Early Events in Chikungunya Virus Infection—From Virus CellBinding to Membrane Fusion
Abstract
:1. Introduction
2. Viral Structure

3. Viral Tropism
3.1. Viremia—Where Is the Virus Produced?
3.2. Arthrotropism of CHIKV
3.3. Less Common Tropism of CHIKV
4. Cell Entry and Membrane Fusion
4.1. Receptor Binding
4.1.1. Prohibitin
4.1.2. Phosphatidylserine (PtdSer)-Mediated Virus Entry-enhancing Receptors
4.1.3. Glycosaminoglycans
4.1.4. ATP Synthase β Subunit
4.1.5. Other CHIKV Receptors Candidates
4.2. CHIKV Cell Entry and Membrane Fusion
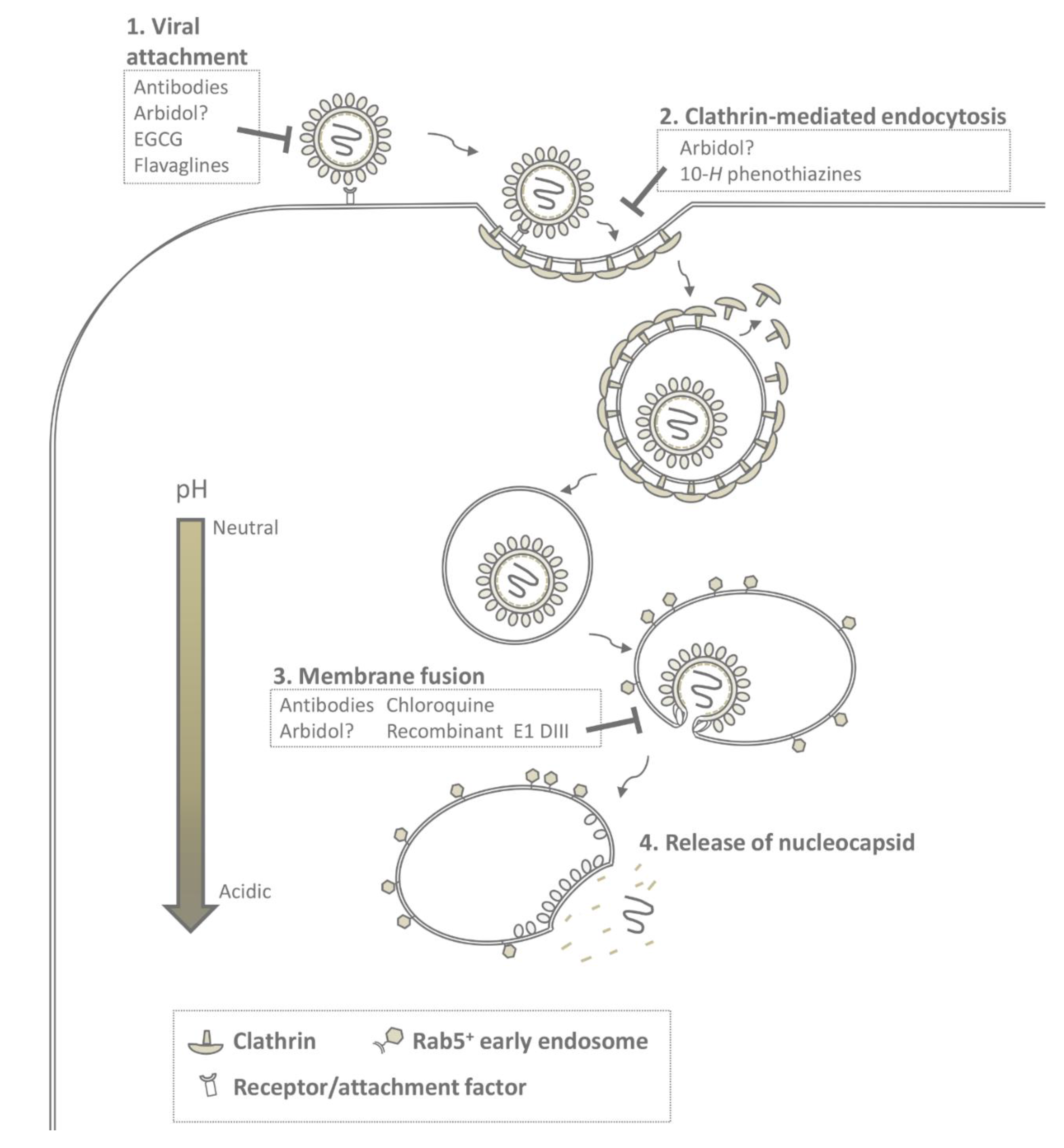
5. Molecular Mechanism of CHIKV Fusion

6. Inhibition of Early Events in Infection
6.1. Interference with Virus-Receptor Binding
6.2. Interference with Endocytosis
6.3. Interference with Membrane Fusion
7. Future Perspectives and Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Thiberville, S.D.; Moyen, N.; Dupuis-Maguiraga, L.; Nougairede, A.; Gould, E.A.; Roques, P.; de Lamballerie, X. Chikungunya Fever: Epidemiology, Clinical Syndrome, Pathogenesis and Therapy. Antiviral Res. 2013, 99, 345–370. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E. Reemergence of Chikungunya Virus. J. Virol. 2014, 88, 11644–11647. [Google Scholar] [CrossRef] [PubMed]
- Dupuis-Maguiraga, L.; Noret, M.; Brun, S.; Le Grand, R.; Gras, G.; Roques, P. Chikungunya Disease: Infection-Associated Markers from the Acute to the Chronic Phase of Arbovirus-Induced Arthralgia. PLoS Negl. Trop. Dis. 2012, 6, e1446. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Jaffar-Bandjee, M.C.; Hoarau, J.J.; Krejbich Trotot, P.; Denizot, M.; Lee-Pat-Yuen, G.; Sahoo, R.; Guiraud, P.; Ramful, D.; Robin, S.; et al. Chikungunya Fever: CNS Infection and Pathologies of a Re-Emerging Arbovirus. Prog. Neurobiol. 2010, 91, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Pellot, A.S.; Alessandri, J.L.; Robin, S.; Samperiz, S.; Attali, T.; Brayer, C.; Pasquet, M.; Jaffar-Bandjee, M.C.; Benhamou, L.S.; Tiran-Rajaofera, I.; et al. Severe Forms of Chikungunya Virus Infection in a Pediatric Intensive Care Unit on Reunion Island. Med. Trop. (Mars) 2012, 72, 88–93. [Google Scholar] [PubMed]
- Staples, J.E.; Breiman, R.F.; Powers, A.M. Chikungunya Fever: An Epidemiological Review of a Re-Emerging Infectious Disease. Clin. Infect. Dis. 2009, 49, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Lecuit, M. Chikungunya Virus and the Global Spread of a Mosquito-Borne Disease. N. Engl. J. Med. 2015, 372, 1231–1239. [Google Scholar] [PubMed]
- Pan American Health Organization. Number of Reported Cases of Chikungunya Fever in the Americas, by Country Or Territory 2013–2015 (to Week Noted) Epidemiological Week/EW 17. (Updated as of 1 May 2015)Available online: https://clinicaltri-als.gov/ct2/show/NCT02230163?term=Chikungunya&rank=5 or http://www.webcita-tion.org/6YE7aebST. (accessed on 2 May 2015).
- Volk, S.M.; Chen, R.; Tsetsarkin, K.A.; Adams, A.P.; Garcia, T.I.; Sall, A.A.; Nasar, F.; Schuh, A.J.; Holmes, E.C.; Higgs, S.; et al. Genome-Scale Phylogenetic Analyses of Chikungunya Virus Reveal Independent Emergences of Recent Epidemics and various Evolutionary Rates. J. Virol. 2010, 84, 6497–6504. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary History and Recent Epidemic Spread. Antiviral Res. 2015, 120, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Chen, R.; Yun, R.; Rossi, S.L.; Plante, K.S.; Guerbois, M.; Forrester, N.; Perng, G.C.; Sreekumar, E.; Leal, G.; et al. Multi-Peaked Adaptive Landscape for Chikungunya Virus Evolution Predicts Continued Fitness Optimization in Aedes Albopictus Mosquitoes. Nat. Commun. 2014, 5, 4084. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A Single Mutation in Chikungunya Virus Affects Vector Specificity and Epidemic Potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C. Arrival of Chikungunya Virus in the New World: Prospects for Spread and Impact on Public Health. PLoS Negl Trop. Dis. 2014, 8, e2921. [Google Scholar] [CrossRef] [PubMed]
- Powers, A.M.; Brault, A.C.; Shirako, Y.; Strauss, E.G.; Kang, W.; Strauss, J.H.; Weaver, S.C. Evolutionary Relationships and Systematics of the Alphaviruses. J. Virol. 2001, 75, 10118–10131. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Snyder, J.E.; Kuhn, R.J. A Structural and Functional Perspective of Alphavirus Replication and Assembly. Future Microbiol. 2009, 4, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Simizu, B.; Yamamoto, K.; Hashimoto, K.; Ogata, T. Structural Proteins of Chikungunya Virus. J. Virol. 1984, 51, 254–258. [Google Scholar] [PubMed]
- Cao, S.; Zhang, W. Characterization of an Early-Stage Fusion Intermediate of Sindbis Virus using Cryoelectron Microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 13362–13367. [Google Scholar] [CrossRef] [PubMed]
- Allan, D.; Quinn, P. Membrane Phospholipid Asymmetry in Semliki Forest Virus Grown in BHK Cells. Biochim. Biophys. Acta Biomembr. 1989, 987, 199–204. [Google Scholar] [CrossRef]
- Renkonen, O.; Kaarainen, L.; Simons, K.; Gahmberg, C.G. The Lipid Class Composition of Semliki Forest Virus and Plasma Membranes of the Host Cells. Virology 1971, 46, 318–326. [Google Scholar] [CrossRef]
- Van Meer, G.; Simons, K.; Op den Kamp, J.A.; van Deenen, L.M. Phospholipid Asymmetry in Semliki Forest Virus Grown on Baby Hamster Kidney (BHK-21) Cells. Biochemistry 1981, 20, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Kalvodova, L.; Sampaio, J.L.; Cordo, S.; Ejsing, C.S.; Shevchenko, A.; Simons, K. The Lipidomes of Vesicular Stomatitis Virus, Semliki Forest Virus, and the Host Plasma Membrane Analyzed by Quantitative Shotgun Mass Spectrometry. J. Virol. 2009, 83, 7996–8003. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xiang, Y.; Akahata, W.; Holdaway, H.; Pal, P.; Zhang, X.; Diamond, M.S.; Nabel, G.J.; Rossmann, M.G. Structural Analyses at Pseudo Atomic Resolution of Chikungunya Virus and Antibodies show Mechanisms of Neutralization. eLife 2013, 2, e00435. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Zhang, W.; Gabler, S.; Chipman, P.R.; Strauss, E.G.; Strauss, J.H.; Baker, T.S.; Kuhn, R.J.; Rossmann, M.G. Mapping the Structure and Function of the E1 and E2 Glycoproteins in Alphaviruses. Structure 2006, 14, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Lescar, J.; Roussel, A.; Wien, M.W.; Navaza, J.; Fuller, S.D.; Wengler, G.; Wengler, G.; Rey, F.A. The Fusion Glycoprotein Shell of Semliki Forest Virus: An Icosahedral Assembly Primed for Fusogenic Activation at Endosomal pH. Cell 2001, 105, 137–148. [Google Scholar] [CrossRef]
- Roussel, A.; Lescar, J.; Vaney, M.C.; Wengler, G.; Wengler, G.; Rey, F.A. Structure and Interactions at the Viral Surface of the Envelope Protein E1 of Semliki Forest Virus. Structure 2006, 14, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Voss, J.E.; Vaney, M.C.; Duquerroy, S.; Vonrhein, C.; Girard-Blanc, C.; Crublet, E.; Thompson, A.; Bricogne, G.; Rey, F.A. Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-ray Crystallography. Nature 2010, 468, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Przybyla, L.; Edwards, T.J.; Perera, R.; Burgner, J.W., 2nd; Kuhn, R.J. Interactions of the Cytoplasmic Domain of Sindbis Virus E2 with Nucleocapsid Cores Promote Alphavirus Budding. J. Virol. 2012, 86, 2585–2599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Hryc, C.F.; Cong, Y.; Liu, X.; Jakana, J.; Gorchakov, R.; Baker, M.L.; Weaver, S.C.; Chiu, W. 4.4 A Cryo-EM Structure of an Enveloped Alphavirus Venezuelan Equine Encephalitis Virus. EMBO J. 2011, 30, 3854–3863. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jose, J.; Xiang, Y.; Kuhn, R.J.; Rossmann, M.G. Structural Changes of Envelope Proteins during Alphavirus Fusion. Nature 2010, 468, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.H.; Provencher, S.W.; von Bonsdorff, C.H.; Adrian, M.; Dubochet, J. Envelope Structure of Semliki Forest Virus Reconstructed from Cryo-Electron Micrographs. Nature 1986, 320, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Pletnev, S.V.; Zhang, W.; Mukhopadhyay, S.; Fisher, B.R.; Hernandez, R.; Brown, D.T.; Baker, T.S.; Rossmann, M.G.; Kuhn, R.J. Locations of Carbohydrate Sites on Alphavirus Glycoproteins show that E1 Forms an Icosahedral Scaffold. Cell 2001, 105, 127–136. [Google Scholar] [CrossRef]
- Cheng, R.H.; Kuhn, R.J.; Olson, N.H.; Rossmann, M.G.; Choi, H.K.; Smith, T.J.; Baker, T.S. Nucleocapsid and Glycoprotein Organization in an Enveloped Virus. Cell 1995, 80, 621–630. [Google Scholar] [CrossRef]
- Reiter, P.; Fontenille, D.; Paupy, C. Aedes Albopictus as an Epidemic Vector of Chikungunya Virus: Another Emerging Problem? Lancet Infect. Dis. 2006, 6, 463–464. [Google Scholar] [CrossRef]
- Ramasubramanian, M.K.; Barham, O.M.; Swaminathan, V. Mechanics of a Mosquito Bite with Applications to Microneedle Design. Bioinspir. Biomim. 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Osorio, J.E.; Livengood, J.A.; Chen, R.; Stinchcomb, D.T. Chikungunya Virus and Prospects for a Vaccine. Expert Rev. Vaccines 2012, 11, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Sourisseau, M.; Schilte, C.; Casartelli, N.; Trouillet, C.; Guivel-Benhassine, F.; Rudnicka, D.; Sol-Foulon, N.; Le Roux, K.; Prevost, M.C.; Fsihi, H.; et al. Characterization of Reemerging Chikungunya Virus. PLoS Pathog. 2007, 3, e89. [Google Scholar] [CrossRef] [PubMed]
- Solignat, M.; Gay, B.; Higgs, S.; Briant, L.; Devaux, C. Replication Cycle of Chikungunya: A Re-Emerging Arbovirus. Virology 2009, 393, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Wikan, N.; Sakoonwatanyoo, P.; Ubol, S.; Yoksan, S.; Smith, D.R. Chikungunya Virus Infection of Cell Lines: Analysis of the East, Central and South African Lineage. PLoS ONE 2012, 7, e31102. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Her, Z.; Ong, E.K.; Chen, J.M.; Dimatatac, F.; Kwek, D.J.; Barkham, T.; Yang, H.; Renia, L.; Leo, Y.S.; et al. Persistent Arthralgia Induced by Chikungunya Virus Infection is Associated with Interleukin-6 and Granulocyte Macrophage Colony-Stimulating Factor. J. Infect. Dis. 2011, 203, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Naze, F.; Le Roux, K.; Schuffenecker, I.; Zeller, H.; Staikowsky, F.; Grivard, P.; Michault, A.; Laurent, P. Simultaneous Detection and Quantitation of Chikungunya, Dengue and West Nile Viruses by Multiplex RT-PCR Assays and Dengue Virus Typing using High Resolution Melting. J. Virol. Methods 2009, 162, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.C.; Tan, L.K.; Tan, C.H.; Tan, S.S.; Hapuarachchi, H.C.; Pok, K.Y.; Lai, Y.L.; Lam-Phua, S.G.; Bucht, G.; Lin, R.T.; et al. Entomologic and Virologic Investigation of Chikungunya, Singapore. Emerg. Infect. Dis. 2009, 15, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Her, Z.; Malleret, B.; Chan, M.; Ong, E.K.; Wong, S.C.; Kwek, D.J.; Tolou, H.; Lin, R.T.; Tambyah, P.A.; Renia, L.; et al. Active Infection of Human Blood Monocytes by Chikungunya Virus Triggers an Innate Immune Response. J. Immunol. 2010, 184, 5903–5913. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.R.; Becker, G.J. Mononuclear Cell Types in Chronic Synovial Effusions of Ross River Virus Disease. Aust. N. Z. J. Med. 1984, 14, 505–506. [Google Scholar] [CrossRef] [PubMed]
- Gardner, C.L.; Burke, C.W.; Tesfay, M.Z.; Glass, P.J.; Klimstra, W.B.; Ryman, K.D. Eastern and Venezuelan Equine Encephalitis Viruses Differ in their Ability to Infect Dendritic Cells and Macrophages: Impact of Altered Cell Tropism on Pathogenesis. J. Virol. 2008, 82, 10634–10646. [Google Scholar] [CrossRef] [PubMed]
- Hidmark, A.S.; McInerney, G.M.; Nordstrom, E.K.; Douagi, I.; Werner, K.M.; Liljestrom, P.; Karlsson Hedestam, G.B. Early Alpha/Beta Interferon Production by Myeloid Dendritic Cells in Response to UV-Inactivated Virus Requires Viral Entry and Interferon Regulatory Factor 3 but Not MyD88. J. Virol. 2005, 79, 10376–10385. [Google Scholar] [CrossRef] [PubMed]
- Shabman, R.S.; Morrison, T.E.; Moore, C.; White, L.; Suthar, M.S.; Hueston, L.; Rulli, N.; Lidbury, B.; Ting, J.P.; Mahalingam, S.; et al. Differential Induction of Type I Interferon Responses in Myeloid Dendritic Cells by Mosquito and Mammalian-Cell-Derived Alphaviruses. J. Virol. 2007, 81, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Schilte, C.; Couderc, T.; Chretien, F.; Sourisseau, M.; Gangneux, N.; Guivel-Benhassine, F.; Kraxner, A.; Tschopp, J.; Higgs, S.; Michault, A.; et al. Type I IFN Controls Chikungunya Virus Via its Action on Nonhematopoietic Cells. J. Exp. Med. 2010, 207, 429–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanlandingham, D.L.; Hong, C.; Klingler, K.; Tsetsarkin, K.; McElroy, K.L.; Powers, A.M.; Lehane, M.J.; Higgs, S. Differential Infectivities of O’Nyong-Nyong and Chikungunya Virus Isolates in Anopheles Gambiae and Aedes Aegypti Mosquitoes. Am. J. Trop. Med. Hyg. 2005, 72, 616–621. [Google Scholar] [PubMed]
- Kumar, S.; Jaffar-Bandjee, M.C.; Giry, C.; Connen de Kerillis, L.; Merits, A.; Gasque, P.; Hoarau, J.J. Mouse Macrophage Innate Immune Response to Chikungunya Virus Infection. Virol. J. 2012, 9. [Google Scholar] [CrossRef] [PubMed]
- Issac, T.H.; Tan, E.L.; Chu, J.J. Proteomic Profiling of Chikungunya Virus-Infected Human Muscle Cells: Reveal the Role of Cytoskeleton Network in CHIKV Replication. J. Proteomics 2014, 108, 445–464. [Google Scholar] [CrossRef] [PubMed]
- Couderc, T.; Lecuit, M. Focus on Chikungunya Pathophysiology in Human and Animal Models. Microbes Infect. 2009, 11, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Noret, M.; Herrero, L.; Rulli, N.; Rolph, M.; Smith, P.N.; Li, R.W.; Roques, P.; Gras, G.; Mahalingam, S. Interleukin 6, RANKL, and Osteoprotegerin Expression by Chikungunya Virus-Infected Human Osteoblasts. J. Infect. Dis. 2012, 206, 457–459. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Foo, S.S.; Rulli, N.E.; Taylor, A.; Sheng, K.C.; Herrero, L.J.; Herring, B.L.; Lidbury, B.A.; Li, R.W.; Walsh, N.C.; et al. Arthritogenic Alphaviral Infection Perturbs Osteoblast Function and Triggers Pathologic Bone Loss. Proc. Natl. Acad. Sci. USA 2014, 111, 6040–6045. [Google Scholar] [CrossRef] [PubMed]
- Ozden, S.; Huerre, M.; Riviere, J.P.; Coffey, L.L.; Afonso, P.V.; Mouly, V.; de Monredon, J.; Roger, J.C.; El Amrani, M.; Yvin, J.L.; et al. Human Muscle Satellite Cells as Targets of Chikungunya Virus Infection. PLoS ONE 2007, 2, e527. [Google Scholar] [CrossRef] [PubMed]
- Couderc, T.; Chretien, F.; Schilte, C.; Disson, O.; Brigitte, M.; Guivel-Benhassine, F.; Touret, Y.; Barau, G.; Cayet, N.; Schuffenecker, I.; et al. A Mouse Model for Chikungunya: Young Age and Inefficient Type-I Interferon Signaling are Risk Factors for Severe Disease. PLoS Pathog. 2008, 4, e29. [Google Scholar] [CrossRef] [PubMed]
- Brighton, S.W.; Simson, I.W. A Destructive Arthropathy Following Chikungunya Virus Arthritis—A Possible Association. Clin. Rheumatol. 1984, 3, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Marimoutou, C.; Ferraro, J.; Javelle, E.; Deparis, X.; Simon, F. Chikungunya Infection: Self-Reported Rheumatic Morbidity and Impaired Quality of Life Persist 6 Years Later. Clin. Microbiol. Infect. 2015, 21, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.J.; Goyal, V.; George, E.; Thekkemuriyil, D.V.; Jayakumar, B.; Chopra, A.; Trivandrum COPCORD Study Group. Rheumatic-Musculoskeletal Pain and Disorders in a Naive Group of Individuals 15 Months Following a Chikungunya Viral Epidemic in South India: A Population Based Observational Study. Int. J. Clin. Pract. 2011, 65, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Foissac, M.; Javelle, E.; Ray, S.; Guerin, B.; Simon, F. Post-Chikungunya Rheumatoid Arthritis, Saint Martin. Emerg. Infect. Dis. 2015, 21, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Queyriaux, B.; Simon, F.; Grandadam, M.; Michel, R.; Tolou, H.; Boutin, J.P. Clinical Burden of Chikungunya Virus Infection. Lancet Infect. Dis. 2008, 8, 2–3. [Google Scholar] [CrossRef]
- Tang, B.L. The Cell Biology of Chikungunya Virus Infection. Cell. Microbiol. 2012, 14, 1354–1363. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Foo, S.S.; Sims, N.A.; Herrero, L.J.; Walsh, N.C.; Mahalingam, S. Arthritogenic Alphaviruses: New Insights into Arthritis and Bone Pathology. Trends Microbiol. 2015, 23, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Labadie, K.; Larcher, T.; Joubert, C.; Mannioui, A.; Delache, B.; Brochard, P.; Guigand, L.; Dubreil, L.; Lebon, P.; Verrier, B.; et al. Chikungunya Disease in Nonhuman Primates Involves Long-Term Viral Persistence in Macrophages. J. Clin. Investig. 2010, 120, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Malvy, D.; Ezzedine, K.; Mamani-Matsuda, M.; Autran, B.; Tolou, H.; Receveur, M.C.; Pistone, T.; Rambert, J.; Moynet, D.; Mossalayi, D. Destructive Arthritis in a Patient with Chikungunya Virus Infection with Persistent Specific IgM Antibodies. BMC Infect. Dis. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Wielanek, A.C.; Monredon, J.D.; Amrani, M.E.; Roger, J.C.; Serveaux, J.P. Guillain-Barre Syndrome Complicating a Chikungunya Virus Infection. Neurology 2007, 69, 2105–2107. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, G.; Chadda, K.; Reboux, A.H.; Martinet, O.; Gauzere, B.A. Guillain-Barre Syndrome After Chikungunya Infection. Emerg. Infect. Dis. 2009, 15, 495–496. [Google Scholar] [CrossRef] [PubMed]
- Chusri, S.; Siripaitoon, P.; Hirunpat, S.; Silpapojakul, K. Case Reports of Neuro-Chikungunya in Southern Thailand. Am. J. Trop. Med. Hyg. 2011, 85, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Grivard, P.; Le Roux, K.; Laurent, P.; Fianu, A.; Perrau, J.; Gigan, J.; Hoarau, G.; Grondin, N.; Staikowsky, F.; Favier, F.; et al. Molecular and Serological Diagnosis of Chikungunya Virus Infection. Pathol. Biol. (Paris) 2007, 55, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Wikan, N.; Kuadkitkan, A.; Jaimipuk, T.; Ubol, S.; Pulmanausahakul, R.; Auewarakul, P.; Kasinrerk, W.; Weng, W.Y.; Panyasrivanit, M.; et al. Identification of Prohibitin as a Chikungunya Virus Receptor Protein. J. Med. Virol. 2012, 84, 1757–1770. [Google Scholar] [CrossRef] [PubMed]
- Abraham, R.; Mudaliar, P.; Padmanabhan, A.; Sreekumar, E. Induction of Cytopathogenicity in Human Glioblastoma Cells by Chikungunya Virus. PLoS ONE 2013, 8, e75854. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.J.; Chu, J.J. A Polarized Cell Model for Chikungunya Virus Infection: Entry and Egress of Virus Occurs at the Apical Domain of Polarized Cells. PLoS Negl. Trop. Dis. 2014, 8, e2661. [Google Scholar] [CrossRef] [PubMed]
- Haywood, A.M. Virus Receptors: Binding, Adhesion Strengthening, and Changes in Viral Structure. J. Virol. 1994, 68, 1–5. [Google Scholar] [PubMed]
- Mercer, J.; Schelhaas, M.; Helenius, A. Virus Entry by Endocytosis. Annu. Rev. Biochem. 2010, 79, 803–833. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Cheng, R.H.; Olson, N.H.; Peterson, P.; Chase, E.; Kuhn, R.J.; Baker, T.S. Putative Receptor Binding Sites on Alphaviruses as Visualized by Cryoelectron Microscopy. Proc. Natl. Acad. Sci. USA 1995, 92, 10648–10652. [Google Scholar] [CrossRef] [PubMed]
- Ashbrook, A.W.; Burrack, K.S.; Silva, L.A.; Montgomery, S.A.; Heise, M.T.; Morrison, T.E.; Dermody, T.S. Residue 82 of the Chikungunya Virus E2 Attachment Protein Modulates Viral Dissemination and Arthritis in Mice. J. Virol. 2014, 88, 12180–12192. [Google Scholar] [CrossRef] [PubMed]
- Asnet Mary, J.; Paramasivan, R.; Tyagi, B.K.; Surender, M.; Shenbagarathai, R. Identification of Structural Motifs in the E2 Glycoprotein of Chikungunya Involved in Virus-Host Interaction. J. Biomol. Struct. Dyn. 2013, 31, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Zhang, M. Structures and Target Recognition Modes of PDZ Domains: Recurring Themes and Emerging Pictures. Biochem. J. 2013, 455, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Vasudevan, S.; Nguyen, H.; Bork, U.; Sitaraman, S.; Merlin, D. Extracellular Interaction between hCD98 and the PDZ Class II Domain of hCASK in Intestinal Epithelia. J. Membr. Biol. 2007, 215, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Moller-Tank, S.; Kondratowicz, A.S.; Davey, R.A.; Rennert, P.D.; Maury, W. Role of the Phosphatidylserine Receptor TIM-1 in Enveloped-Virus Entry. J. Virol. 2013, 87, 8327–8341. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.A.; Khomandiak, S.; Ashbrook, A.W.; Weller, R.; Heise, M.T.; Morrison, T.E.; Dermody, T.S. A Single-Amino-Acid Polymorphism in Chikungunya Virus E2 Glycoprotein Influences Glycosaminoglycan Utilization. J. Virol. 2014, 88, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Fongsaran, C.; Jirakanwisal, K.; Kuadkitkan, A.; Wikan, N.; Wintachai, P.; Thepparit, C.; Ubol, S.; Phaonakrop, N.; Roytrakul, S.; Smith, D.R. Involvement of ATP Synthase Beta Subunit in Chikungunya Virus Entry into Insect Cells. Arch. Virol. 2014, 159, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Marsh, M.; Helenius, A. Virus Entry: Open Sesame. Cell 2006, 124, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Salvador, B.; Zhou, Y.; Michault, A.; Muench, M.O.; Simmons, G. Characterization of Chikungunya Pseudotyped Viruses: Identification of Refractory Cell Lines and Demonstration of Cellular Tropism Differences Mediated by Mutations in E1 Glycoprotein. Virology 2009, 393, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, I.; Thompson, W.E.; Thomas, K. Prohibitins Role in Cellular Survival through Ras-Raf-MEK-ERK Pathway. J. Cell. Physiol. 2014, 229, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Hossen, M.N.; Kajimoto, K.; Akita, H.; Hyodo, M.; Harashima, H. Vascular-Targeted Nanotherapy for Obesity: Unexpected Passive Targeting Mechanism to Obese Fat for the Enhancement of Active Drug Delivery. J. Control. Release 2012, 163, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Kolonin, M.G.; Saha, P.K.; Chan, L.; Pasqualini, R.; Arap, W. Reversal of Obesity by Targeted Ablation of Adipose Tissue. Nat. Med. 2004, 10, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Merkwirth, C.; Langer, T. Prohibitin Function within Mitochondria: Essential Roles for Cell Proliferation and Cristae Morphogenesis. Biochim. Biophys. Acta 2009, 1793, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Thuaud, F.; Basmadjian, C.; Roytrakul, S.; Ubol, S.; Desaubry, L.; Smith, D.R. Assessment of Flavaglines as Potential Chikungunya Virus Entry Inhibitors. Microbiol. Immunol. 2015, 59, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Umetsu, S.E.; Lee, W.L.; McIntire, J.J.; Downey, L.; Sanjanwala, B.; Akbari, O.; Berry, G.J.; Nagumo, H.; Freeman, G.J.; Umetsu, D.T.; et al. TIM-1 Induces T Cell Activation and Inhibits the Development of Peripheral Tolerance. Nat. Immunol. 2005, 6, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 Ligand Galectin-9 Negatively Regulates T Helper Type 1 Immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Karisola, P.; Pena-Cruz, V.; Dorfman, D.M.; Jinushi, M.; Umetsu, S.E.; Butte, M.J.; Nagumo, H.; Chernova, I.; Zhu, B.; et al. TIM-1 and TIM-4 Glycoproteins Bind Phosphatidylserine and Mediate Uptake of Apoptotic Cells. Immunity 2007, 27, 927–940. [Google Scholar] [CrossRef] [PubMed]
- Kondratowicz, A.S.; Lennemann, N.J.; Sinn, P.L.; Davey, R.A.; Hunt, C.L.; Moller-Tank, S.; Meyerholz, D.K.; Rennert, P.; Mullins, R.F.; Brindley, M.; et al. T-Cell Immunoglobulin and Mucin Domain 1 (TIM-1) is a Receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA 2011, 108, 8426–8431. [Google Scholar] [CrossRef] [PubMed]
- Moller-Tank, S.; Albritton, L.M.; Rennert, P.D.; Maury, W. Characterizing Functional Domains for TIM-Mediated Enveloped Virus Entry. J. Virol. 2014, 88, 6702–6713. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.S.; Mancera, R.L. The Structure of Glycosaminoglycans and their Interactions with Proteins. Chem. Biol. Drug Des. 2008, 72, 455–482. [Google Scholar] [CrossRef] [PubMed]
- Klimstra, W.B.; Ryman, K.D.; Johnston, R.E. Adaptation of Sindbis Virus to BHK Cells Selects for use of Heparan Sulfate as an Attachment Receptor. J. Virol. 1998, 72, 7357–7366. [Google Scholar] [PubMed]
- Bernard, K.A.; Klimstra, W.B.; Johnston, R.E. Mutations in the E2 Glycoprotein of Venezuelan Equine Encephalitis Virus Confer Heparan Sulfate Interaction, Low Morbidity, and Rapid Clearance from Blood of Mice. Virology 2000, 276, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Gardner, C.L.; Choi-Nurvitadhi, J.; Sun, C.; Bayer, A.; Hritz, J.; Ryman, K.D.; Klimstra, W.B. Natural Variation in the Heparan Sulfate Binding Domain of the Eastern Equine Encephalitis Virus E2 Glycoprotein Alters Interactions with Cell Surfaces and Virulence in Mice. J. Virol. 2013, 87, 8582–8590. [Google Scholar] [CrossRef] [PubMed]
- Smit, J.M.; Waarts, B.L.; Kimata, K.; Klimstra, W.B.; Bittman, R.; Wilschut, J. Adaptation of Alphaviruses to Heparan Sulfate: Interaction of Sindbis and Semliki Forest Viruses with Liposomes Containing Lipid-Conjugated Heparin. J. Virol. 2002, 76, 10128–10137. [Google Scholar] [CrossRef] [PubMed]
- Bear, J.S.; Byrnes, A.P.; Griffin, D.E. Heparin-Binding and Patterns of Virulence for Two Recombinant Strains of Sindbis Virus. Virology 2006, 347, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Brault, A.C.; Powers, A.M.; Kang, W.; Weaver, S.C. Glycosaminoglycan Binding Properties of Natural Venezuelan Equine Encephalitis Virus Isolates. J. Virol. 2003, 77, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.L.; Albee, A.; Strauss, J.H.; Kuhn, R.J. An Amino Acid Substitution in the Coding Region of the E2 Glycoprotein Adapts Ross River Virus to Utilize Heparan Sulfate as an Attachment Moiety. J. Virol. 2001, 75, 6303–6309. [Google Scholar] [CrossRef] [PubMed]
- Gardner, C.L.; Burke, C.W.; Higgs, S.T.; Klimstra, W.B.; Ryman, K.D. Interferon-Alpha/Beta Deficiency Greatly Exacerbates Arthritogenic Disease in Mice Infected with Wild-Type Chikungunya Virus but Not with the Cell Culture-Adapted Live-Attenuated 181/25 Vaccine Candidate. Virology 2012, 425, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Levitt, N.H.; Ramsburg, H.H.; Hasty, S.E.; Repik, P.M.; Cole, F.E., Jr.; Lupton, H.W. Development of an Attenuated Strain of Chikungunya Virus for use in Vaccine Production. Vaccine 1986, 4, 157–162. [Google Scholar] [CrossRef]
- Gardner, C.L.; Hritz, J.; Sun, C.; Vanlandingham, D.L.; Song, T.Y.; Ghedin, E.; Higgs, S.; Klimstra, W.B.; Ryman, K.D. Deliberate Attenuation of Chikungunya Virus by Adaptation to Heparan Sulfate-Dependent Infectivity: A Model for Rational Arboviral Vaccine Design. PLoS Negl. Trop. Dis. 2014, 8, e2719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorchakov, R.; Wang, E.; Leal, G.; Forrester, N.L.; Plante, K.; Rossi, S.L.; Partidos, C.D.; Adams, A.P.; Seymour, R.L.; Weger, J.; et al. Attenuation of Chikungunya Virus Vaccine Strain 181/Clone 25 is Determined by Two Amino Acid Substitutions in the E2 Envelope Glycoprotein. J. Virol. 2012, 86, 6084–6096. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. ATP5B ATP Synthase, H+ Transporting, Mitochondrial F1 Complex, Beta Polypeptide [Homo Sapiens (Human)]; National Center for Biotechnology Information: Bethesda, MD, USA, 2015; Available online: http://www.ncbi.nlm.nih.gov/gene/506 or http://www.webcitation.org/6YE9a0jmy; (accessed on 2 May 2015). [Google Scholar]
- Fu, Y.; Hou, Y.; Fu, C.; Gu, M.; Li, C.; Kong, W.; Wang, X.; Shyy, J.Y.; Zhu, Y. A Novel Mechanism of Gamma/Delta T-Lymphocyte and Endothelial Activation by Shear Stress: The Role of Ecto-ATP Synthase Beta Chain. Circ. Res. 2011, 108, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Champagne, E.; Martinez, L.O.; Collet, X.; Barbaras, R. Ecto-F1Fo ATP Synthase/F1 ATPase: Metabolic and Immunological Functions. Curr. Opin. Lipidol. 2006, 17, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Ye, X.; Simon, S. The Integrins. Genome Biol. 2007, 8, e215. [Google Scholar] [CrossRef] [PubMed]
- Fraisier, C.; Koraka, P.; Belghazi, M.; Bakli, M.; Granjeaud, S.; Pophillat, M.; Lim, S.M.; Osterhaus, A.; Martina, B.; Camoin, L.; et al. Kinetic Analysis of Mouse Brain Proteome Alterations Following Chikungunya Virus Infection before and After Appearance of Clinical Symptoms. PLoS ONE 2014, 9, e91397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathias, P.; Wickham, T.; Moore, M.; Nemerow, G. Multiple Adenovirus Serotypes use Alpha V Integrins for Infection. J. Virol. 1994, 68, 6811–6814. [Google Scholar] [PubMed]
- Li, E.; Brown, S.L.; Stupack, D.G.; Puente, X.S.; Cheresh, D.A.; Nemerow, G.R. Integrin Alpha(V)Beta1 is an Adenovirus Coreceptor. J. Virol. 2001, 75, 5405–5409. [Google Scholar] [CrossRef] [PubMed]
- La Linn, M.; Eble, J.A.; Lubken, C.; Slade, R.W.; Heino, J.; Davies, J.; Suhrbier, A. An Arthritogenic Alphavirus Uses the alpha1beta1 Integrin Collagen Receptor. Virology 2005, 336, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, K.; Keller, M.; Bader, B.L.; Korytar, T.; Finke, S.; Ziegler, U.; Groschup, M.H. Integrins Modulate the Infection Efficiency of West Nile Virus into Cells. J. Gen. Virol. 2013, 94, 1723–1733. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Minegishi, H. HSP60 as a Drug Target. Curr. Pharm. Des. 2013, 19, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Soltys, B.J.; Gupta, R.S. Cell Surface Localization of the 60 kDa Heat Shock Chaperonin Protein (hsp60) in Mammalian Cells. Cell Biol. Int. 1997, 21, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Belles, C.; Kuhl, A.; Nosheny, R.; Carding, S.R. Plasma Membrane Expression of Heat Shock Protein 60 in vivo in Response to Infection. Infect. Immun. 1999, 67, 4191–4200. [Google Scholar] [PubMed]
- Apte-Deshpande, A.D.; Paingankar, M.S.; Gokhale, M.D.; Deobagkar, D.N. Serratia Odorifera Mediated Enhancement in Susceptibility of Aedes Aegypti for Chikungunya Virus. Indian J. Med. Res. 2014, 139, 762–768. [Google Scholar] [PubMed]
- Padwad, Y.S.; Mishra, K.P.; Jain, M.; Chanda, S.; Karan, D.; Ganju, L. RNA Interference Mediated Silencing of Hsp60 Gene in Human Monocytic Myeloma Cell Line U937 Revealed Decreased Dengue Virus Multiplication. Immunobiology 2009, 214, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Vancini, R.; Wang, G.; Ferreira, D.; Hernandez, R.; Brown, D.T. Alphavirus Genome Delivery Occurs Directly at the Plasma Membrane in a Time- and Temperature-Dependent Process. J. Virol. 2013, 87, 4352–4359. [Google Scholar] [CrossRef] [PubMed]
- Paredes, A.M.; Ferreira, D.; Horton, M.; Saad, A.; Tsuruta, H.; Johnston, R.; Klimstra, W.; Ryman, K.; Hernandez, R.; Chiu, W.; et al. Conformational Changes in Sindbis Virions Resulting from Exposure to Low pH and Interactions with Cells Suggest that Cell Penetration may Occur at the Cell Surface in the Absence of Membrane Fusion. Virology 2004, 324, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Benmerah, A.; Lamaze, C. Clathrin-Coated Pits: Vive La Difference? Traffic 2007, 8, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Kirchhausen, T.; Owen, D.; Harrison, S.C. Molecular Structure, Function, and Dynamics of Clathrin-Mediated Membrane Traffic. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Colpitts, T.M.; Moore, A.C.; Kolokoltsov, A.A.; Davey, R.A. Venezuelan Equine Encephalitis Virus Infection of Mosquito Cells Requires Acidification as Well as Mosquito Homologs of the Endocytic Proteins Rab5 and Rab7. Virology 2007, 369, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Marsh, M.; Bolzau, E.; Helenius, A. Penetration of Semliki Forest Virus from Acidic Prelysosomal Vacuoles. Cell 1983, 32, 931–940. [Google Scholar] [CrossRef]
- Kolokoltsov, A.A.; Fleming, E.H.; Davey, R.A. Venezuelan Equine Encephalitis Virus Entry Mechanism Requires Late Endosome Formation and Resists Cell Membrane Cholesterol Depletion. Virology 2006, 347, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Helenius, A. Virus Entry at a Glance. J. Cell. Sci. 2013, 126, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Gold, E.S.; Underhill, D.M.; Morrissette, N.S.; Guo, J.; McNiven, M.A.; Aderem, A. Dynamin 2 is Required for Phagocytosis in Macrophages. J. Exp. Med. 1999, 190, 1849–1856. [Google Scholar] [CrossRef] [PubMed]
- Bernard, E.; Solignat, M.; Gay, B.; Chazal, N.; Higgs, S.; Devaux, C.; Briant, L. Endocytosis of Chikungunya Virus into Mammalian Cells: Role of Clathrin and Early Endosomal Compartments. PLoS ONE 2010, 5, e11479. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Woelk, T.; Puri, C.; Maspero, E.; Tacchetti, C.; Transidico, P.; Di Fiore, P.P.; Polo, S. Clathrin-Independent Endocytosis of Ubiquitinated Cargos. Proc. Natl. Acad. Sci. USA 2005, 102, 2760–2765. [Google Scholar] [CrossRef] [PubMed]
- Ooi, Y.S.; Stiles, K.M.; Liu, C.Y.; Taylor, G.M.; Kielian, M. Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry. PLoS Pathog. 2013, 9, e1003835. [Google Scholar] [CrossRef] [PubMed]
- Hoornweg, T.E.; van Duijl-Richter, M.K.S.; Ayala Nuñez, N.V.; van Hemert, M.J.; Smit, J.M. Dynamics of Chikungunya Virus Cell Entry Unraveled by Single Virus Tracking in Living Cells. Submitted.
- Lee, R.C.; Hapuarachchi, H.C.; Chen, K.C.; Hussain, K.M.; Chen, H.; Low, S.L.; Ng, L.C.; Lin, R.; Ng, M.M.; Chu, J.J. Mosquito Cellular Factors and Functions in Mediating the Infectious Entry of Chikungunya Virus. PLoS Negl. Trop. Dis. 2013, 7, e2050. [Google Scholar] [CrossRef] [PubMed]
- The Murphy Lab. Differences in Early Endosomal pH between Cell Types. Available online: http://murphylab.web.cmu.edu/projects/endosomal-pH-references.html or http://www.webcita-tion.org/6YFhQFTB1. (accessed on 3 May 2015).
- Van Duijl-Richter, M.; Blijleven, J.; van Oijen, A.; Smit, J. Chikungunya Virus Fusion Properties Elucidated by Single-Particle and Bulk Approaches. J. Gen. Virol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Mukhopadhyay, S.; Brooks, C.L., 3rd. Residue-Level Resolution of Alphavirus Envelope Protein Interactions in pH-Dependent Fusion. Proc. Natl. Acad. Sci. USA 2015, 112, 2034–2039. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Helenius, A. pH-Dependent Fusion between the Semliki Forest Virus Membrane and Liposomes. Proc. Natl. Acad. Sci. USA 1980, 77, 3273–3277. [Google Scholar] [CrossRef] [PubMed]
- Bron, R.; Wahlberg, J.M.; Garoff, H.; Wilschut, J. Membrane Fusion of Semliki Forest Virus in a Model System: Correlation between Fusion Kinetics and Structural Changes in the Envelope Glycoprotein. EMBO J. 1993, 12, 693–701. [Google Scholar] [PubMed]
- Smit, J.M.; Bittman, R.; Wilschut, J. Low-pH-Dependent Fusion of Sindbis Virus with Receptor-Free Cholesterol- and Sphingolipid-Containing Liposomes. J. Virol. 1999, 73, 8476–8484. [Google Scholar] [PubMed]
- Wahlberg, J.M.; Boere, W.A.; Garoff, H. The Heterodimeric Association between the Membrane Proteins of Semliki Forest Virus Changes its Sensitivity to Low pH during Virus Maturation. J. Virol. 1989, 63, 4991–4997. [Google Scholar] [PubMed]
- Kampmann, T.; Mueller, D.S.; Mark, A.E.; Young, P.R.; Kobe, B. The Role of Histidine Residues in Low-pH-Mediated Viral Membrane Fusion. Structure 2006, 14, 1481–1487. [Google Scholar] [CrossRef] [PubMed]
- Fields, W.; Kielian, M. A Key Interaction between the Alphavirus Envelope Proteins Responsible for Initial Dimer Dissociation during Fusion. J. Virol. 2013, 87, 3774–3781. [Google Scholar] [CrossRef] [PubMed]
- Hammar, L.; Markarian, S.; Haag, L.; Lankinen, H.; Salmi, A.; Cheng, R.H. Prefusion Rearrangements Resulting in Fusion Peptide Exposure in Semliki Forest Virus. J. Biol. Chem. 2003, 278, 7189–7198. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, D.L.; Ahn, A.; Liao, M.; Hammar, L.; Cheng, R.H.; Kielian, M. Multistep Regulation of Membrane Insertion of the Fusion Peptide of Semliki Forest Virus. J. Virol. 2004, 78, 3312–3318. [Google Scholar] [CrossRef] [PubMed]
- Kielian, M.; Rey, F.A. Virus Membrane-Fusion Proteins: More than One Way to make a Hairpin. Nat. Rev. Microbiol. 2006, 4, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Ahn, A.; Gibbons, D.L.; Kielian, M. The Fusion Peptide of Semliki Forest Virus Associates with Sterol-Rich Membrane Domains. J. Virol. 2002, 76, 3267–3275. [Google Scholar] [CrossRef] [PubMed]
- Klimjack, M.R.; Jeffrey, S.; Kielian, M. Membrane and Protein Interactions of a Soluble Form of the Semliki Forest Virus Fusion Protein. J. Virol. 1994, 68, 6940–6946. [Google Scholar] [PubMed]
- Nieva, J.L.; Bron, R.; Corver, J.; Wilschut, J. Membrane Fusion of Semliki Forest Virus Requires Sphingolipids in the Target Membrane. EMBO J. 1994, 13, 2797–2804. [Google Scholar] [PubMed]
- Vashishtha, M.; Phalen, T.; Marquardt, M.T.; Ryu, J.S.; Ng, A.C.; Kielian, M. A Single Point Mutation Controls the Cholesterol Dependence of Semliki Forest Virus Entry and Exit. J. Cell Biol. 1998, 140, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.K.; Vashishtha, M.; Kielian, M. Biochemical Consequences of a Mutation that Controls the Cholesterol Dependence of Semliki Forest Virus Fusion. J. Virol. 2000, 74, 1623–1631. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.E.; Cassese, T.; Kielian, M. The Cholesterol Requirement for Sindbis Virus Entry and Exit and Characterization of a Spike Protein Region Involved in Cholesterol Dependence. J. Virol. 1999, 73, 4272–4278. [Google Scholar] [PubMed]
- Gay, B.; Bernard, E.; Solignat, M.; Chazal, N.; Devaux, C.; Briant, L. pH-Dependent Entry of Chikungunya Virus into Aedes Albopictus Cells. Infect. Genet. Evol. 2012, 12, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Weaver, S.C. Sequential Adaptive Mutations Enhance Efficient Vector Switching by Chikungunya Virus and its Epidemic Emergence. PLoS Pathog. 2011, 7, e1002412. [Google Scholar] [CrossRef] [PubMed]
- Kuo, S.C.; Chen, Y.J.; Wang, Y.M.; Tsui, P.Y.; Kuo, M.D.; Wu, T.Y.; Lo, S.J. Cell-Based Analysis of Chikungunya Virus E1 Protein in Membrane Fusion. J. Biomed. Sci. 2012, 19, e44. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.K.; Eng, C.H.; Kielian, M. Novel Mutations that Control the Sphingolipid and Cholesterol Dependence of the Semliki Forest Virus Fusion Protein. J. Virol. 2002, 76, 12712–12722. [Google Scholar] [CrossRef] [PubMed]
- Teissier, E.; Pecheur, E.I. Lipids as Modulators of Membrane Fusion Mediated by Viral Fusion Proteins. Eur. Biophys. J. 2007, 36, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Wahlberg, J.M.; Bron, R.; Wilschut, J.; Garoff, H. Membrane Fusion of Semliki Forest Virus Involves Homotrimers of the Fusion Protein. J. Virol. 1992, 66, 7309–7318. [Google Scholar] [PubMed]
- Zheng, Y.; Sanchez-San Martin, C.; Qin, Z.L.; Kielian, M. The Domain I-Domain III Linker Plays an Important Role in the Fusogenic Conformational Change of the Alphavirus Membrane Fusion Protein. J. Virol. 2011, 85, 6334–6342. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.L.; Zheng, Y.; Kielian, M. Role of Conserved Histidine Residues in the Low-pH Dependence of the Semliki Forest Virus Fusion Protein. J. Virol. 2009, 83, 4670–4677. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, D.L.; Vaney, M.C.; Roussel, A.; Vigouroux, A.; Reilly, B.; Lepault, J.; Kielian, M.; Rey, F.A. Conformational Change and Protein-Protein Interactions of the Fusion Protein of Semliki Forest Virus. Nature 2004, 427, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-San Martin, C.; Sosa, H.; Kielian, M. A Stable Prefusion Intermediate of the Alphavirus Fusion Protein Reveals Critical Features of Class II Membrane Fusion. Cell Host Microbe 2008, 4, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-San Martin, C.; Nanda, S.; Zheng, Y.; Fields, W.; Kielian, M. Cross-Inhibition of Chikungunya Virus Fusion and Infection by Alphavirus E1 Domain III Proteins. J. Virol. 2013, 87, 7680–7687. [Google Scholar] [CrossRef] [PubMed]
- Wengler, G.; Koschinski, A.; Wengler, G.; Repp, H. During Entry of Alphaviruses, the E1 Glycoprotein Molecules Probably Form Two Separate Populations that Generate either a Fusion Pore Or Ion-Permeable Pores. J. Gen. Virol. 2004, 85, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Chu, J.J. Chikungunya Virus: An Update on Antiviral Development and Challenges. Drug Discov. Today 2013, 18, 969–983. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Sliva, K.; von Rhein, C.; Kummerer, B.M.; Schnierle, B.S. The Green Tea Catechin, Epigallocatechin Gallate Inhibits Chikungunya Virus Infection. Antiviral Res. 2015, 113, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Colpitts, C.C.; Schang, L.M. A Small Molecule Inhibits Virion Attachment to Heparan Sulfate- or Sialic Acid-Containing Glycans. J. Virol. 2014, 88, 7806–7817. [Google Scholar] [CrossRef] [PubMed]
- Couderc, T.; Khandoudi, N.; Grandadam, M.; Visse, C.; Gangneux, N.; Bagot, S.; Prost, J.F.; Lecuit, M. Prophylaxis and Therapy for Chikungunya Virus Infection. J. Infect. Dis. 2009, 200, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Fric, J.; Bertin-Maghit, S.; Wang, C.I.; Nardin, A.; Warter, L. Use of Human Monoclonal Antibodies to Treat Chikungunya Virus Infection. J. Infect. Dis. 2013, 207, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Fong, R.H.; Banik, S.S.; Mattia, K.; Barnes, T.; Tucker, D.; Liss, N.; Lu, K.; Selvarajah, S.; Srinivasan, S.; Mabila, M.; et al. Exposure of Epitope Residues on the Outer Face of the Chikungunya Virus Envelope Trimer Determines Antibody Neutralizing Efficacy. J. Virol. 2014, 88, 14364–14379. [Google Scholar] [CrossRef] [PubMed]
- Pohjala, L.; Utt, A.; Varjak, M.; Lulla, A.; Merits, A.; Ahola, T.; Tammela, P. Inhibitors of Alphavirus Entry and Replication Identified with a Stable Chikungunya Replicon Cell Line and Virus-Based Assays. PLoS ONE 2011, 6, e28923. [Google Scholar] [CrossRef] [PubMed]
- Gastaminza, P.; Whitten-Bauer, C.; Chisari, F.V. Unbiased Probing of the Entire Hepatitis C Virus Life Cycle Identifies Clinical Compounds that Target Multiple Aspects of the Infection. Proc. Natl. Acad. Sci. USA 2010, 107, 291–296. [Google Scholar] [CrossRef] [PubMed]
- De Lamballerie, X.; Boisson, V.; Reynier, J.C.; Enault, S.; Charrel, R.N.; Flahault, A.; Roques, P.; Le Grand, R. On Chikungunya Acute Infection and Chloroquine Treatment. Vector Borne Zoonotic Dis. 2008, 8, 837–839. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Santhosh, S.R.; Tiwari, M.; Lakshmana Rao, P.V.; Parida, M. Assessment of in vitro Prophylactic and Therapeutic Efficacy of Chloroquine Against Chikungunya Virus in Vero Cells. J. Med. Virol. 2010, 82, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Delogu, I.; de Lamballerie, X. Chikungunya Disease and Chloroquine Treatment. J. Med. Virol. 2011, 83, 1058–1059. [Google Scholar] [CrossRef] [PubMed]
- Nuckols, J.T.; McAuley, A.J.; Huang, Y.J.; Horne, K.M.; Higgs, S.; Davey, R.A.; Vanlandingham, D.L. pH-Dependent Entry of Chikungunya Virus Fusion into Mosquito Cells. Virol. J. 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Kielian, M. Domain III from Class II Fusion Proteins Functions as a Dominant-Negative Inhibitor of Virus Membrane Fusion. J. Cell Biol. 2005, 171, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Delogu, I.; Pastorino, B.; Baronti, C.; Nougairede, A.; Bonnet, E.; de Lamballerie, X. In vitro Antiviral Activity of Arbidol Against Chikungunya Virus and Characteristics of a Selected Resistant Mutant. Antiviral Res. 2011, 90, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Di Mola, A.; Peduto, A.; La Gatta, A.; Delang, L.; Pastorino, B.; Neyts, J.; Leyssen, P.; de Rosa, M.; Filosa, R. Structure-Activity Relationship Study of Arbidol Derivatives as Inhibitors of Chikungunya Virus Replication. Bioorg. Med. Chem. 2014, 22, 6014–6025. [Google Scholar] [CrossRef] [PubMed]
- Villalain, J. Membranotropic Effects of Arbidol, a Broad Anti-Viral Molecule, on Phospholipid Model Membranes. J. Phys. Chem. B 2010, 114, 8544–8554. [Google Scholar] [CrossRef] [PubMed]
- Boriskin, Y.S.; Leneva, I.A.; Pecheur, E.I.; Polyak, S.J. Arbidol: A Broad-Spectrum Antiviral Compound that Blocks Viral Fusion. Curr. Med. Chem. 2008, 15, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Selvarajah, S.; Sexton, N.R.; Kahle, K.M.; Fong, R.H.; Mattia, K.A.; Gardner, J.; Lu, K.; Liss, N.M.; Salvador, B.; Tucker, D.F.; et al. A Neutralizing Monoclonal Antibody Targeting the Acid-Sensitive Region in Chikungunya Virus E2 Protects from Disease. PLoS Negl. Trop. Dis. 2013, 7, e2423. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Dowd, K.A.; Brien, J.D.; Edeling, M.A.; Gorlatov, S.; Johnson, S.; Lee, I.; Akahata, W.; Nabel, G.J.; Richter, M.K.; et al. Development of a Highly Protective Combination Monoclonal Antibody Therapy Against Chikungunya Virus. PLoS Pathog. 2013, 9, e1003312. [Google Scholar] [CrossRef] [PubMed]
- Long, K.M.; Whitmore, A.C.; Ferris, M.T.; Sempowski, G.D.; McGee, C.; Trollinger, B.; Gunn, B.; Heise, M.T. Dendritic Cell Immunoreceptor Regulates Chikungunya Virus Pathogenesis in Mice. J. Virol. 2013, 87, 5697–5706. [Google Scholar] [CrossRef] [PubMed]
- Linn, M.L.; Aaskov, J.G.; Suhrbier, A. Antibody-Dependent Enhancement and Persistence in Macrophages of an Arbovirus Associated with Arthritis. J. Gen. Virol. 1996, 77, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Ryman, K.D.; Klimstra, W.B. Closing the Gap between Viral and Noninfectious Arthritis. Proc. Natl. Acad. Sci. USA 2014, 111, 5767–5768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sheng, J.; Austin, S.K.; Hoornweg, T.E.; Smit, J.M.; Kuhn, R.J.; Diamond, M.S.; Rossmann, M.G. Structure of Acidic pH Dengue Virus Showing the Fusogenic Glycoprotein Trimers. J. Virol. 2015, 89, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Hawman, D.W.; Stoermer, K.A.; Montgomery, S.A.; Pal, P.; Oko, L.; Diamond, M.S.; Morrison, T.E. Chronic Joint Disease Caused by Persistent Chikungunya Virus Infection is Controlled by the Adaptive Immune Response. J. Virol. 2013, 87, 13878–13888. [Google Scholar] [CrossRef] [PubMed]
- U.S. National Institutes of Health. Clinical Evaluation of Anti-CHIKV Hyperimmune Intravenous Immunoglobulins (CHIKVIVG-01). Available online: https://clinicaltri-als.gov/ct2/show/NCT02230163?term=Chikungunya&rank=5 or http://www.webcita-tion.org/6YE7aebST . (accessed on 2 May 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Duijl-Richter, M.K.S.; Hoornweg, T.E.; Rodenhuis-Zybert, I.A.; Smit, J.M. Early Events in Chikungunya Virus Infection—From Virus CellBinding to Membrane Fusion. Viruses 2015, 7, 3647-3674. https://doi.org/10.3390/v7072792
Van Duijl-Richter MKS, Hoornweg TE, Rodenhuis-Zybert IA, Smit JM. Early Events in Chikungunya Virus Infection—From Virus CellBinding to Membrane Fusion. Viruses. 2015; 7(7):3647-3674. https://doi.org/10.3390/v7072792
Chicago/Turabian StyleVan Duijl-Richter, Mareike K. S., Tabitha E. Hoornweg, Izabela A. Rodenhuis-Zybert, and Jolanda M. Smit. 2015. "Early Events in Chikungunya Virus Infection—From Virus CellBinding to Membrane Fusion" Viruses 7, no. 7: 3647-3674. https://doi.org/10.3390/v7072792




