Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus Infection
2.3. Generation of the Recombinant Viruses TB40E/IE1-mCherry and TB40E/IE2-EYFP
| Name | Sequence in 5′-3′ |
|---|---|
| IE1-mCherry fw | CTGGAGGCAAGAGCACCCACCCTATGGTGACTAGAAGCAAGGCTGACCAGATGGTGAGCAAGGGCGAGGAGGAT |
| IE1-mCherry rev | TAGTGACGTGGGATCCATAACAGTAACTGATATATATATACAATAGTTTACTTGTACAGCTCGTCCATGCCGCC |
| IE2-EYFP-fw | TGAGCCTGGCCATCGAGGCAGCCATCCAGGACCTGAGGAACAAGTCTCAGATGGTGAGCAAGGGCGAGGAGCTG |
| IE2-EYFP-rev | GGGGAATCACTATGTACAAGAGTCCATGTCTCTTTCCAGTTTTTCACTTACTTGTACAGCTCGTCCATGCCGAG |
2.4. Fluorescence-Activated Cell Sorter (FACS) Analysis
2.5. Indirect Immunofluorescence Analysis and Western Blotting
2.6. Antibodies
3. Results
3.1. Analysis of the Subnuclear Localization and the Expression Pattern of the Major ND10 Proteins PML, Sp100 and hDaxx in Undifferentiated THP-1 Monocytes and THP-1 Derived Macrophages
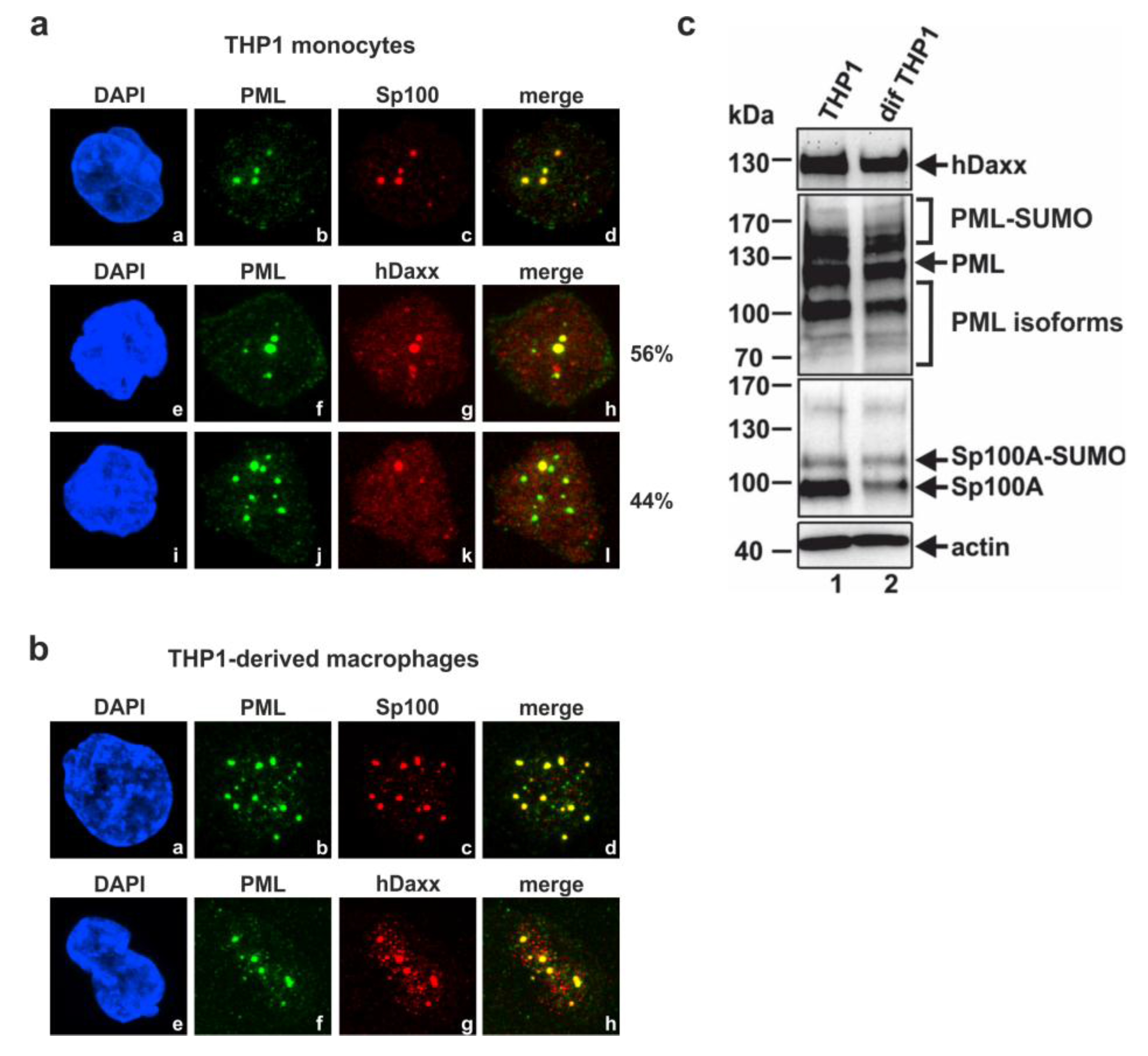
3.2. Generation of THP-1 Cells with a Stable Knockdown of PML, hDaxx or Sp100
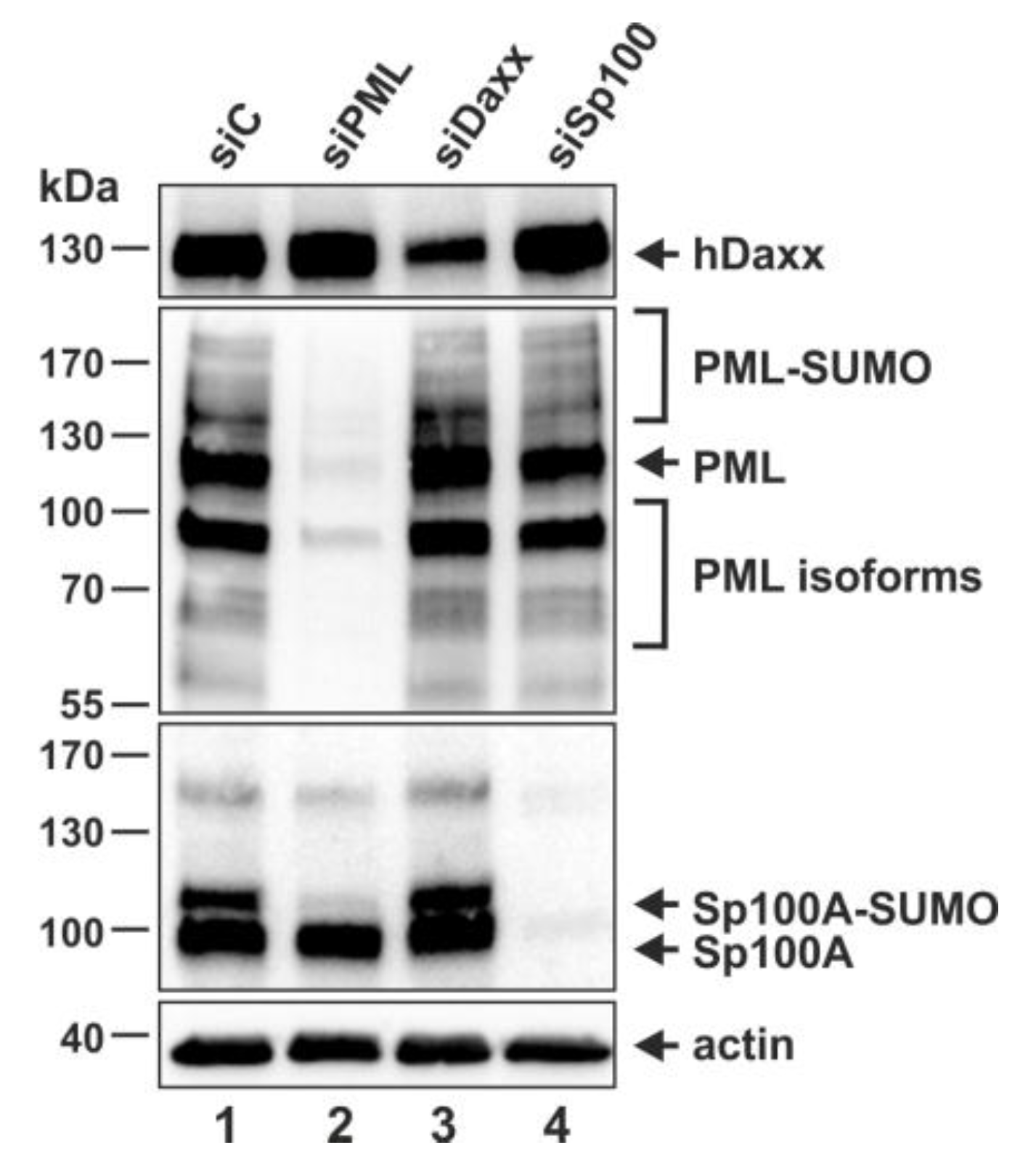
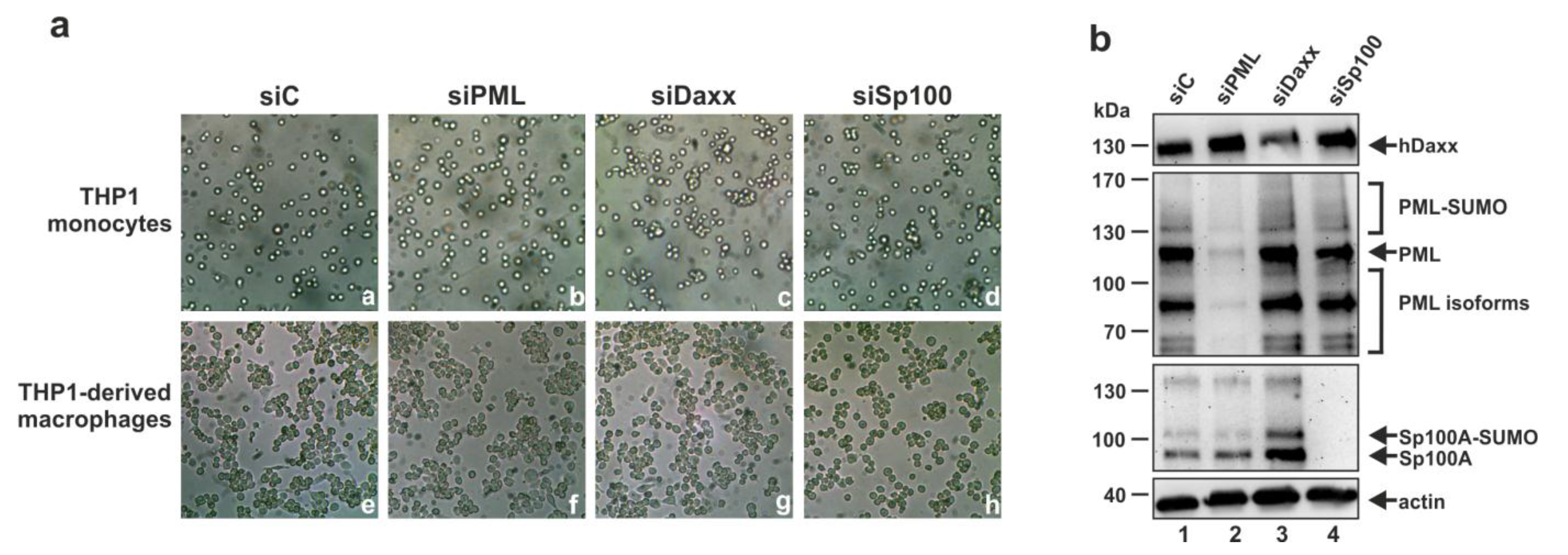
3.3. Analysis of the Role of PML, hDaxx or Sp100 for the Differentiation of THP-1 Monocytes
3.4. Construction and Characterization of Recombinant Cytomegaloviruses Expressing the Major Immediate-Early Proteins IE1 or IE2 in Fusion with Autofluorescent Proteins
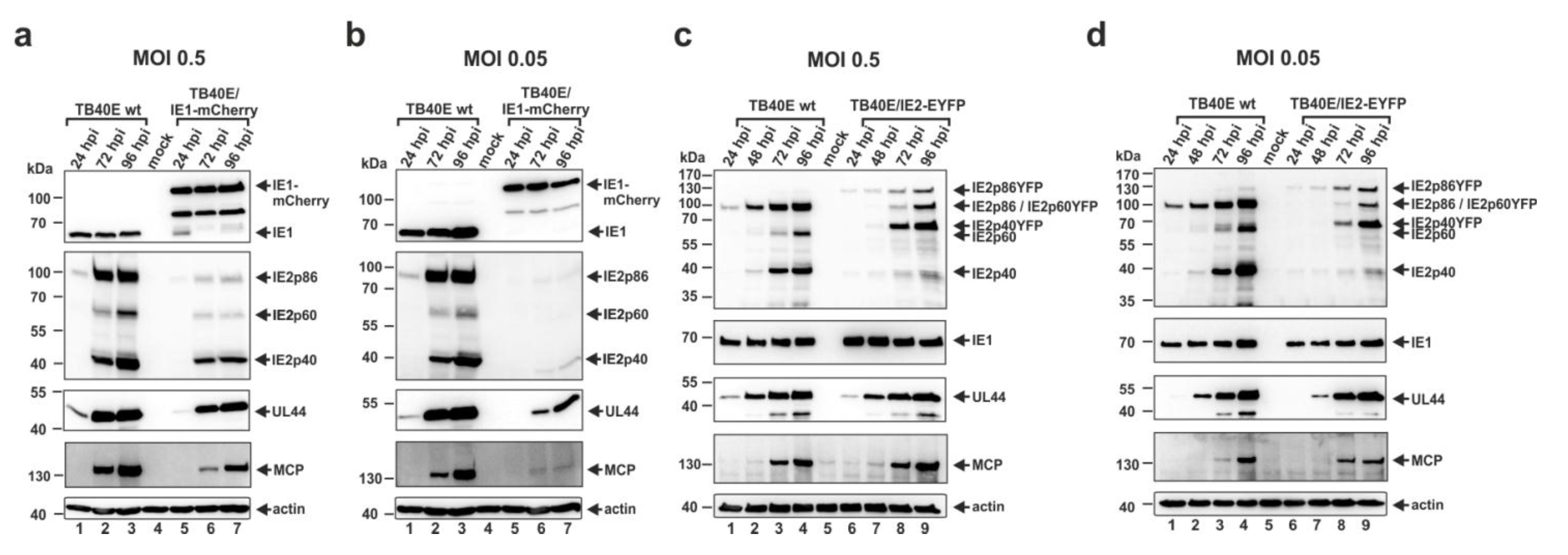
3.5. Infection of Non-Differentiated and Differentiated THP-1 Cells with Recombinant Virus TB40E/IE2-EYFP
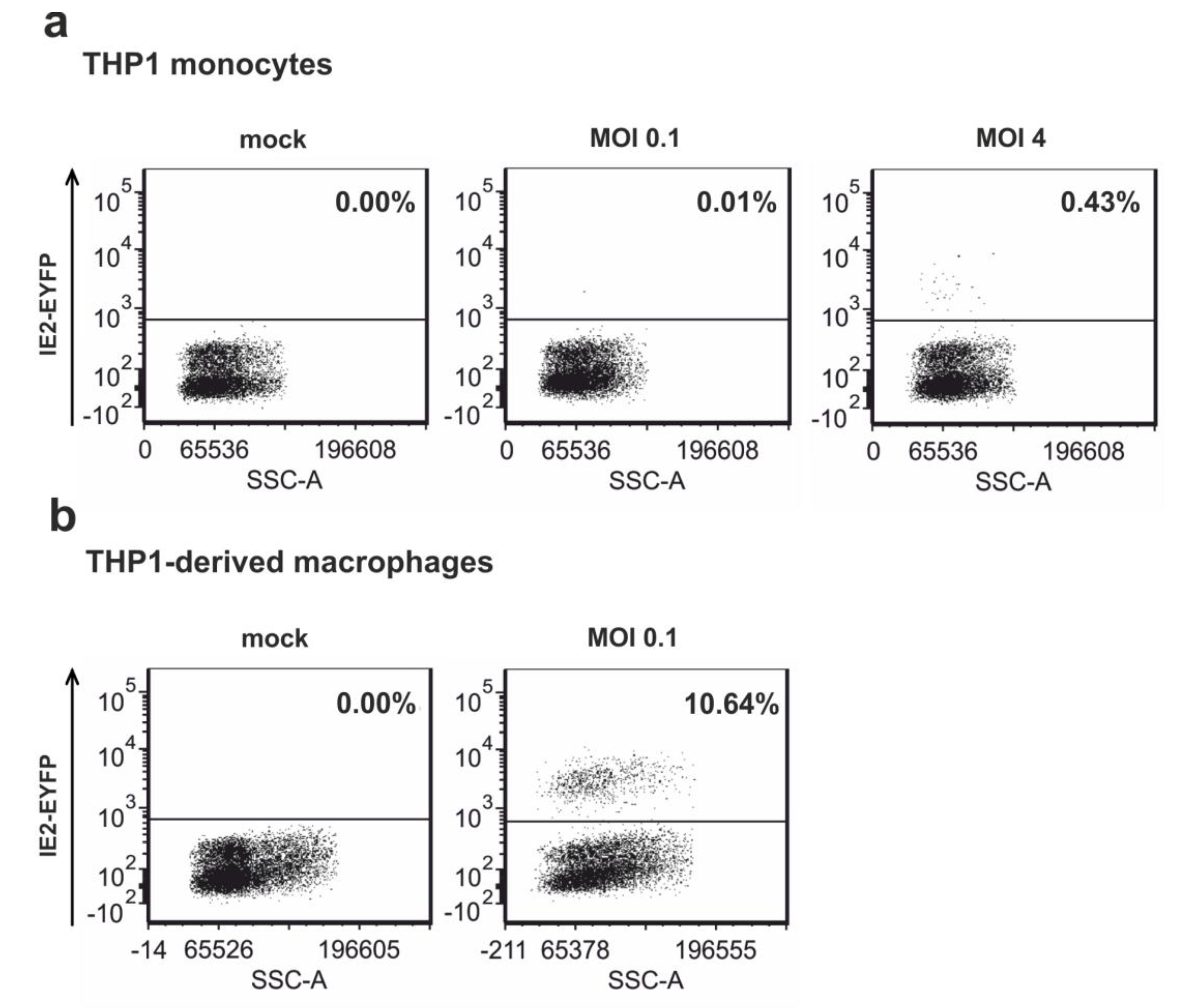
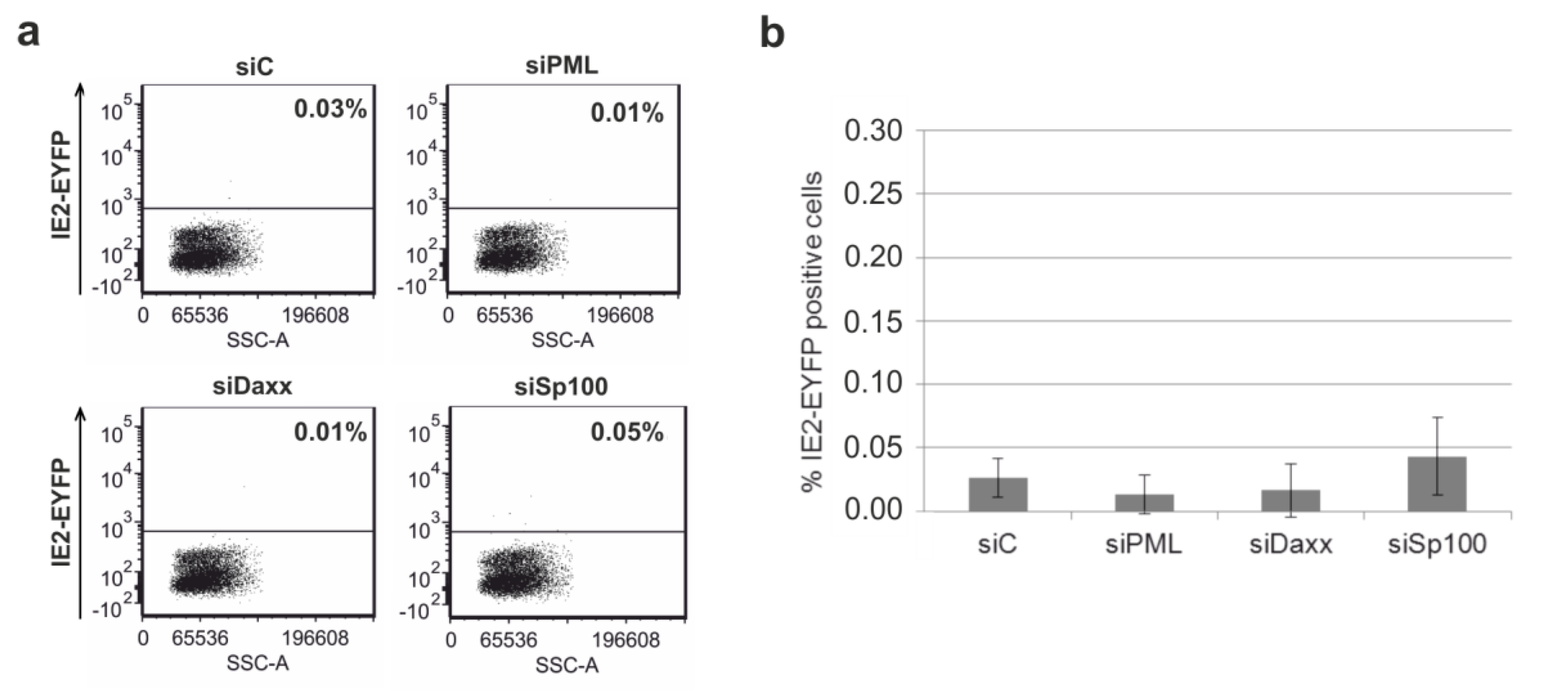
3.6. Depletion of the Major ND10 Proteins PML, hDaxx or Sp100 Does Not Affect the Establishment of HCMV Latency in Undifferentiated THP-1 Monocytes
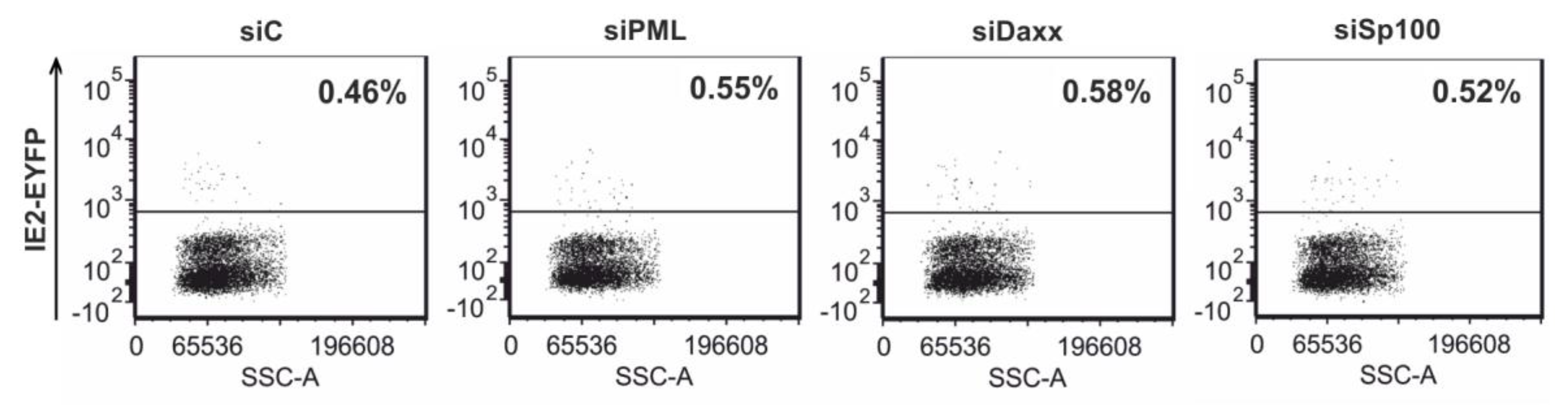
3.7. Analysis of HCMV IE Gene Expression after Differentiation of PML, hDaxx or Sp100 Knockdown Cells to THP-1 Derived Macrophages
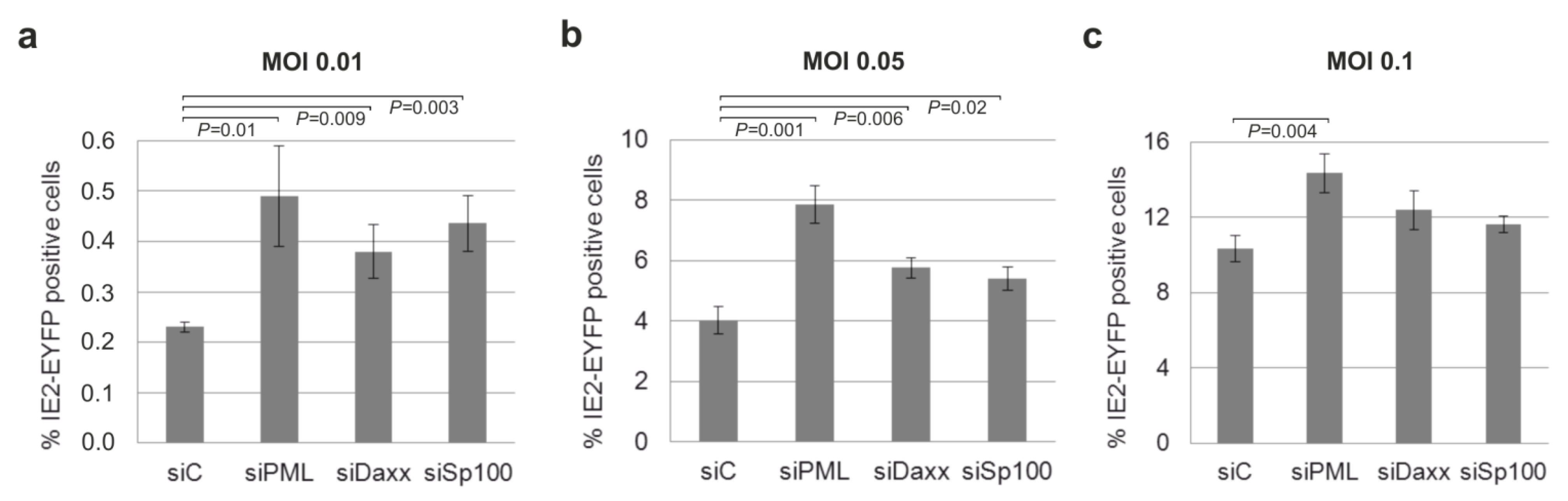
3.8. Knockdown of PML, hDaxx or Sp100 Facilitates HCMV Reactivation from Latency

4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bolovan-Fritts, C.A.; Mocarski, E.S.; Wiedeman, J.A. Peripheral blood CD14(+) cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood 1999, 93, 394–398. [Google Scholar] [PubMed]
- Khaiboullina, S.F.; Maciejewski, J.P.; Crapnell, K.; Spallone, P.A.; Dean, S.A.; Pari, G.S.; Zanjani, E.D.; Jeor, S.S. Human cytomegalovirus persists in myeloid progenitors and is passed to the myeloid progeny in a latent form. Br. J. Haematol. 2004, 126, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Sindre, H.; Tjoonnfjord, G.E.; Rollag, H.; Ranneberg-Nilsen, T.; Veiby, O.P.; Beck, S.; Degre, M.; Hestdal, K. Human cytomegalovirus suppression of and latency in early hematopoietic progenitor cells. Blood 1996, 88, 4526–4533. [Google Scholar] [PubMed]
- Sinclair, J. Human cytomegalovirus: Latency and reactivation in the myeloid lineage. J. Clin. Virol. 2008, 41, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, J.; Sissons, P. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 2006, 87, 1763–1779. [Google Scholar] [CrossRef] [PubMed]
- Mendelson, M.; Monard, S.; Sissons, P.; Sinclair, J. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 1996, 77, 3099–3102. [Google Scholar] [CrossRef] [PubMed]
- Soderberg-Naucler, C.; Fish, K.N.; Nelson, J.A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 1997, 91, 119–126. [Google Scholar] [CrossRef]
- Reeves, M.B.; Lehner, P.J.; Sissons, J.G.; Sinclair, J.H. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 2005, 86, 2949–2954. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; MacAry, P.A.; Lehner, P.J.; Sissons, J.G.; Sinclair, J.H. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 2005, 102, 4140–4145. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; Sinclair, J.H. Circulating dendritic cells isolated from healthy seropositive donors are sites of human cytomegalovirus reactivation in vivo. J. Virol. 2013, 87, 10660–10667. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M.; Cliffe, A. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 2008, 6, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Tempera, I.; Lieberman, P.M. Chromatin organization of gammaherpesvirus latent genomes. Biochim. Biophys. Acta 2010, 1799, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Stamminger, T. Interplay between Herpesvirus Infection and Host Defense by PML Nuclear Bodies. Viruses 2009, 1, 1240–1264. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D.; Chelbi-Alix, M.K. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie 2007, 89, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Woodhall, D.L.; Groves, I.J.; Reeves, M.B.; Wilkinson, G.; Sinclair, J.H. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 2006, 281, 37652–37660. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Lee, J.H.; Kim, E.T.; Shin, H.J.; Gu, S.Y.; Seol, H.S.; Ling, P.D.; Lee, C.H.; Ahn, J.H. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J. Virol. 2011, 85, 11928–11937. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.; Laukens, K.; Dang, T.H.; Van Ostade, X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 2010, 6, 51–67. [Google Scholar]
- Lallemand-Breitenbach, V.; Zhu, J.; Puvion, F.; Koken, M.; Honore, N.; Doubeikovsky, A.; Duprez, E.; Pandolfi, P.P.; Puvion, E.; Freemont, P.; de, T.H. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 2001, 193, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Ishov, A.M.; Sotnikov, A.G.; Negorev, D.; Vladimirova, O.V.; Neff, N.; Kamitani, T.; Yeh, E.T.H.; Strauss, J.F.; Maul, G.G. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999, 147, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.Y.; Huang, Y.S.; Jeng, J.C.; Kuo, H.Y.; Chang, C.C.; Chao, T.T.; Ho, C.C.; Chen, Y.C.; Lin, T.P.; Fang, H.I.; et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 2006, 24, 341–354. [Google Scholar]
- Chang, C.C.; Naik, M.T.; Huang, Y.S.; Jeng, J.C.; Liao, P.H.; Kuo, H.Y.; Ho, C.C.; Hsieh, Y.L.; Lin, C.H.; Huang, N.J.; et al. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 2011, 42, 62–74. [Google Scholar]
- Ullmann, R.; Chien, C.D.; Avantaggiati, M.L.; Muller, S. An acetylation switch regulates SUMO-dependent protein interaction networks. Mol. Cell 2012, 46, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Glass, M.; Everett, R.D. Components of Promyelocytic Leukemia Nuclear Bodies (ND10) Act Cooperatively To Repress Herpesvirus Infection. J. Virol. 2013, 87, 2174–2185. [Google Scholar] [CrossRef] [PubMed]
- Adler, M.; Tavalai, N.; Muller, R.; Stamminger, T. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J. Gen. Virol. 2011, 92, 1532–1538. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Papior, P.; Rechter, S.; Stamminger, T. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 2008, 82, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Saffert, R.T.; Kalejta, R.F. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 2006, 80, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Brignole, E.J., III; Hayward, G.S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell Biol. 1998, 18, 4899–4913. [Google Scholar]
- Lee, H.R.; Kim, D.J.; Lee, J.M.; Choi, C.Y.; Ahn, B.Y.; Hayward, G.S.; Ahn, J.H. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 2004, 78, 6527–6542. [Google Scholar] [CrossRef] [PubMed]
- Saffert, R.T.; Kalejta, R.F. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 2007, 81, 9109–9120. [Google Scholar] [CrossRef] [PubMed]
- Saffert, R.T.; Penkert, R.R.; Kalejta, R.F. Cellular and viral control over the initial events of human cytomegalovirus experimental latency in CD34+ cells. J. Virol. 2010, 84, 5594–5604. [Google Scholar] [CrossRef] [PubMed]
- Albright, E.R.; Kalejta, R.F. Myeloblastic cell lines mimic some but not all aspects of human cytomegalovirus experimental latency defined in primary CD34+ cell populations. J. Virol. 2013, 87, 9802–9812. [Google Scholar] [CrossRef] [PubMed]
- Groves, I.J.; Sinclair, J.H. Knockdown of hDaxx in normally non-permissive undifferentiated cells does not permit human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 2007, 88, 2935–2940. [Google Scholar] [CrossRef] [PubMed]
- Beisser, P.S.; Laurent, L.; Virelizier, J.L.; Michelson, S. Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J. Virol. 2001, 75, 5949–5957. [Google Scholar] [CrossRef] [PubMed]
- Ioudinkova, E.; Arcangeletti, M.C.; Rynditch, A.; De, C.F.; Motta, F.; Covan, S.; Pinardi, F.; Razin, S.V.; Chezzi, C. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 2006, 384, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Lee, G.C.; Chan, Y.J.; Chiou, C.J.; Ahn, J.H.; Hayward, G.S. Factors affecting human cytomegalovirus gene expression in human monocyte cell lines. Mol. Cells 1999, 9, 37–44. [Google Scholar] [PubMed]
- Keller, M.J.; Wu, A.W.; Andrews, J.I.; McGonagill, P.W.; Tibesar, E.E.; Meier, J.L. Reversal of human cytomegalovirus major immediate-early enhancer/promoter silencing in quiescently infected cells via the cyclic AMP signaling pathway. J. Virol. 2007, 81, 6669–6681. [Google Scholar] [CrossRef] [PubMed]
- Turtinen, L.W.; Seufzer, B.J. Selective permissiveness of TPA differentiated THP-1 myelomonocytic cells for human cytomegalovirus strains AD169 and Towne. Microb. Pathog. 1994, 16, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Yee, L.F.; Lin, P.L.; Stinski, M.F. Ectopic expression of HCMV IE72 and IE86 proteins is sufficient to induce early gene expression but not production of infectious virus in undifferentiated promonocytic THP-1 cells. Virology 2007, 363, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Jahn, G.; Knust, E.; Schmolla, H.; Sarre, T.; Nelson, J.A.; McDougall, J.K.; Fleckenstein, B. Predominant immediate-early transcripts of human cytomegalovirus AD 169. J. Virol. 1984, 49, 363–370. [Google Scholar] [PubMed]
- Lorz, K.; Hofmann, H.; Berndt, A.; Tavalai, N.; Mueller, R.; Schlotzer-Schrehardt, U.; Stamminger, T. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 2006, 80, 5423–5434. [Google Scholar] [CrossRef] [PubMed]
- Andreoni, M.; Faircloth, M.; Vugler, L.; Britt, W.J. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 1989, 23, 157–167. [Google Scholar] [CrossRef]
- Tischer, B.K.; Smith, G.A.; Osterrieder, N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 2010, 634, 421–430. [Google Scholar] [PubMed]
- Berndt, A.; Hofmann-Winkler, H.; Tavalai, N.; Hahn, G.; Stamminger, T. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection. J. Virol. 2009, 83, 12881–12894. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Floss, S.; Stamminger, T. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 2000, 74, 2510–2524. [Google Scholar] [CrossRef] [PubMed]
- Waldo, F.B.; Britt, W.J.; Tomana, M.; Julian, B.A.; Mestecky, J. Non-specific mesangial staining with antibodies against cytomegalovirus in immunoglobulin-A nephropathy. Lancet 1989, 1, 129–131. [Google Scholar] [CrossRef]
- Everett, R.D.; Rechter, S.; Papior, P.; Tavalai, N.; Stamminger, T.; Orr, A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 2006, 80, 7995–8005. [Google Scholar] [CrossRef] [PubMed]
- Khalfin-Rabinovich, Y.; Weinstein, A.; Levi, B.Z. PML is a key component for the differentiation of myeloid progenitor cells to macrophages. Int. Immunol. 2011, 23, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Sinzger, C.; Hahn, G.; Digel, M.; Katona, R.; Sampaio, K.L.; Messerle, M.; Hengel, H.; Koszinowski, U.; Brune, W.; Adler, B. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 2008, 89, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, R.M.; Thomsen, D.R.; Stinski, M.F. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 1984, 49, 190–199. [Google Scholar] [PubMed]
- Stenberg, R.M.; Witte, P.R.; Stinski, M.F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J. Virol. 1985, 56, 665–675. [Google Scholar] [PubMed]
- Tavalai, N.; Papior, P.; Rechter, S.; Leis, M.; Stamminger, T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 2006, 80, 8006–8018. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Zegerman, P.; Partridge, J.F.; Miska, E.A.; Thomas, J.O.; Allshire, R.C.; Kouzarides, T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001, 410, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.C.; Fischle, W.; Verdin, E.; Sinclair, J.H. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 2002, 21, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.; Sissons, P.; Sinclair, J. Reactivation of human cytomegalovirus in dendritic cells. Discov. Med. 2005, 5, 170–174. [Google Scholar] [PubMed]
- Rossetto, C.C.; Tarrant-Elorza, M.; Pari, G.S. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013, 9, e1003366. [Google Scholar] [CrossRef] [PubMed]
- Catez, F.; Picard, C.; Held, K.; Gross, S.; Rousseau, A.; Theil, D.; Sawtell, N.; Labetoulle, M.; Lomonte, P. HSV-1 genome subnuclear positioning and associations with host-cell PML-NBs and centromeres regulate LAT locus transcription during latency in neurons. PLoS Pathog. 2012, 8, e1002852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lusic, M.; Marini, B.; Ali, H.; Lucic, B.; Luzzati, R.; Giacca, M. Proximity to PML nuclear bodies regulates HIV-1 latency in CD4+ T cells. Cell Host Microbe 2013, 13, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Weinshenker, B.G.; Wilton, S.; Rice, G.P. Phorbol ester-induced differentiation permits productive human cytomegalovirus infection in a monocytic cell line. J. Immunol. 1988, 140, 1625–1631. [Google Scholar] [PubMed]
- Hsu, W.L.; Everett, R.D. Human neuron-committed teratocarcinoma NT2 cell line has abnormal ND10 structures and is poorly infected by herpes simplex virus type 1. J. Virol. 2001, 75, 3819–3831. [Google Scholar] [CrossRef] [PubMed]
- Tavalai, N.; Stamminger, T. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res. 2011, 157, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Dag, F.; Dolken, L.; Holzki, J.; Drabig, A.; Weingartner, A.; Schwerk, J.; Lienenklaus, S.; Conte, I.; Geffers, R.; Davenport, C.; et al. Reversible silencing of cytomegalovirus genomes by type I interferon governs virus latency. PLoS Pathog. 2014, 10, e1003962. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagenknecht, N.; Reuter, N.; Scherer, M.; Reichel, A.; Müller, R.; Stamminger, T. Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1. Viruses 2015, 7, 2884-2907. https://doi.org/10.3390/v7062751
Wagenknecht N, Reuter N, Scherer M, Reichel A, Müller R, Stamminger T. Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1. Viruses. 2015; 7(6):2884-2907. https://doi.org/10.3390/v7062751
Chicago/Turabian StyleWagenknecht, Nadine, Nina Reuter, Myriam Scherer, Anna Reichel, Regina Müller, and Thomas Stamminger. 2015. "Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1" Viruses 7, no. 6: 2884-2907. https://doi.org/10.3390/v7062751





