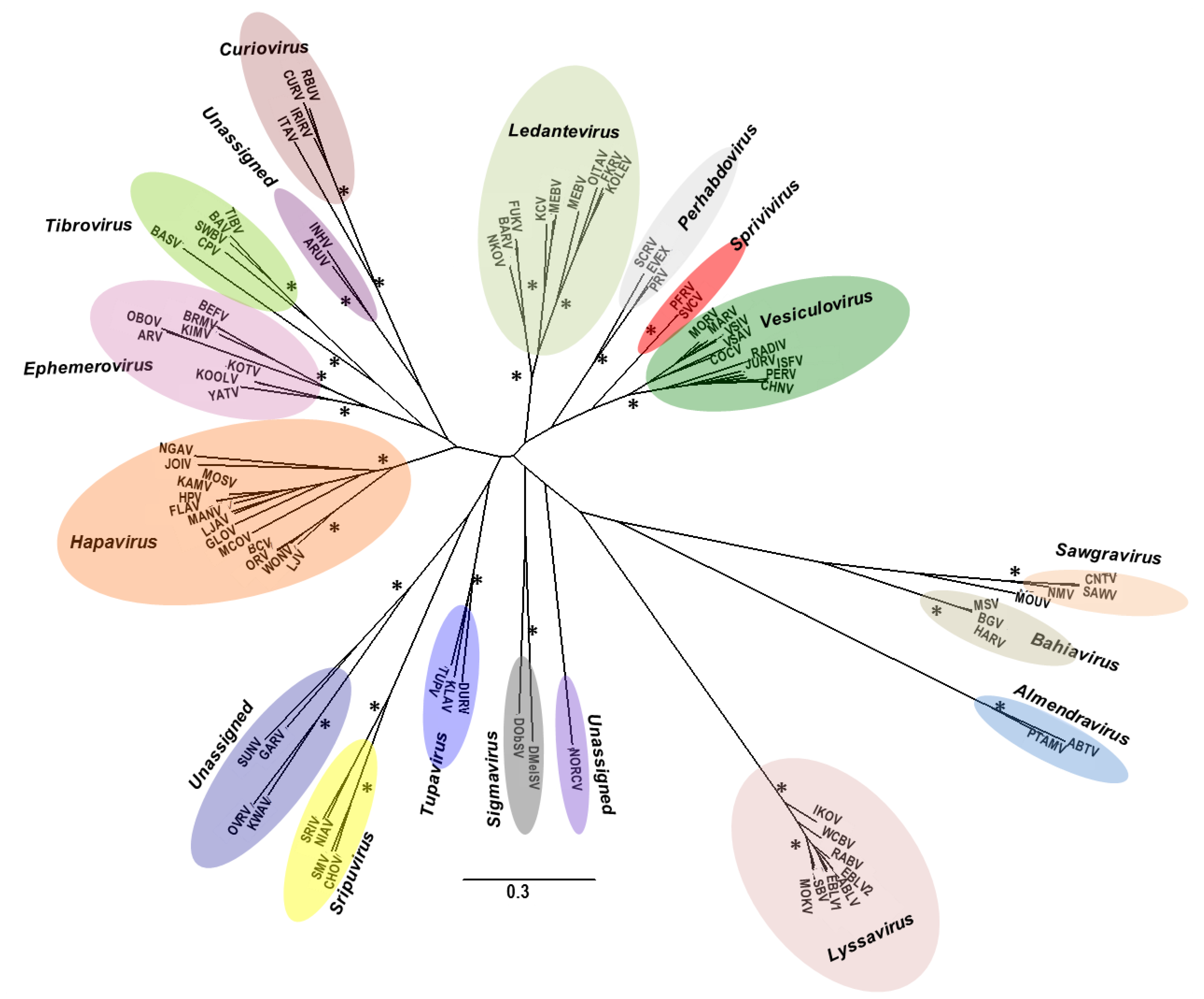
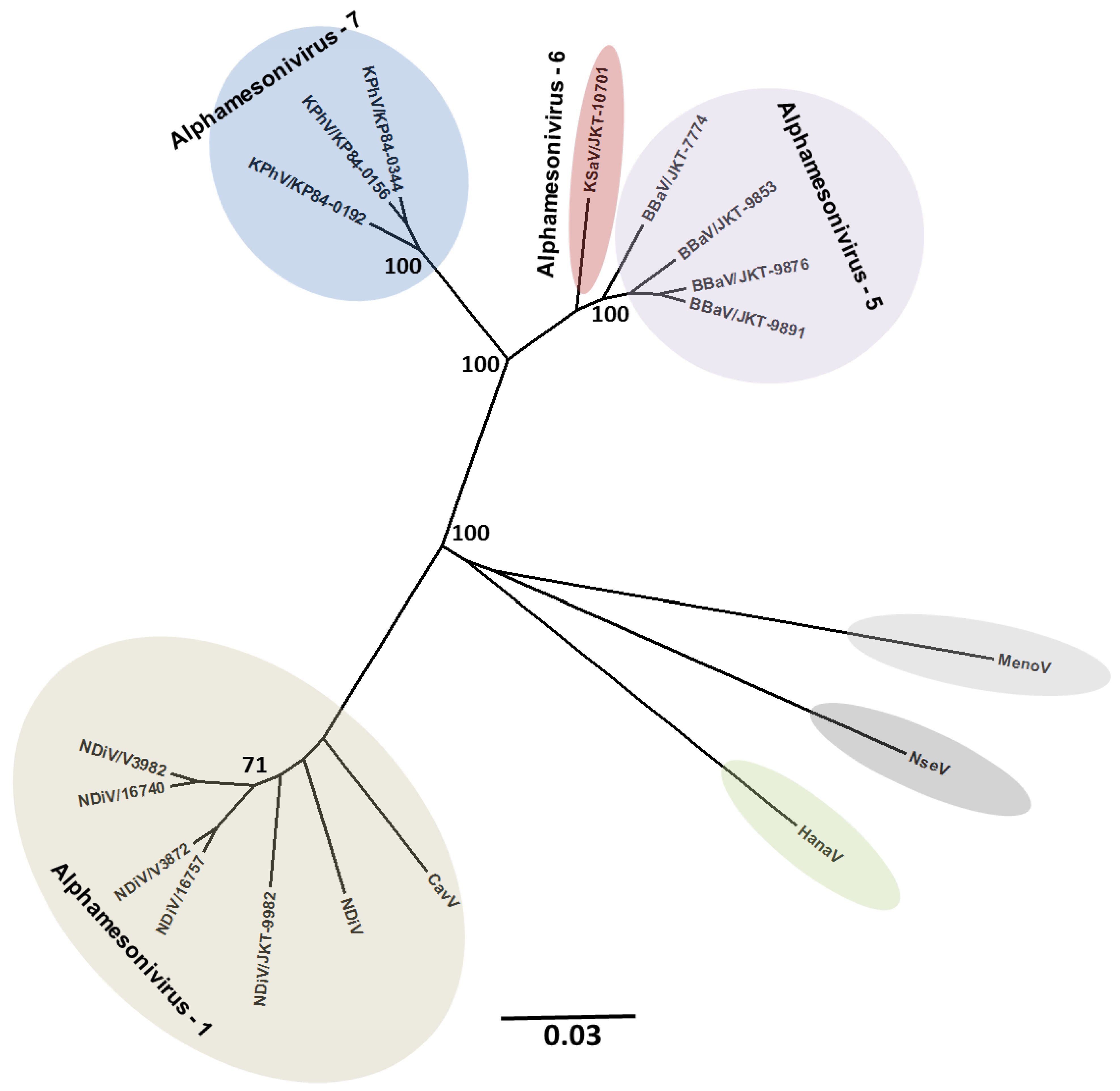
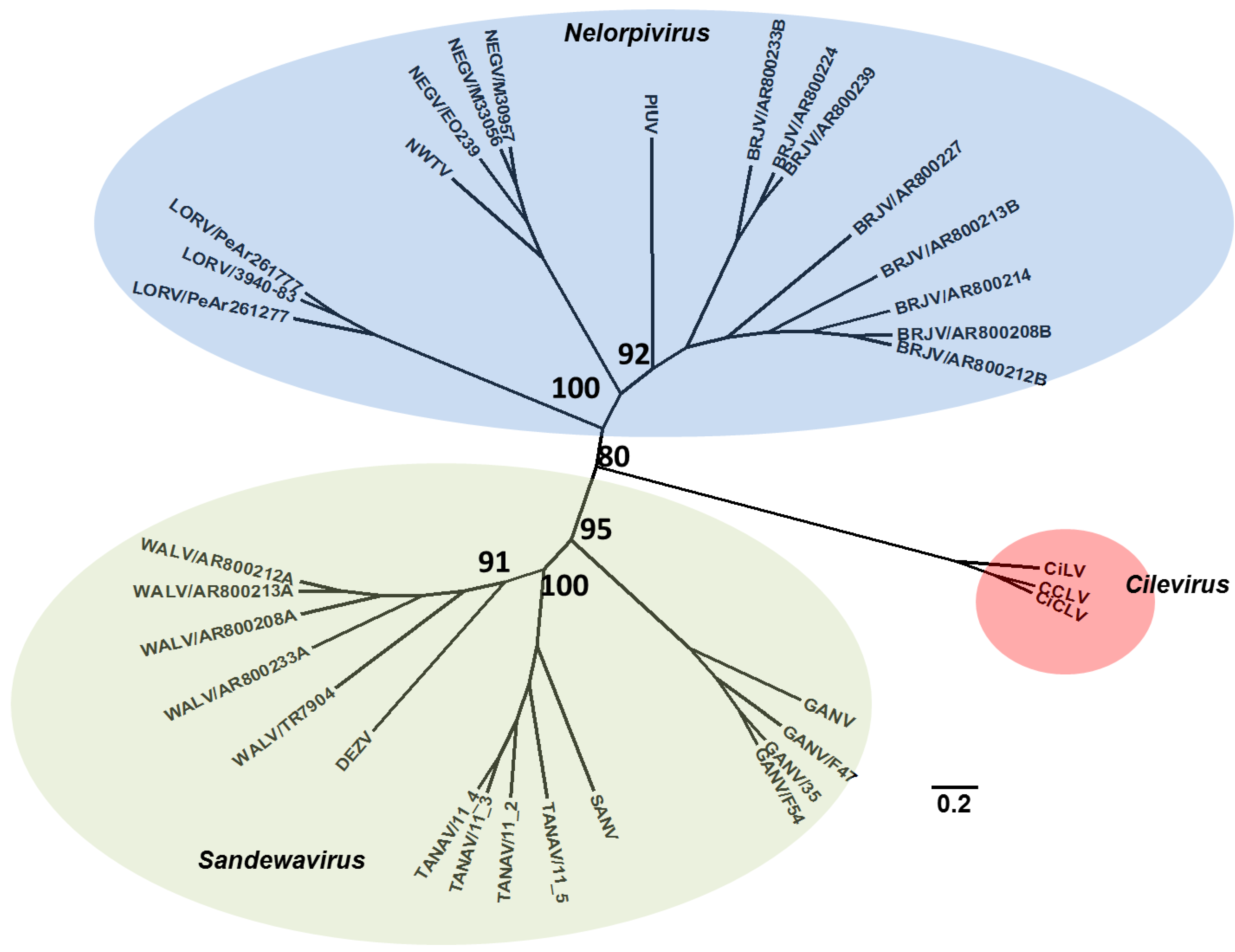

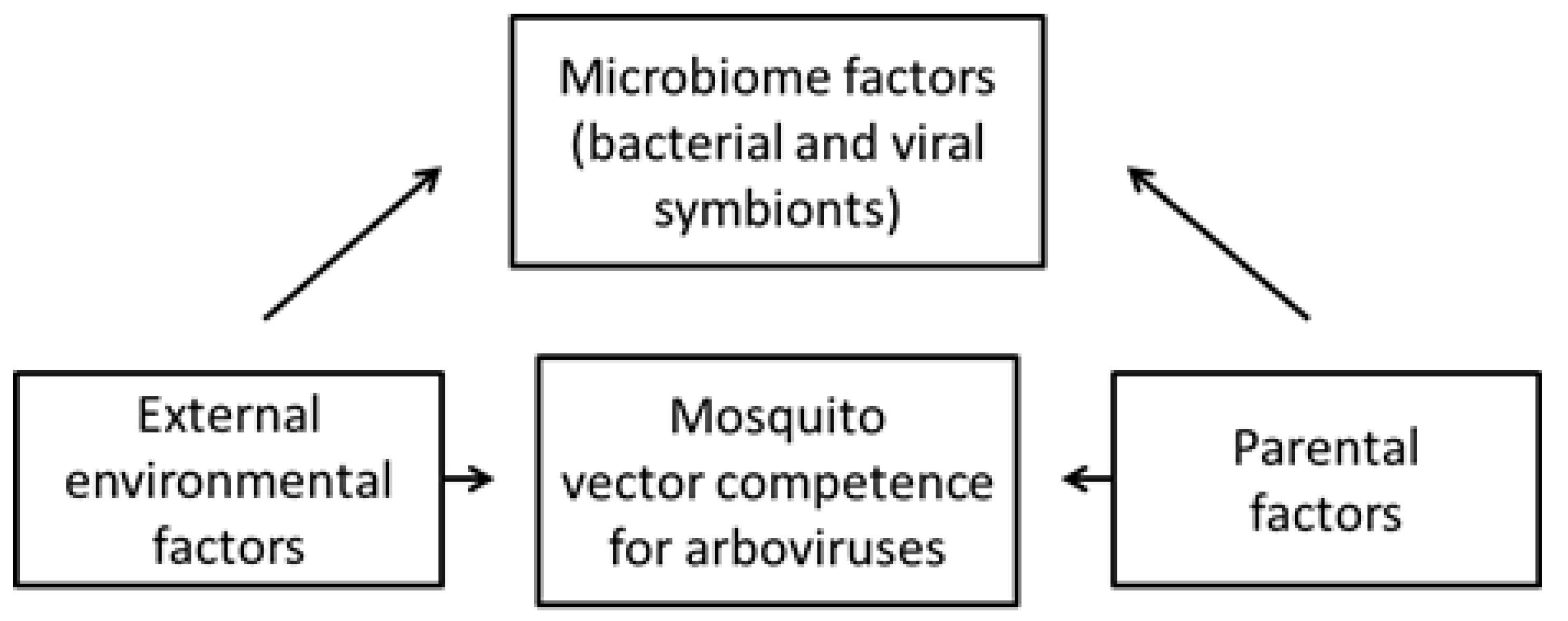
5. Effects on Vector Competence for Arboviruses
6. Potential Novel Applications of Insect-Specific Viruses
6.1. Use of Insect-Specific Viruses as Biological Control Agents
6.2. Use of Insect-Specific Viruses as Vaccine and Diagnostic Platforms
7. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stollar, V.; Thomas, L. An Agent in the Aedes aegypti Cell Line (Peleg) which Causes Fusion of Aedes albopictus Cells. Virology 1975, 64, 367–377. [Google Scholar] [CrossRef]
- Cammisa-Parks, H.; Cisar, L.A.; Kane, A.; Stollar, V. The complete nucleotide sequence of cell fusing agent (CFA): Homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology 1992, 189, 511–524. [Google Scholar] [CrossRef]
- Cook, S.; Bennett, S.N.; Holmes, E.C.; De Chesse, R.; Moureau, G.; de Lamballerie, X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J. Gen. Virol. 2006, 87, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, K.; Isawa, H.; Tsuda, Y.; Sawabe, K.; Kobayashi, M. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology 2009, 391, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, A.; Thongrungkiat, S.; Ramasoota, P.; Konishi, E. Genetic and evolutionary analysis of cell-fusing agent virus based on Thai strains isolated in 2008 and 2012. Infect. Genet. Evol. 2013, 19, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Gómez, F.; López-Lemus, A.U.; Rodriguez-Sanchez, I.P.; Martinez-Fierro, M.L.; Newton-Sánchez, O.A.; Chávez-Flores, E.; Delgado-Enciso, I. Detection of sequences from a potentially novel strain of cell fusing agent virus in Mexican Stegomyia (Aedes) aegypti mosquitoes. Arch. Virol. 2011, 156, 1263–1267. [Google Scholar] [CrossRef] [PubMed]
- Marin, M.S.; Zanotto, P.M.; Gritsun, T.S.; Gould, E. a Phylogeny of TYU, SRE, and CFA virus: Different evolutionary rates in the genus Flavivirus. Virology 1995, 206, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, M.B.; Sang, R.C.; Stollar, V.; Dunster, L.M.; Miller, B.R. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch. Virol. 2003, 148, 1095–1118. [Google Scholar] [CrossRef] [PubMed]
- Sang, R.C.; Gichogo, A.; Gachoya, J.; Dunster, M.D.; Ofula, V.; Hunt, A.R.; Crabtree, M.B.; Miller, B.R.; Dunster, L.M. Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch. Virol. 2003, 148, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Crochu, S.; Cook, S.; Attoui, H.; Charrel, R.N.; De Chesse, R.; Belhouchet, M.; Lemasson, J.J.; de Micco, P.; de Lamballerie, X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J. Gen. Virol. 2004, 85, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Roiz, D.; Vázquez, A.; Seco, M.P.S.; Tenorio, A.; Rizzoli, A. Detection of novel insect flavivirus sequences integrated in Aedes albopictus (Diptera: Culicidae) in Northern Italy. Virol. J. 2009, 6. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, K.; Isawa, H.; Tsuda, Y.; Yano, K.; Sasaki, T.; Yuda, M.; Takasaki, T.; Kobayashi, M.; Sawabe, K. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology 2007, 359, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Morales-Betoulle, M.E.; Monzón Pineda, M.L.; Sosa, S.M.; Panella, N.; López, M.R.B.; Cordón-Rosales, C.; Komar, N.; Powers, A.; Johnson, B.W. Culex flavivirus isolates from mosquitoes in Guatemala. J. Med. Entomol. 2008, 45, 1187–1190. [Google Scholar] [CrossRef] [PubMed]
- Cook, S.; Moureau, G.; Harbach, R.E.; Mukwaya, L.; Goodger, K.; Ssenfuka, F.; Gould, E.; Holmes, E.C.; de Lamballerie, X. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J. Gen. Virol. 2009, 90, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- Farfan-Ale, J.A.; Loroño-Pino, M.A.; Garcia-Rejon, J.E.; Hovav, E.; Powers, A.M.; Lin, M.; Dorman, K.S.; Platt, K.B.; Bartholomay, L.C.; Soto, V.; et al. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am. J. Trop. Med. Hyg. 2009, 80, 85–95. [Google Scholar] [PubMed]
- Bolling, B.G.; Eisen, L.; Moore, C.G.; Blair, C.D. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am. J. Trop. Med. Hyg. 2011, 85, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Guzman, H.; Bueno, R.; Dennett, J.A.; Auguste, A.J.; Carrington, C.V.F.; Popov, V.L.; Weaver, S.C.; Beasley, D.W.C.; Tesh, R.B. Characterization of Culex Flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology 2009, 386, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Huanyu, W.; Haiyan, W.; Shihong, F.; Guifang, L.; Hong, L.; Xiaoyan, G.; Lizhi, S.; Rayner, S.; Aiqiang, X.; Guodong, L. Isolation and identification of a distinct strain of Culex Flavivirus from mosquitoes collected in Mainland China. Virol. J. 2012, 9. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.C.; Mondini, A.; dos Santos Santana, V.; Yonamine, P.T.K.; Chiaravalloti Neto, F.; Zanotto, P.M.D.A.; Nogueira, M.L. First identification of Culex flavivirus (Flaviviridae) in Brazil. Intervirology 2012, 55, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Lin, J.W.; Fan, Y.C.; Tu, W.C.; Chang, G.J.J.; Chiou, S.S. First detection of the Africa/Caribbean/Latin American subtype of Culex flavivirus in Asian country, Taiwan. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Marklewitz, M.; Gloza-Rausch, F.; Kurth, A.; Kümmerer, B.M.; Drosten, C.; Junglen, S. First isolation of an Entomobirnavirus from free-living insects. J. Gen. Virol. 2012, 93, 2431–2435. [Google Scholar] [CrossRef] [PubMed]
- Vancini, R.; Paredes, A.; Ribeiro, M.; Blackburn, K.; Ferreira, D.; Kononchik, J.P.; Hernandez, R.; Brown, D. Espirito Santo virus: A new birnavirus that replicates in insect cells. J. Virol. 2012, 86, 2390–2399. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mi, Z.; Zhuang, L.; Ma, M.; An, X.; Liu, W.; Cao, W.; Tong, Y. Presence of entomobirnaviruses in Chinese mosquitoes in the absence of Dengue virus co-infection. J. Gen. Virol. 2013, 94, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Auguste, A.J.; Carrington, C.V.F.; Forrester, N.L.; Popov, V.L.; Guzman, H.; Widen, S.G.; Wood, T.G.; Weaver, S.C.; Tesh, R.B. Characterization of a novel Negevirus and a novel Bunyavirus isolated from Culex (Culex) declarator mosquitoes in Trinidad. J. Gen. Virol. 2014, 95, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Marklewitz, M.; Handrick, S.; Grasse, W.; Kurth, A.; Lukashev, A.; Drosten, C.; Ellerbrok, H.; Leendertz, F.H.; Pauli, G.; Junglen, S. Gouleako virus isolated from West African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J. Virol. 2011, 85, 9227–9234. [Google Scholar] [CrossRef] [PubMed]
- Marklewitz, M.; Zirkel, F.; Rwego, I.B.; Heidemann, H.; Trippner, P.; Kurth, A.; Kallies, R.; Briese, T.; Lipkin, W.I.; Drosten, C.; et al. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J. Virol. 2013, 87, 12850–12865. [Google Scholar] [CrossRef] [PubMed]
- Yamao, T.; Eshita, Y.; Kihara, Y.; Satho, T.; Kuroda, M.; Sekizuka, T.; Nishimura, M.; Sakai, K.; Watanabe, S.; Akashi, H.; et al. Novel virus discovery in field-collected mosquito larvae using an improved system for rapid determination of viral RNA sequences (RDV ver4.0). Arch. Virol. 2009, 154, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, M.; Zé-Zé, L.; Růžek, D.; Vázquez, A.; Jeffries, C.; Defilippo, F.; Osório, H.C.; Kilian, P.; Ruíz, S.; Fooks, A.R.; et al. Detection of mosquito-only flaviviruses in Europe. J. Gen. Virol. 2012, 93, 1215–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, K.; Takahashi-Nakaguchi, A.; Isawa, H.; Sasaki, T.; Higa, Y.; Kasai, S.; Tsuda, Y.; Sawabe, K.; Kobayashi, M. Entomological surveillance for flaviviruses at migratory bird stopover sites in Hokkaido, Japan, and a new insect flavivirus detected in Aedes galloisi (Diptera: Culicidae). J. Med. Entomol. 2012, 49, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejek, J.; Pachler, K.; Bin, H.; Mendelson, E.; Shulman, L.; Orshan, L.; Nowotny, N. Barkedji virus, a novel mosquito-borne flavivirus identified in Culex perexiguus mosquitoes, Israel, 2011. J. Gen. Virol. 2013, 94, 2449–2457. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Grubaugh, N.D.; Kondig, J.P.; Turell, M.J.; Kim, H.C.; Klein, T.A.; O’Guinn, M.L. Isolation and genomic characterization of Chaoyang virus strain ROK144 from Aedes vexans nipponii from the Republic of Korea. Virology 2013, 435, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Parreira, R.; Cook, S.; Lopes, Â.; de Matos, A.P.; de Almeida, A.P.G.; Piedade, J.; Esteves, A. Genetic characterization of an insect-specific flavivirus isolated from Culex theileri mosquitoes collected in southern Portugal. Virus Res. 2012, 167, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Huhtamo, E.; Moureau, G.; Cook, S.; Julkunen, O.; Putkuri, N.; Kurkela, S.; Uzcátegui, N.Y.; Harbach, R.E.; Gould, E.A.; Vapalahti, O.; et al. Novel insect-specific flavivirus isolated from northern Europe. Virology 2012, 433, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Huhtamo, E.; Putkuri, N.; Kurkela, S.; Manni, T.; Vaheri, A.; Vapalahti, O.; Uzcátegui, N.Y. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J. Virol. 2009, 83, 9532–9540. [Google Scholar] [CrossRef] [PubMed]
- Carrera, J.; Guzman, H.; Beltran, D.; Diaz, Y.; Lopez-Verges, S.; Torres, R.; Popov, V.; Widen, S.; Wood, T.; Weaver, S.; et al. Mercadeo Virus: A Novel Mosquito-Specific Flavivirus from Panama. Am. J. Trop. Med. Hyg. 2015. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, A.; Sánchez-Seco, M.P.; Palacios, G.; Molero, F.; Reyes, N.; Ruiz, S.; Aranda, C.; Marqués, E.; Escosa, R.; Moreno, J.; et al. Novel Flaviviruses Detected in Different Species of Mosquitoes in Spain. Vector Borne Zoonotic Dis. 2012, 12, 223–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evangelista, J.; Cruz, C.; Guevara, C.; Astete, H.; Carey, C.; Kochel, T.J.; Morrison, A.C.; Williams, M.; Halsey, E.S.; Forshey, B.M. Characterization of a novel flavivirus isolated from Culex (Melanoconion) ocossa mosquitoes from Iquitos, Peru. J. Gen. Virol. 2013, 94, 1266–1272. [Google Scholar] [CrossRef] [PubMed]
- Pauvolid-Corrêa, A.; Solberg, O.; Couto-Lima, D.; Kenney, J.; Serra-Freire, N.; Brault, A.; Nogueira, R.; Langevin, S.; Komar, N. Nhumirim virus, a novel flavivirus isolated from mosquitoes from the Pantanal, Brazil. Arch. Virol. 2014, 160, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Junglen, S.; Kopp, A.; Kurth, A.; Pauli, G.; Ellerbrok, H.; Leendertz, F.H. A new flavivirus and a new vector: Characterization of a novel flavivirus isolated from uranotaenia mosquitoes from a tropical rain forest. J. Virol. 2009, 83, 4462–4468. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.D.; Cook, S.; Lopes, A.; de Matos, A.P.; Esteves, A.; Abecasis, A.; de Almeida, A.P.G.; Piedade, J.; Parreira, R. Characterization of an insect-specific flavivirus (OCFVPT) co-isolated from Ochlerotatus caspius collected in southern Portugal along with a putative new Negev-like virus. Virus Genes 2013, 47, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Hobson-Peters, J.; Yam, A.W.Y.; Lu, J.W.F.; Setoh, Y.X.; May, F.J.; Kurucz, N.; Walsh, S.; Prow, N.A.; Davis, S.S.; Weir, R.; et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE 2013, 8, e56534. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, M.B.; Nga, P.T.; Miller, B.R. Isolation and characterization of a new mosquito flavivirus, Quang Binh virus, from Vietnam. Arch. Virol. 2009, 154, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Zhao, Q.; Guo, X.; Zhou, H.; Cao, W.; Zhang, J. Detection of Quang Binh virus from mosquitoes in China. Virus Res. 2014, 180, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Guzman, H.; Firth, C.; Forrester, N.L.; Widen, S.G.; Wood, T.G.; Rossi, S.L.; Ghedin, E.; Popov, V.; Blasdell, K.R.; et al. Mesoniviruses are mosquito-specific viruses with extensive geographic distribution and host range. Virol. J. 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Warrilow, D.; Watterson, D.; Hall, R.A.; Davis, S.S.; Weir, R.; Kurucz, N.; Whelan, P.; Allcock, R.; Hall-Mendelin, S.; O’Brien, C.A.; et al. A New Species of Mesonivirus from the Northern Territory, Australia. PLoS ONE 2014, 9, e91103. [Google Scholar] [CrossRef] [PubMed]
- Zirkel, F.; Kurth, A.; Quan, P.; Briese, T.; Ellerbrok, H.; Pauli, G.; Leendertz, F.H. An Insect Nidovirus Emerging from a Primary Tropical Rainforest. mBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, R.; Satho, T.; Isawa, H.; Yen, N.T.; Phong, T.V.; Nga, P.T.; Kurashige, T.; Hiramatsu, Y.; Fukumitsu, Y.; Hoshino, K.; et al. Characterization of Dak Nong virus, an insect nidovirus isolated from Culex mosquitoes in Vietnam. Arch. Virol. 2013, 158, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Zirkel, F.; Roth, H.; Kurth, A.; Drosten, C.; Ziebuhr, J.; Junglen, S. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J. Virol. 2013, 87, 6346–6358. [Google Scholar] [CrossRef] [PubMed]
- Nga, P.T.; Parquet, M.D.C.; Lauber, C.; Parida, M.; Nabeshima, T.; Yu, F.; Thuy, N.T.; Inoue, S.; Ito, T.; Okamoto, K.; et al. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Forrester, N.L.; Palacios, G.; Nasar, F.; Savji, N.; Rossi, S.L.; Guzman, H.; Wood, T.G.; Popov, V.; Gorchakov, R.; et al. Negevirus: A proposed new taxon of insect-specific viruses with wide geographic distribution. J. Virol. 2013, 87, 2475–2488. [Google Scholar] [CrossRef] [PubMed]
- Kallies, R.; Kopp, A.; Zirkel, F.; Estrada, A.; Gillespie, T.; Drosten, C.; Junglen, S. Genetic Characterization of Goutanap Virus, a Novel Virus Related to Negeviruses, Cileviruses and Higreviruses. Viruses 2014, 6, 4346–4357. [Google Scholar] [CrossRef] [PubMed]
- Nabeshima, T.; Inoue, S.; Okamoto, K.; Posadas-Herrera, G.; Yu, F.; Uchida, L.; Ichinose, A.; Sakaguchi, M.; Sunahara, T.; Buerano, C.; et al. Tanay virus, a new species of virus isolated from mosquitoes in the Philippines. J. Gen. Virol. 2014, 95, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Zirkel, F.; Kurth, A.; van Cleef, K.W.R.; Drosten, C.; van Rij, R.P.; Junglen, S. A Unique Nodavirus with Novel Features: Mosinovirus Expresses Two Subgenomic RNAs, a Capsid Gene of Unknown Origin, and a Suppressor of the Antiviral RNA Interference Pathway. J. Virol. 2014, 88, 13447–13459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attoui, H.; Mohd Jaafar, F.; Belhouchet, M.; Biagini, P.; Cantaloube, J.F.; de Micco, P.; de Lamballerie, X. Expansion of family Reoviridae to include nine-segmented dsRNA viruses: Isolation and characterization of a new virus designated Aedes pseudoscutellaris reovirus assigned to a proposed genus (Dinovernavirus). Virology 2005, 343, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, K.; Zirkel, F.; Kurth, A.; Drosten, C.; Junglen, S. Cimodo virus belongs to a novel lineage of reoviruses isolated from African mosquitoes. J. Gen. Virol. 2014, 95, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Auguste, A.J.; Kaelber, J.; Fokam, E.; Guzman, H.; Carrington, C.; Erasmus, J.; Kamgang, B.; Popov, V.; Jakana, J.; Liu, X.; et al. A newly isolated reovirus has the simplest genomic and structural organization of any reovirus. J. Virol. 2015, 89, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Castro-Llanos, F.; Widen, S.G.; Aguilar, P.V.; Guzman, H.; Guevara, C.; Fernandez, R.; Auguste, A.J.; Wood, T.G.; Popov, V.; et al. Arboretum and Puerto Almendras viruses: Two novel rhabdoviruses isolated from mosquitoes in Peru. J. Gen. Virol. 2014, 95, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, R.; Isawa, H.; Hoshino, K.; Tsuda, Y.; Yanase, T.; Sasaki, T.; Kobayashi, M.; Sawabe, K. RNA splicing in a new rhabdovirus from Culex mosquitoes. J. Virol. 2011, 85, 6185–6196. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Junglen, S.; Tashmukhamedova, A.; Conlan, S.; Hutchison, S.K.; Kurth, A.; Ellerbrok, H.; Egholm, M.; Briese, T.; Leendertz, F.H.; et al. Moussa virus: A new member of the Rhabdoviridae family isolated from Culex decens mosquitoes in Côte d’Ivoire. Virus Res. 2010, 147, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Nasar, F.; Palacios, G.; Gorchakov, R.V.; Guzman, H.; Da Rosa, A.P.T.; Savji, N.; Popov, V.L.; Sherman, M.B.; Lipkin, W.I.; Tesh, R.B.; et al. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc. Natl. Acad. Sci. USA 2012, 109, 14622–14627. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lv, X.; Zhai, Y.; Fu, S.; Wang, D.; Rayner, S.; Tang, Q.; Liang, G. Genomic characterization of a novel virus of the family Tymoviridae isolated from mosquitoes. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.J.J. Biological transmission of arboviruses: Reexamination of and new insights into components, mechanisms, and unique traits as well as their evolutionary trends. Clin. Microbiol. Rev. 2005, 18, 608–637. [Google Scholar] [CrossRef] [PubMed]
- Higgs, S.; Beaty, B.J. Natural cycles of vector-borne pathogens. In Biology of Disease Vectors; Marquardt, W., Ed.; Elsevier Academic Press: Burlington, MA, USA, 2005. [Google Scholar]
- Nasar, F.; Gorchakov, R.V.; Tesh, R.B.; Weaver, S.C. Eilat Virus Host Range Restriction Is Present at Multiple Levels of the Virus Life Cycle. J. Virol. 2015, 89, 1404–1418. [Google Scholar] [CrossRef] [PubMed]
- Lutomiah, J.J.L.; Mwandawiro, C.; Magambo, J.; Sang, R.C. Infection and vertical transmission of Kamiti river virus in laboratory bred Aedes aegypti mosquitoes. J. Insect Sci. 2007, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Saiyasombat, R.; Bolling, B.G.; Brault, A.C.; Bartholomay, L.C.; Blitvich, B.J. Evidence of Efficient Transovarial Transmission of Culex Flavivirus by Culex pipiens (Diptera: Culicidae). J. Med. Entomol. 2011, 48, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.G.; Olea-Popelka, F.J.; Eisen, L.; Moore, C.G.; Blair, C.D. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 2012, 427, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Haddow, A.D.; Guzman, H.; Popov, V.L.; Wood, T.G.; Widen, S.G.; Haddow, A.D.; Tesh, R.B.; Weaver, S.C. First isolation of Aedes flavivirus in the Western Hemisphere and evidence of vertical transmission in the mosquito Aedes (Stegomyia) albopictus (Diptera: Culicidae). Virology 2013, 440, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Shi, M.; Tian, J.; Lin, X.; Kang, Y.; Chen, L.; Qin, X.; Xu, J.; Holmes, E.C.; Zhang, Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Junglen, S.; Drosten, C. Virus discovery and recent insights into virus diversity in arthropods. Curr. Opin. Microbiol. 2013, 16, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.G.; Vasilakis, N.; Guzman, H.; Widen, S.G.; Wood, T.G.; Popov, V.L.; Thangamani, S.; Tesh, R.B. Insect-Specific Viruses Detected in Laboratory Mosquito Colonies and Their Potential Implications for Experiments Evaluating Arbovirus Vector Competence. Am. J. Trop. Med. Hyg. 2014, 92, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Cook, S.; Chung, B.Y.W.; Bass, D.; Moureau, G.; Tang, S.; McAlister, E.; Culverwell, C.L.; Glücksman, E.; Wang, H.; Brown, T.D.K.; et al. Novel Virus Discovery and Genome Reconstruction from Field RNA Samples Reveals Highly Divergent Viruses in Dipteran Hosts. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Firth, C.; Widen, S.G.; Blasdell, K.R.; Guzman, H.; Wood, T.G.; Paradkar, P.N.; Holmes, E.C.; Tesh, R.B.; Vasilakis, N. Evolution of Genome Size and Complexity in the Rhabdoviridae. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Marklewitz, M.; Zirkel, F.; Kurth, A.; Drosten, C.; Junglen, S. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl. Acad. Sci. USA 2015, 112, 7536–7541. [Google Scholar] [CrossRef] [PubMed]
- Dudas, G.; Obbard, D.J. Are arthropods at the heart of virus evolution? Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Fort, P.; Albertini, A.; Van-Hua, A.; Berthomieu, A.; Roche, S.; Delsuc, F.; Pasteur, N.; Capy, P.; Gaudin, Y.; Weill, M. Fossil rhabdoviral sequences integrated into arthropod genomes: Ontogeny, evolution, and potential functionality. Mol. Biol. Evol. 2012, 29, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Ballinger, M.J.; Bruenn, J.A.; Kotov, A.A.; Taylor, D.J. Selectively maintained paleoviruses in Holarctic water fleas reveal an ancient origin for phleboviruses. Virology 2013, 446, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Maramorosch, K. Multiplication of plant viruses in insect vectors. Adv. Virus Res. 1955, 3, 221–249. [Google Scholar] [PubMed]
- Andrewes, C. Factors in virus evolution. Adv. Virus Res. 1957, 4, 1–24. [Google Scholar] [PubMed]
- Mattingly, P. Ecological aspects of the evolution of mosquito-borne virus diseases. Trans. R. Soc. Trop. Med. Hyg. 1960, 54, 97–112. [Google Scholar] [CrossRef]
- O’Neal, S.; Samuel, G.; Adelman, Z.; Myles, K. Mosquito-Borne Viruses and Suppressors of Invertebrate Antiviral RNA Silencing. Viruses 2014, 6, 4314–4331. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Cherry, S. Viruses and antiviral immunity in Drosophila. Dev. Comp. Immunol. 2014, 42, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.L.; Souza-Neto, J.; Torres Cosme, R.; Rovira, J.; Ortiz, A.; Pascale, J.M.; Dimopoulos, G. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 2012, 6, e1561. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhou, G.; Wu, J.; Bian, G.; Lu, P.; Raikhel, A.S.; Xi, Z. PNAS Plus: Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. 2012, 109, E23–E31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Hussain, M.; O’Neill, S.L.; Asgari, S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 10276–10281. [Google Scholar] [CrossRef] [PubMed]
- Karpf, A.R.; Lenches, E.; Strauss, E.G.; Strauss, J.H.; Brown, D.T. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 1997, 71, 7119–7123. [Google Scholar]
- Burivong, P.; Pattanakitsakul, S.N.; Thongrungkiat, S.; Malasit, P.; Flegel, T.W. Markedly reduced severity of Dengue virus infection in mosquito cell cultures persistently infected with Aedes albopictus densovirus (AalDNV). Virology 2004, 329, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Pepin, K.M.; Lambeth, K.; Hanley, K. a Asymmetric competitive suppression between strains of dengue virus. BMC Microbiol. 2008, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Newman, C.M.; Cerutti, F.; Anderson, T.K.; Hamer, G.L.; Walker, E.D.; Kitron, U.D.; Ruiz, M.O.; Brawn, J.D.; Goldberg, T.L. Culex flavivirus and West Nile virus mosquito coinfection and positive ecological association in Chicago, United States. Vector Borne Zoonotic Dis. 2011, 11, 1099–1105. [Google Scholar] [CrossRef] [PubMed]
- Crockett, R.K.; Burkhalter, K.; Mead, D.; Kelly, R.; Brown, J.; Varnado, W.; Roy, A.; Horiuchi, K.; Biggerstaff, B.J.; Miller, B.; Nasci, R. Culex flavivirus and West Nile virus in Culex quinquefasciatus populations in the southeastern United States. J. Med. Entomol. 2012, 49, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Kent, R.J.; Crabtree, M.B.; Miller, B.R. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex Flavivirus Izabal. PLoS Negl. Trop. Dis. 2010, 4, e671. [Google Scholar] [CrossRef] [PubMed]
- Kenney, J.L.; Solberg, O.D.; Langevin, S.A.; Brault, A.C. Characterization of a novel insect-specific flavivirus from Brazil: potential for inhibition of infection of arthropod cells with medically important flaviviruses. J. Gen. Virol. 2014, 95, 2796–2808. [Google Scholar] [CrossRef] [PubMed]
- Brackney, D.E.; Scott, J.C.; Sagawa, F.; Woodward, J.E.; Miller, N.A.; Schilkey, F.D.; Mudge, J.; Wilusz, J.; Olson, K.E.; Blair, C.D.; Ebel, G.D. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl. Trop. Dis. 2010, 4, e856. [Google Scholar] [CrossRef] [PubMed]
- Kuwata, R.; Isawa, H.; Hoshino, K.; Sasaki, T.; Kobayashi, M.; Maeda, K.; Sawabe, K. Analysis of Mosquito-Borne Flavivirus Superinfection in Culex tritaeniorhynchus (Diptera: Culicidae) Cells Persistently Infected with Culex Flavivirus (Flaviviridae). J. Med. Entomol. 2015, 52, 222–229. [Google Scholar] [CrossRef]
- Walton, W.E. Larvivorous fish including Gambusia. J. Am. Mosq. Control Assoc. 2007, 23, 184–220. [Google Scholar] [CrossRef]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.S.; Soberón, M. Bacillus thuringiensis : A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Laven, H. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 1967, 216, 383–384. [Google Scholar] [CrossRef] [PubMed]
- Hedges, L.M.; Brownlie, J.C.; O’Neill, S.L.; Johnson, K.N. Wolbachia and virus protection in insects. Science 2008, 322. [Google Scholar] [CrossRef] [PubMed]
- Glaser, R.L.; Meola, M.A. The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Kambris, Z.; Blagborough, A.M.; Pinto, S.B.; Blagrove, M.S.C.; Godfray, H.C.J.; Sinden, R.E.; Sinkins, S.P. Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynaud, J.M.; Kim, D.Y.; Atasheva, S.; Rasalouskaya, A.; White, J.P.; Diamond, M.S.; Weaver, S.C.; Frolova, E.I.; Frolov, I. IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolling, B.G.; Weaver, S.C.; Tesh, R.B.; Vasilakis, N. Insect-Specific Virus Discovery: Significance for the Arbovirus Community. Viruses 2015, 7, 4911-4928. https://doi.org/10.3390/v7092851
Bolling BG, Weaver SC, Tesh RB, Vasilakis N. Insect-Specific Virus Discovery: Significance for the Arbovirus Community. Viruses. 2015; 7(9):4911-4928. https://doi.org/10.3390/v7092851
Chicago/Turabian StyleBolling, Bethany G., Scott C. Weaver, Robert B. Tesh, and Nikos Vasilakis. 2015. "Insect-Specific Virus Discovery: Significance for the Arbovirus Community" Viruses 7, no. 9: 4911-4928. https://doi.org/10.3390/v7092851







