The Interaction between Cyclin B1 and Cytomegalovirus Protein Kinase pUL97 is Determined by an Active Kinase Domain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs
2.2. Cell Culture, HCMV Infections and Plasmid Transfection
2.3. Antibodies
2.4. Coimmunoprecipitation (CoIP)
2.5. Confocal Imaging
2.6. In Vitro Kinase Assay (IVKA)
2.7. Homology Modeling of pUL97
3. Results
3.1. HCMV Protein Kinase pUL97 Shows a Particularly Strong Interaction with Cyclin B1
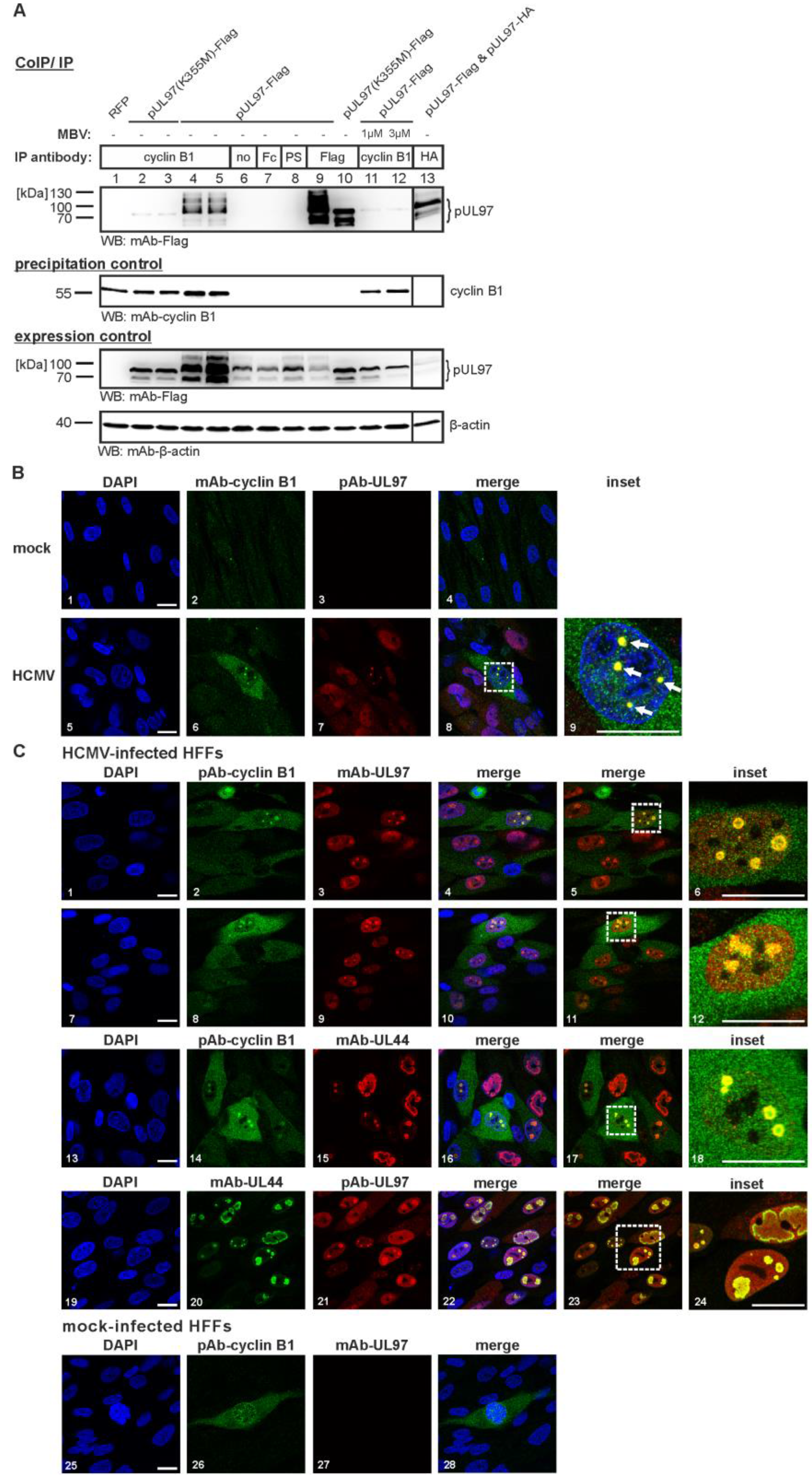
3.2. The pUL97-Cyclin B1 Interaction is Dependent on Kinase Activity
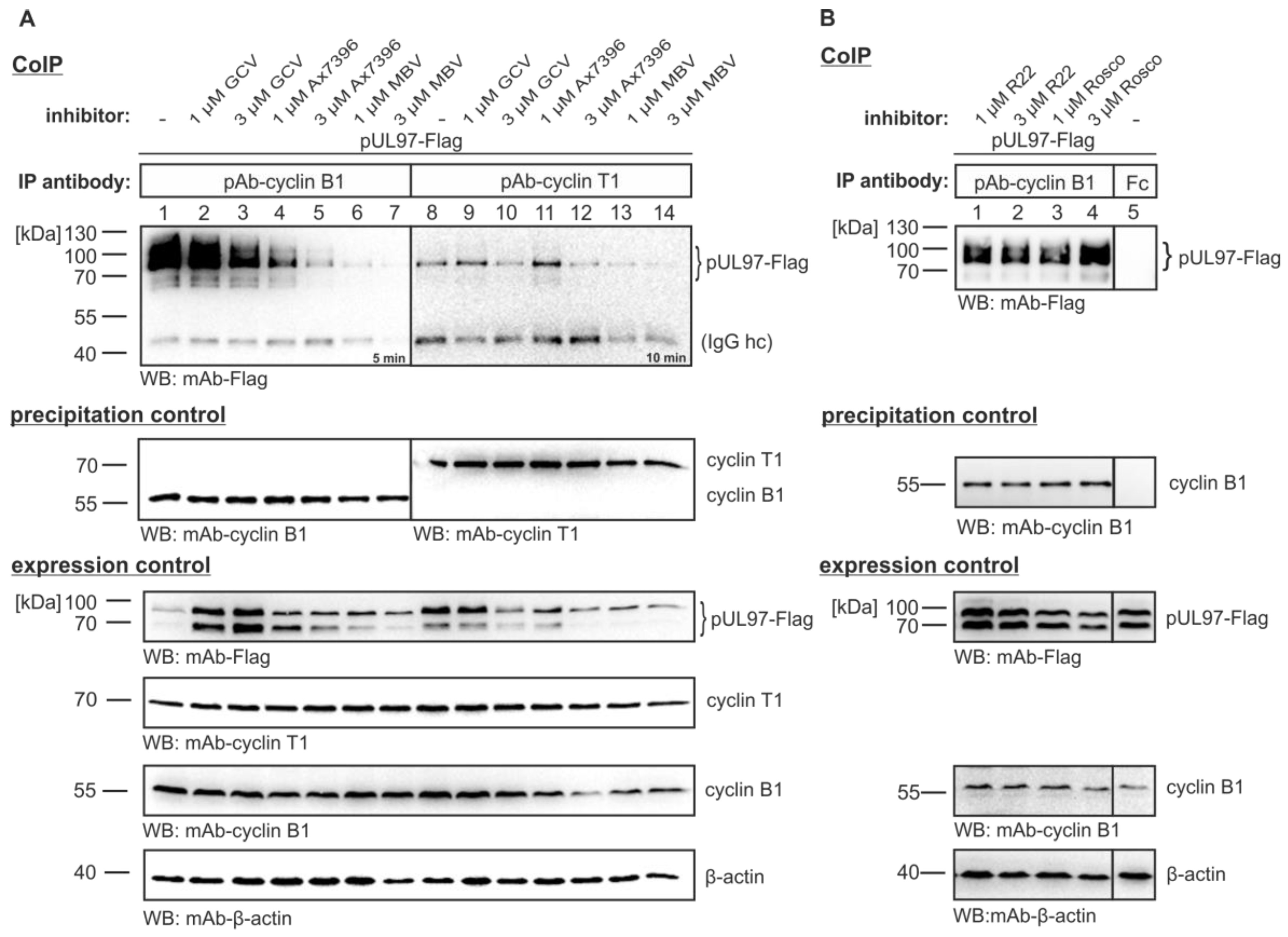
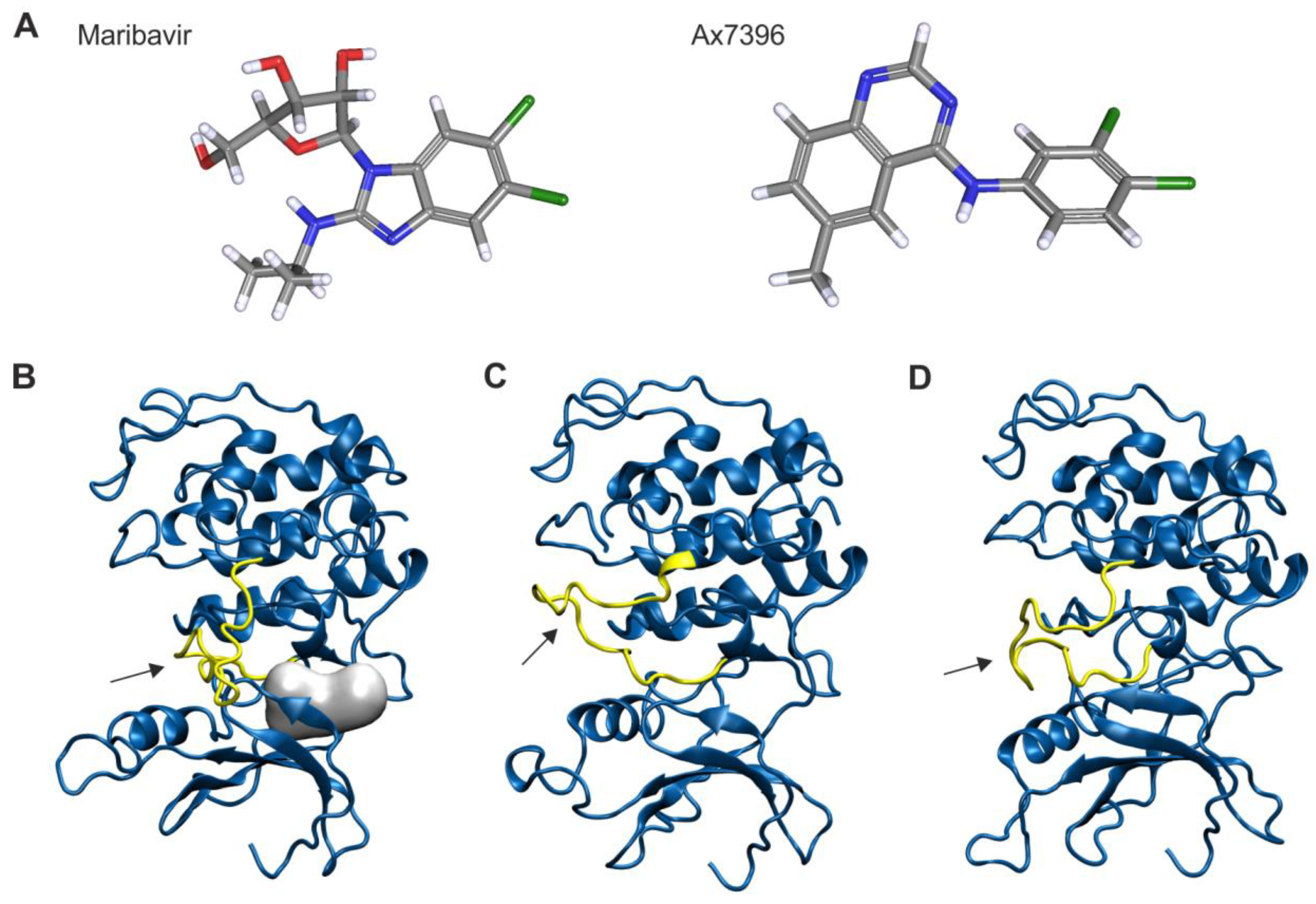
3.3. Serial Deletions of the pUL97 C-Terminus Abrogate Cyclin B1 Interaction
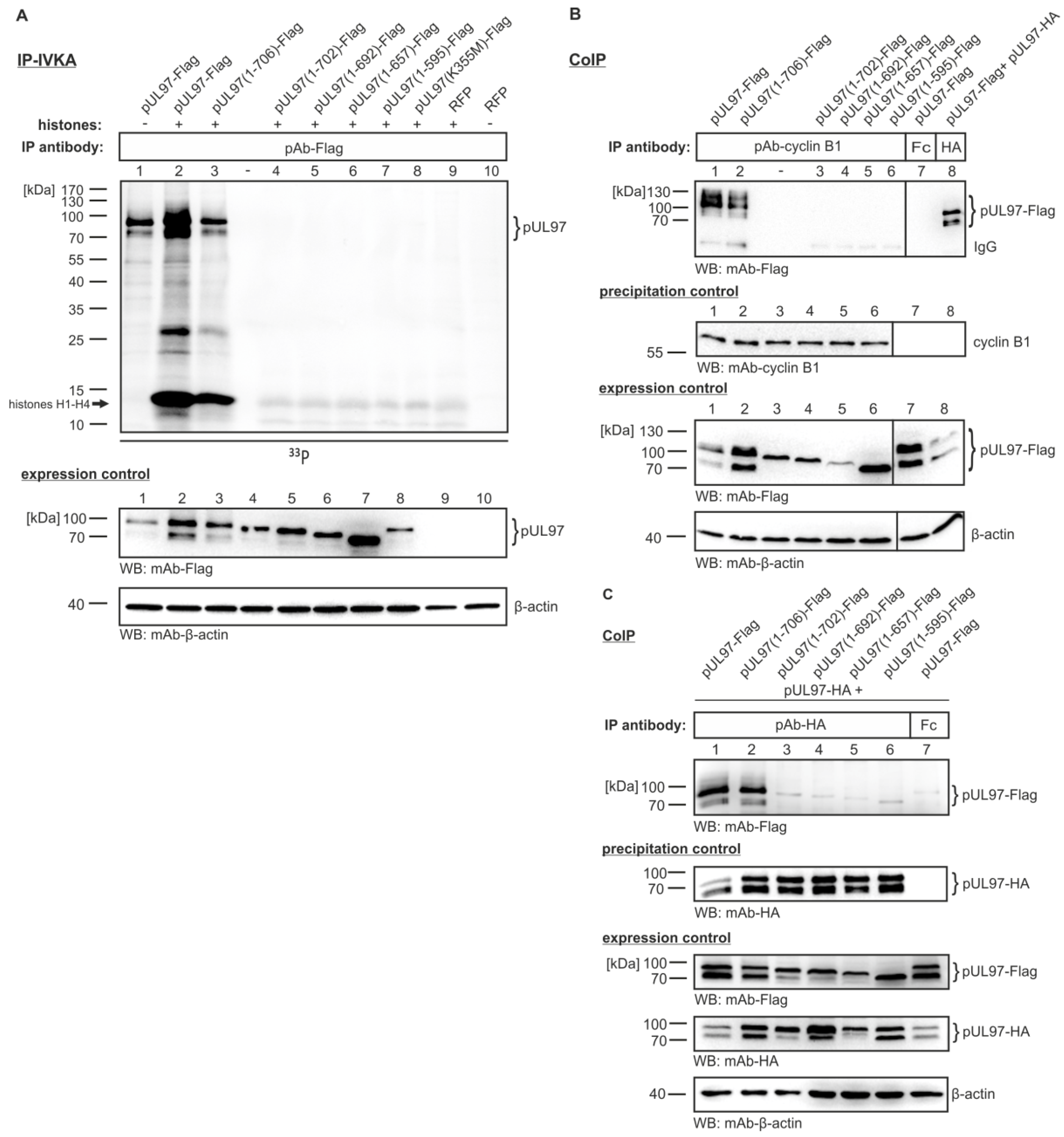
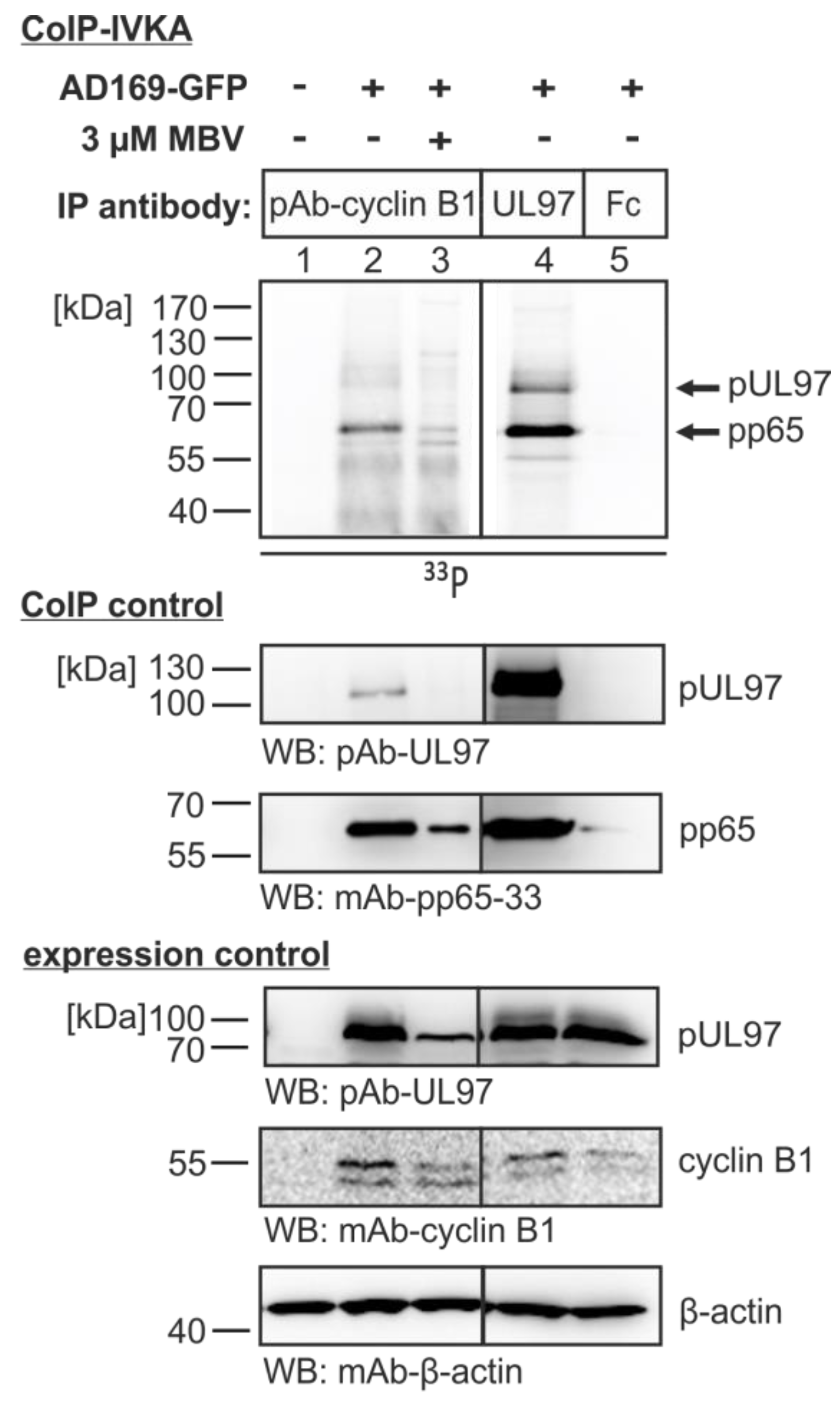
3.4. The Cyclin B1-pUL97 Complex Precipitated from HCMV-Infected Cells Comprises Substrate-Binding and Substrate-Phosphorylating Activity
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Mocarski, E.S.; Shenk, T.; Griffiths, P.D.; Pass, R.F. Cytomegaloviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 2, pp. 1960–2014. [Google Scholar]
- Schreiber, A.; Härter, G.; Schubert, A.; Bunjes, D.; Mertens, T.; Michel, D. Antiviral treatment of cytomegalovirus infection and resistant strains. Expert Opin. Pharmacother. 2009, 10, 191–209. [Google Scholar]
- Marschall, M.; Feichtinger, S.; Milbradt, J. Regulatory roles of protein kinases in cytomegalovirus replication. Adv. Virus Res. 2011, 80, 69–101. [Google Scholar]
- Marschall, M.; Stamminger, T. Molecular targets for antiviral therapy of cytomegalovirus infections. Future Microbiol. 2009, 4, 731–742. [Google Scholar]
- Prichard, M.N. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 2009, 19, 215–229. [Google Scholar]
- Chou, S. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev. Med. Virol. 2008, 18, 233–246. [Google Scholar]
- Herget, T.; Marschall, M. Recent developments in anti-herpesviral combination therapy based on protein kinase inhibitors. In New Concepts of Antiviral Therapy; Bogner, E., Holzenburg, A., Eds.; Springer: London, UK, 2006; pp. 351–371. [Google Scholar]
- Schang, L.M.; St. Vincent, M.R.; Lacasse, J.J. Five years of progress on cyclindependent kinases and other cellular proteins as potential targets for antiviral drugs. Antivir. Chem. Chemother. 2006, 17, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Feichtinger, S.; Stamminger, T.; Müller, R.; Graf, L.; Klebl, B.; Eickhoff, J.; Marschall, M. Recruitment of cyclin-dependent kinase 9 to nuclear compartments during cytomegalovirus late replication: Importance of an interaction between viral pUL69 and cyclin T1. J. Gen. Virol. 2011, 92, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Hertel, L.; Chou, S.; Mocarski, E.S. Viral and cell cycle-regulated kinases in cytomegalovirusinduced pseudomitosis and replication. PLoS Pathog. 2007, 3, e6. [Google Scholar]
- Hutterer, C.; Wandinger, S.K.; Wagner, S.; Müller, R.; Stamminger, T.; Zeitträger, I.; Godl, K.; Baumgartner, R.; Strobl, S.; Marschall, M. Profiling of the kinome of cytomegalovirus-infected cells reveals the functional importance of host kinases Aurora A, ABL and AMPK. Antivir. Res. 2013, 99, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Kapasi, A.J.; Spector, D.H. Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 2008, 82, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, V.; Spector, D.H. Cyclin-dependent kinase activity is required for efficient expression and posttranslational modification of human cytomegalovirus proteins and for production of extracellular particles. J. Virol. 2006, 80, 5886–5896. [Google Scholar] [CrossRef] [PubMed]
- Tamrakar, S.; Kapasi, A.J.; Spector, D.H. Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J. Virol. 2005, 79, 15477–15493. [Google Scholar] [CrossRef] [PubMed]
- Romaker, D.; Schregel, V.; Maurer, K.; Auerochs, S.; Marzi, A.; Sticht, H.; Marschall, M. Analysis of the structure-activity relationship of four herpesviral UL97 subfamily protein kinases reveals partial but not full functional conservation. J. Med. Chem. 2006, 49, 7044–7053. [Google Scholar] [CrossRef] [PubMed]
- Hume, A.J.; Finkel, J.S.; Kamil, J.P.; Coen, D.M.; Culbertson, M.R.; Kalejta, R.F. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 2008, 320, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Marzi, A.; aus dem Siepen, P.; Jochmann, R.; Kalmer, M.; Auerochs, S.; Lischka, P.; Leis, M.; Stamminger, T. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 2005, 280, 33357–33367. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Rechter, S.; Milbradt, J.; Auerochs, S.; Müller, R.; Stamminger, T.; Marschall, M. The cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J. Gen. Virol. 2009, 90, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Rechter, S.; Scott, G.M.; Eickhoff, J.; Zielke, K.; Auerochs, S.; Müller, R.; Stamminger, T.; Rawlinson, W.D.; Marschall, M. Cyclin-dependent kinases phosphorylate the cytomegalovirus RNA export protein pUL69 to modulate its nuclear localization and activity. J. Biol. Chem. 2009, 284, 8605–8613. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Kato, K.; Tanaka, M.; Kanamori, M.; Nishiyama, Y.; Yamanashi, Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 2013, 77, 2359–2368. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Matsumura, T.; Roizman, B.; Hirai, K. Cellular elongation factor 1δ is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J. Virol. 1999, 73, 4456–4460. [Google Scholar] [PubMed]
- Milbradt, J.; Webel, R.; Auerochs, S.; Sticht, H.; Marschall, M. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus . J. Biol. Chem. 2010, 285, 13979–13989. [Google Scholar]
- Milbradt, J.; Auerochs, S.; Sticht, H.; Marschall, M. Cytomegaloviral proteins that associate with the nuclear lamina: Components of a postulated nuclear egress complex. J. Gen. Virol. 2009, 90, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Hamirally, S.; Kamil, J.P.; Ndassa-Colday, Y.M.; Lin, A.J.; Jahng, W.J.; Baek, M.C.; Noton, S.; Silva, L.A.; Simpson-Holley, M.; Knipe, D.M.; et al. Viral mimicry of Cdc2/cyclin-dependent kinase mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS. Pathog. 2009, 5, e1000275. [Google Scholar] [CrossRef] [PubMed]
- Prichard, M.N.; Sztul, E.; Daily, S.L.; Perry, A.L.; Frederick, S.L.; Gill, R.B.; Hartline, C.B.; Streblow, D.N.; Varnum, S.M.; Smith, R.D.; et al. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J. Virol. 2008, 82, 5054–5067. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.C.; Krosky, P.M.; Pearson, A.; Coen, D.M. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology 2004, 324, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Stein-Gerlach, M.; Freitag, M.; Kupfer, R.; van den Bogaard, M.; Stamminger, T. Direct targeting of human cytomegalovirus protein kinase pUL97 by kinase inhibitors is a novel principle of antiviral therapy. J. Gen. Virol. 2002, 83, 1013–1023. [Google Scholar] [PubMed]
- Prichard, M.N.; Gao, N.; Jairath, S.; Mulamba, G.; Krosky, P.; Coen, D.M.; Parker, B.O.; Pari, G.S. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 1999, 73, 5663–5670. [Google Scholar] [PubMed]
- Marschall, M.; Stein-Gerlach, M.; Freitag, M.; Kupfer, R.; van den Bogaard, M.; Stamminger, T. Inhibitors of human cytomegalovirus replication drastically reduce the activity of the viral protein kinase pUL97. J. Gen. Virol. 2001, 82, 1439–1450. [Google Scholar] [PubMed]
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science 1988, 241, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Webel, R.; Milbradt, J.; Auerochs, S.; Schregel, V.; Held, C.; Nöbauer, K.; Razzazi-Fazeli, E.; Jardin, C.; Wittenberg, T.; Sticht, H.; Marschall, M. Two isoforms of the protein kinase pUL97 of human cytomegalovirus are differentially regulated in their nuclear translocation. J. Gen. Virol. 2011, 92, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Webel, R.; Solbak, S.M.Ø.; Held, C.; Milbradt, J.; Groß, A.; Eichler, J.; Wittenberg, T.; Jardin, C.; Sticht, H.; Fossen, T.; et al. The nuclear import of isoforms of the cytomegalovirus kinase pUL97 is mediated by differential activity of NLS1 and NLS2 both acting through classical importin-α binding. J. Gen. Virol. 2012, 93, 1756–1768. [Google Scholar] [PubMed]
- Webel, R.; Hakki, M.; Prichard, M.N.; Rawlinson, W.D.; Marschall, M.; Chou, S. Differential properties of cytomegalovirus pUL97 kinase isoforms affect viral replication and maribavir susceptibility. J. Virol. 2014, 88, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Schregel, V.; Auerochs, S.; Jochmann, R.; Maurer, K.; Stamminger, T.; Marschall, M. Mapping of a self-interaction domain of the cytomegalovirus protein kinase pUL97. J. Virol. 2007, 88, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.C.; Krosky, P.M.; Coen, D.M. Relationship between Autophosphorylation and Phosphorylation of Exogenous Substrates by the Human Cytomegalovirus UL97 Protein Kinase. J. Virol. 2002, 76, 11943–11952. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; He, Y.S.; Kim, Y.; Chu, L.; Ohmstede, C.; Biron, K.K.; Coen, D.M. The human cytomegalovirus UL97 protein is a protein kinase that autophosphorylates on serines and threonines. J. Virol. 1997, 71, 405–411. [Google Scholar] [PubMed]
- Marschall, M.; Freitag, M.; Weiler, S.; Sorg, G.; Stamminger, T. Recombinant green fluorescent protein-expressing human cytomegalovirus as a tool for screening antiviral agents. Antimicrob. Agents Chemother. 2000, 44, 1588–1597. [Google Scholar] [CrossRef] [PubMed]
- Sielecki, T.M.; Johnson, T.L.; Liu, J.; Muckelbauer, J.K.; Grafstrom, R.H.; Cox, S.; Boylan, J.; Burton, C.R.; Chen, H.; Smallwood, A.; et al. Quinazolines as cyclin dependent kinase inhibitors. Bioorg. Med. Chem. Lett. 2001, 11, 1157–1160. [Google Scholar] [CrossRef]
- Martin, M.P.; Olesen, S.H.; Georg, G.I.; Schonbrunn, E. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem. Biol. 2013, 8, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.R.; Lowe, E.D.; Petri, E.; Skamnaki, V.; Antrobus, R.; Johnson, L.N. Cyclin B and cyclin A confer different substrate recognition properties on CDK2. Cell Cycle 2007, 6, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.R.; Korolchuk, S.; Martin, M.P.; Stanley, W.A.; Moukhametzianov, R.; Noble, M.E.M.; Endicott, J.A. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat. Commun. 2015, 6, e6769. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2014, 47. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Graf, L.; Webel, R.; Wagner, S.; Hamilton, S.T.; Rawlinson, W.D.; Sticht, H.; Marschall, M. The cyclin-dependent kinase ortholog pUL97 of human cytomegalovirus interacts with cyclins. Viruses 2013, 18, 3213–3230. [Google Scholar]
- Marschall, M.; Freitag, M.; Suchy, P.; Romaker, D.; Kupfer, R.; Hanke, M.; Stamminger, T. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 2003, 311, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Herget, T.; Freitag, M.; Morbitzer, M.; Kupfer, R.; Stamminger, T.; Marschall, M. Novel chemical class of pUL97 protein kinase-specific inhibitors with strong anticytomegaloviral activity. Antimicrob. Agents Chemother. 2004, 48, 4154–4162. [Google Scholar]
- Schleiss, M.; Eickhoff, J.; Auerochs, S.; Leis, M.; Abele, S.; Rechter, S.; Choi, Y.; Anderson, J.; Scott, G.; Rawlinson, W.; et al. Protein kinase inhibitors of the quinazoline class exert anticytomegaloviral activity in vitro and in vivo. Antivir. Res. 2008, 79, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Biron, K.K.; Harvey, R.J.; Chamberlain, S.C.; Good, S.S.; Smith, A.A., 3rd; Davis, M.G.; Talarico, C.L.; Miller, W.H.; Ferris, R.; Dornsife, R.E.; et al. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole L-riboside with a unique mode of action. Antimicrob. Agents Chemother. 2002, 46, 2365–2372. [Google Scholar] [CrossRef] [PubMed]
- Price, N.B.; Prichard, M.N. Progress in the development of new therapies for herpesvirus infections. Curr. Opin. Virol. 2011, 1, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Chou, S. Cytomegalovirus UL97 mutations in the era of ganciclovir and maribavir. Rev. Med. Virol. 2008, 18, 233–246. [Google Scholar]
- Becke, S.; Fabre-Mersseman, V.; Aue, S.; Auerochs, S.; Sedmak, T.; Wolfrum, U.; Strand, D.; Marschall, M.; Plachter, B.; Reyda, S. Modification of the major tegument protein pp 65 of human cytomegalovirus inhibits viral growth and leads to the enhancement of a protein complex with pUL69 and pUL97 in infected cells. J. Gen. Virol. 2010, 91, 2531–2541. [Google Scholar] [CrossRef] [PubMed]
- Milbradt, J.; Kraut, A.; Hutterer, C.; Sonntag, E.; Schmeiser, C.; Ferro, M.; Wagner, S.; Lenac, T.; Claus, C.; Pinkert, S.; et al. Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol. Cell Proteomics 2014, 13, 2132–2146. [Google Scholar] [CrossRef] [PubMed]
- Hutterer, C.; Eickhoff, J.; Milbradt, J.; Korn, K.; Zeitträger, I.; Bahsi, H.; Wagner, S.; Zischinsky, G.; Wolf, A.; Degenhart, C.; et al. A novel CDK7 inhibitor of the pyrazolo-triazine class exerts broad-spectrum antiviral activity at nanomolar concentrations. Antimicrob. Agents Chemother. 2015, 59, 2062–2071. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steingruber, M.; Socher, E.; Hutterer, C.; Webel, R.; Bergbrede, T.; Lenac, T.; Sticht, H.; Marschall, M. The Interaction between Cyclin B1 and Cytomegalovirus Protein Kinase pUL97 is Determined by an Active Kinase Domain. Viruses 2015, 7, 4582-4601. https://doi.org/10.3390/v7082834
Steingruber M, Socher E, Hutterer C, Webel R, Bergbrede T, Lenac T, Sticht H, Marschall M. The Interaction between Cyclin B1 and Cytomegalovirus Protein Kinase pUL97 is Determined by an Active Kinase Domain. Viruses. 2015; 7(8):4582-4601. https://doi.org/10.3390/v7082834
Chicago/Turabian StyleSteingruber, Mirjam, Eileen Socher, Corina Hutterer, Rike Webel, Tim Bergbrede, Tihana Lenac, Heinrich Sticht, and Manfred Marschall. 2015. "The Interaction between Cyclin B1 and Cytomegalovirus Protein Kinase pUL97 is Determined by an Active Kinase Domain" Viruses 7, no. 8: 4582-4601. https://doi.org/10.3390/v7082834




