Profiling of Measles-Specific Humoral Immunity in Individuals Following Two Doses of MMR Vaccine Using Proteome Microarrays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Proteome Microarray
2.3. Plaque Reduction Microneutralization Assay (PRMN)
2.4. Measles-Specific IFNγ ELISPOT Assay
2.5. Measles-Specific Secreted Cytokines
2.6. Statistical Analyses
3. Results
3.1. Characterization of the Study Cohort
| Overall (N = 150) | High Ab Responders (N = 75) | Low Ab Responders (N = 75) | p-Value b | |
|---|---|---|---|---|
| Median age at enrollment, years (IQR a) | 15.5 (13.0–17.0) | 15.0 (13.0–17.0) | 16.0 (14.0–17.0) | 0.28 |
| Median age at first measles immunization, months (IQR) | 15.0 (15.0–18.0) | 15.0 (15.0–18.0) | 15.0 (15.0–21.0) | 0.89 |
| Median age at second measles immunization, years (IQR) | 6.0 (5.0–12.0) | 5.0 (4.0–12.0) | 7.0 (5.0–11.0) | 0.75 |
| Median time from second measles immunization to enrollment, years, (IQR) | 7.2 (5.2–9.5) | 7.3 (5.2–9.1) | 7.1 (4.9–10.5) | 0.61 |
| Gender, N(%) | ||||
| Male | 84 (56.0%) | 42 (56.0%) | 42 (56.0%) | N/A |
| Female | 66 (44.0%) | 33 (44.0%) | 33 (44.0%) | |
| Race, N(%) | 0.32 | |||
| White | 110 (73.3%) | 57 (76.0%) | 53 (70.7%) | |
| African-Americans | 31 (20.7%) | 13 (17.3%) | 18 (24.0%) | |
| Other | 9 (6.0%) | 5 (6.7%) | 4 (5.3%) | |
| Ethnicity, N(%) | 0.04 | |||
| Not Hispanic or Latino | 146 (97.3%) | 71 (94.7%) | 75 (100.0%) | |
| Hispanic or Latino | 2 (1.3%) | 2 (2.7%) | 0 (0.0%) | |
| Don’t Know/Other | 2 (1.3%) | 2 (2.7%) | 0 (0.0%) | |
| IFNα c (IQR, pg/mL) | 547 (275–912) | 512 (235–839) | 589 (374–945) | 0.07 |
| IFNγ c (IQR, pg/mL) | 62 (31–123) | 64 (34–150) | 60 (26–95) | 0.20 |
| IFNλ1 c (IQR, pg/mL) | 32 (12–65) | 29 (5–65) | 36 (17–64) | 0.18 |
| IL-10 c (IQR, pg/mL) | 19 (11–28) | 20 (11–29) | 19 (11–24) | 0.48 |
| IL-2 c (IQR, pg/mL) | 35 (18–59) | 42 (26–73) | 32 (17–54) | 0.08 |
| IL-6 c (IQR, pg/mL) | 359 (263–472) | 335 (253–422) | 376 (286–515) | 0.07 |
| TNFα c (IQR, pg/mL) | 13 (8–18) | 14 (9–18) | 13 (8–20) | 0.61 |
| IFNγ ELISPOT c (IQR, SFUs per 2 × 105 cells) | 32 (12–62) | 30 (16–50) | 38 (10–72) | 0.72 |
| PRMN antibody titer c (IQR, mIU/mL) | 1546 (168–3730) | 3730 (3114–4333) | 168 (115–191) | <0.0001 |
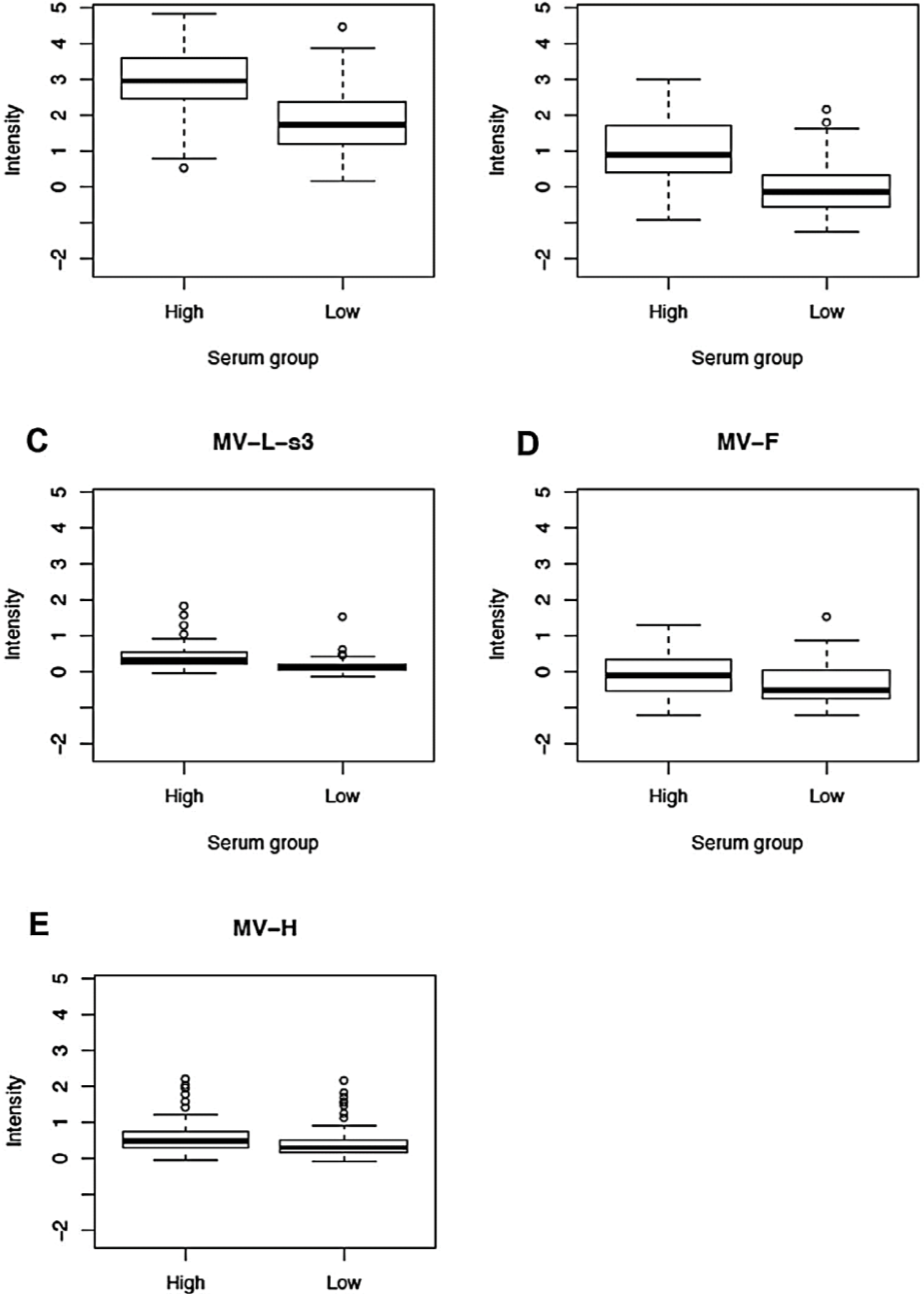
3.2. Proteomic Profiling of Measles-Specific Antibody Responses (Measures/Summaries for All MV Proteins)
| Immune Measure | Response Category (PRMN) | Median (IQR) a | Median Difference b | p-Value c |
|---|---|---|---|---|
| Neut. antibody mIU/mL | Lowest | 168 (115–191) | ||
| Highest | 3730 (3114–4333) | 3562 | N/A d | |
| Whole Cohort | 1546 (168–3730) | |||
| Anti-H microarray reactivity | Lowest | 0.30 (0.16, 0.49) | ||
| Highest | 0.48 (0.3, 0.74) | 0.18 | 0.002 | |
| Whole Cohort | 0.40 (0.21, 0.67) | |||
| Anti-F microarray reactivity | Lowest | −0.51 (−0.74, 0.05) | ||
| Highest | −0.09 (−0.53, 0.35) | 0.42 | 9E-04 | |
| Whole Cohort | −0.30 (−0.67, 0.15) | |||
| Anti-N microarray reactivity | Lowest | −0.13 (−0.52, 0.36) | ||
| Highest | 0.91 (0.42, 1.7) | 1.04 | 2.7E-11 | |
| Whole Cohort | 0.41 (−0.18, 1.11) | |||
| Anti-P microarray reactivity | Lowest | 1.74 (1.22, 2.37) | ||
| Highest | 2.96 (2.44, 3.57) | 1.23 | 1.5E-11 | |
| Whole Cohort | 2.41 (1.53, 3.19) | |||
| Anti-V microarray reactivity | Lowest | −0.22 (−0.67, 0.57) | ||
| Highest | −0.21 (−0.71, 0.38) | 0.01 | 0.62 | |
| Whole Cohort | −0.21 (−0.7, 0.46) | |||
| Anti-C microarray reactivity e | Lowest | −1.20 (−1.48, −0.92) | ||
| Highest | −1.25 (−1.52, −0.98) | −0.05 | 0.77 | |
| Whole Cohort | −1.24 (−1.5, −0.93) | |||
| Anti-M microarray reactivity e | Lowest | −0.79 (−0.97, −0.57) | ||
| Highest | −0.80 (−0.93, −0.57) | −0.008 | 0.95 | |
| Whole Cohort | −0.80 (−0.97, −0.57) | |||
| Anti-L-s1 microarray reactivity | Lowest | −0.34 (−0.67, 0.08) | ||
| Highest | −0.44 (−0.82, 0.05) | −0.10 | 0.49 | |
| Whole Cohort | −0.39 (−0.72, 0.07) | |||
| Anti-L-s2 microarray reactivity e | Lowest | −0.92 (−1.1, −0.66) | ||
| Highest | −0.95 (−1.12, −0.49) | −0.02 | 0.92 | |
| Whole Cohort | −0.94 (−1.12, −0.54) | |||
| Anti-L-s3 microarray reactivity | Lowest | 0.14 (0.06, 0.22) | ||
| Highest | 0.33 (0.2, 0.53) | 0.19 | 6.6E-09 | |
| Whole Cohort | 0.22 (0.11, 0.39) | |||
| Anti-L-s4 microarray reactivity | Lowest | −0.29 (−0.52, 0.22) | ||
| Highest | −0.33 (−0.57, 0.22) | −0.03 | 0.79 | |
| Whole Cohort | −0.32 (−0.55, 0.23) |
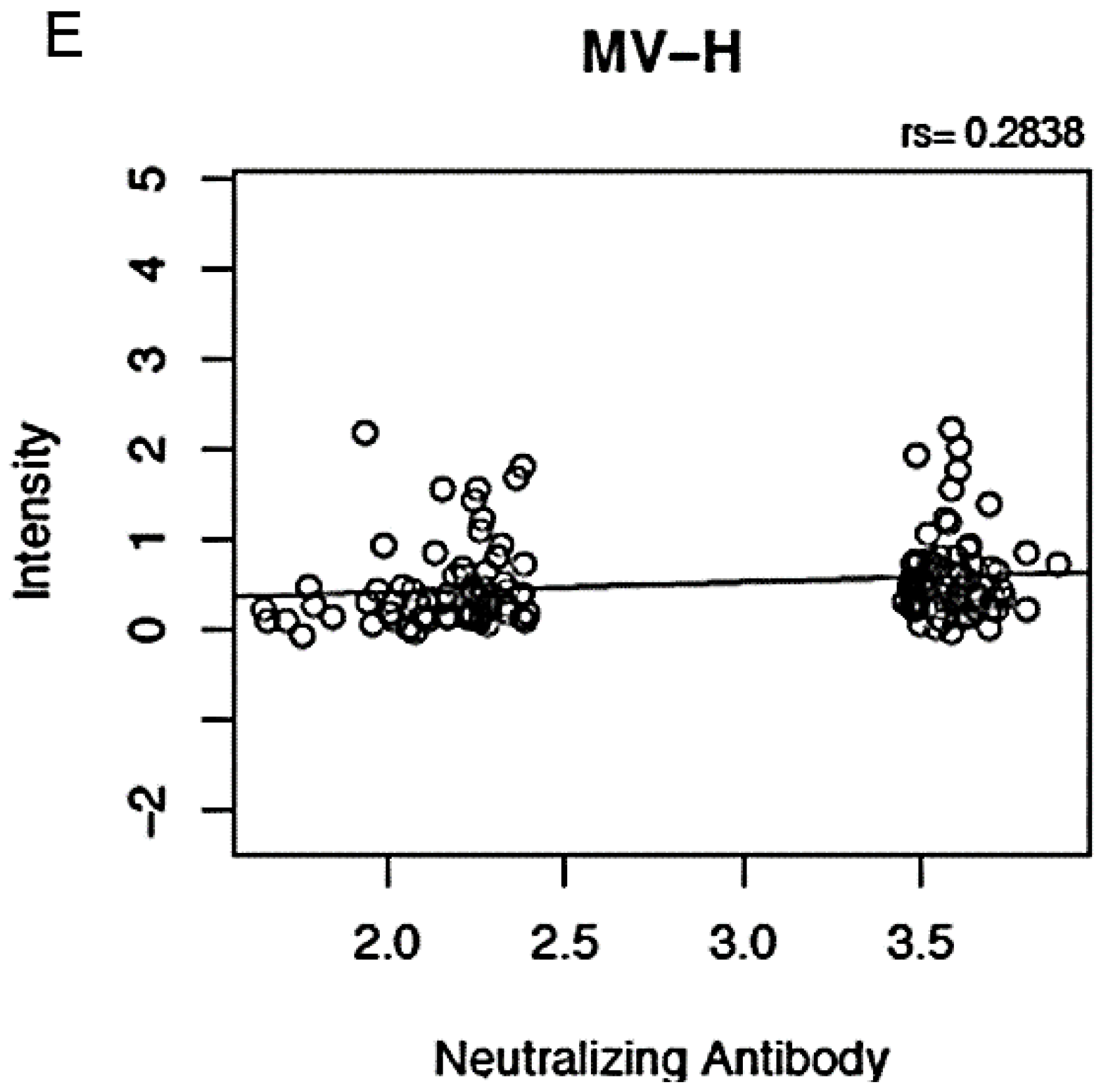
3.3. Correlations between Proteome Microarray Antibody Reactivities and Other MV Immune Response Outcomes

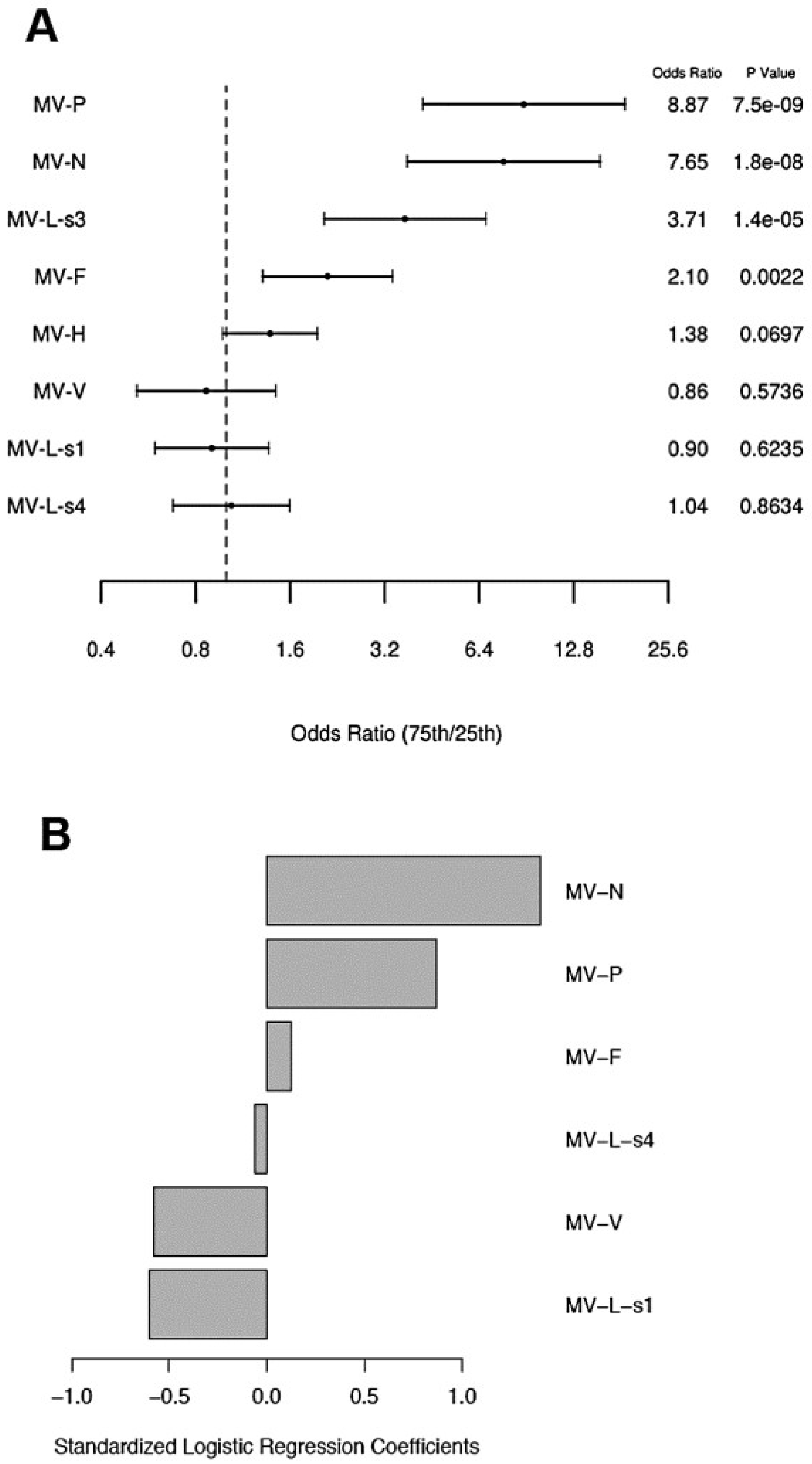
3.4. Proteomic Modeling of Antibody Responses after Measles Vaccination
4. Discussion
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Perry, R.T.; Gacic-Dobo, M.; Dabbagh, A.; Mulders, M.N.; Strebel, P.M.; Okwo-Bele, J.M.; Rota, P.A.; Goodson, J.L. Global control and regional elimination of measles, 2000–2012. Morb. Mortal. Wkly. Rep. 2014, 63, 103–107. [Google Scholar]
- Fields, R.; Dabbagh, A.; Jain, M.; Sagar, K.S. Moving forward with strengthening routine immunization delivery as part of measles and rubella elimination activities. Vaccine 2013, 31, B115–B121. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Jacobson, R.M. Failure to reach the goal of measles elimination. Apparent paradox of measles infections in immunized persons. Arch. Int. Med. 1994, 154, 1815–1820. [Google Scholar] [CrossRef]
- Elliman, D.; Sengupta, N. Measles. Curr. Opin. Infect. Dis. 2005, 18, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, W.A.; Herrmann, K.; Albrecht, P.; Bernier, R.; Holmgreen, P.; Bart, K.J.; Hinman, A.R. Immunity against measles and rubella in Massachusetts schoolchildren. Dev. Biol. Stand. 1986, 65, 75–83. [Google Scholar] [PubMed]
- Poland, G.A.; Jacobson, R.M.; Schaid, D.J.; Moore, S.B.; Jacobsen, S.J. The association between HLA class I alleles and measles vaccine-induced antibody response: Evidence of a significant association. Vaccine 1998, 16, 1869–1871. [Google Scholar] [CrossRef] [PubMed]
- Sheppeard, V.; Forssman, B.; Ferson, M.J.; Moreira, C.; Campbell-Lloyd, S.; Dwyer, D.E.; McAnulty, J.M. Vaccine failures and vaccine effectiveness in children during measles outbreaks in New South Wales, March-May 2006. Commun. Dis. Intel. 2009, 33, 21–26. [Google Scholar]
- Zipprich, J.; Winter, K.; Hacker, J.; Xia, D.; Watt, J.; Harriman, K. Measles outbreak-California, December 2014–February 2015. Morb. Mortal. Wkly. Rep. 2015, 64, 153–154. [Google Scholar]
- He, H.; Chen, E.F.; Li, Q.; Wang, Z.; Yan, R.; Fu, J.; Pan, J. Waning immunity to measles in young adults and booster effects of revaccination in secondary school students. Vaccine 2013, 31, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Santisteve, P.; Lopalco, P.L. Measles still spreads in Europe: Who is responsible for the failure to vaccinate? Clin. Microbiol. Infect. 2012, 18, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Jacobson, R.M. The re-emergence of measles in developed countries: Time to develop the next-generation measles vaccines? Vaccine 2012, 30, 103–104. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, J.A.; Poland, G.A. Measles and mumps outbreaks in the United States: solid think globally, vaccinate locally. Vaccine 2014, 32, 4703–4704. [Google Scholar] [CrossRef] [PubMed]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Pankratz, V.S.; Kennedy, R.B.; Jacobson, R.M.; Poland, G.A. The genetic basis for interindividual immune response variation to measles vaccine: New understanding and new vaccine approaches. Exp. Rev. Vaccines 2013, 12, 57–70. [Google Scholar] [CrossRef]
- Rosen, J.B.; Rota, J.S.; Hickman, C.J.; Sowers, S.B.; Mercader, S.; Rota, P.A.; Bellini, W.J.; Huang, A.J.; Doll, M.K.; Zucker, J.R.; et al. Outbreak of measles among persons with prior evidence of immunity, New York City, 2011. Clin. Infect. Dis. 2014, 58, 1205–1210. [Google Scholar] [CrossRef]
- De Serres, G.; Boulianne, N.; Defay, F.; Brousseau, N.; Benoit, M.; Lacoursiere, S.; Guillemette, F.; Soto, J.; Ouakki, M.; Ward, B.J.; et al. Higher risk of measles when the first dose of a 2-dose schedule of measles vaccine is given at 12–14 months versus 15 months of age. Clin. Infect. Dis. 2012, 55, 394–402. [Google Scholar] [CrossRef]
- Haralambieva, I.H.; Ovsyannikova, I.G.; O’Byrne, M.; Pankratz, V.S.; Jacobson, R.M.; Poland, G.A. A large observational study to concurrently assess persistence of measles specific B-cell and T-cell immunity in individuals following two doses of MMR vaccine. Vaccine 2011, 29, 4485–4491. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Jacobson, R.M.; Thampy, A.M.; Colbourne, S.A.; Wollan, P.C.; Lipsky, J.J.; Jacobson, S.J. Measles re-immunization in children seronegative after initial immunization. JAMA 1997, 277, 1156–1158. [Google Scholar] [CrossRef] [PubMed]
- Paunio, M.; Peltola, H.; Valle, M.; Davidkin, I.; Virtanen, M.; Heinonen, O.P. Explosive school-based measles outbreak: Intense exposure may have resulted in high risk, even among revaccinees. Am. J. Epidemiol. 1998, 148, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Pannuti, C.S.; Morello, R.J.; Moraes, J.C.; Curti, S.P.; Afonso, A.M.S.; Camargo, M.C.; Souza, V.A. Identification of primary and secondary measles vaccine failures by measurement of immunoglobulin G avidity in measles cases during the 1997 Sao Paulo epidemic. Clin. Diagn. Lab. Immunol. 2004, 11, 119–122. [Google Scholar] [PubMed]
- Hickman, C.J.; Hyde, T.B.; Sowers, S.B.; Mercader, S.; McGrew, M.; Williams, N.J.; Beeler, J.A.; Audet, S.; Kiehl, B.; Nandy, R.; et al. Laboratory characterization of measles virus infection in previously vaccinated and unvaccinated individuals. J. Infect. Dis. 2011, 204, S549–S558. [Google Scholar] [CrossRef]
- Glass, K.; Grenfell, B.T. Waning immunity and subclinical measles infections in England. Vaccine 2004, 22, 4110–4116. [Google Scholar] [CrossRef] [PubMed]
- Mossong, J.; Nokes, D.J.; Edmunds, W.J.; Cox, M.J.; Ratnam, S.; Muller, C.P. Modeling the impact of subclinical measles transmission in vaccinated populations with waning immunity. Am. J. Epidemiol. 1999, 150, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.; Burgess, C.; Garrison, L.P., Jr.; Bauch, C.; Babigumira, J.; Simons, E.; Dabbagh, A. Global eradication of measles: An epidemiologic and economic evaluation. J. Infect. Dis. 2011, 204, S98–S106. [Google Scholar] [CrossRef] [PubMed]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Vierkant, R.A.; Poland, G.A. Development of a novel efficient fluorescence-based plaque reduction microneutralization assay for measles immunity. Clin. Vaccine Immunol. 2008, 15, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Wild, T.F.; Malvoisin, E.; Buckland, R. Measles virus: Both the haemagglutinin and fusion glycoproteins are required for fusion. J. Gen. Virol. 1991, 72, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, R.; Rose, J.K. Cell fusion by the envelope glycoproteins of persistent measles viruses which caused lethal human brain disease. J. Virol. 1993, 67, 1493–1502. [Google Scholar] [PubMed]
- Navaratnarajah, C.K.; Leonard, V.H.; Cattaneo, R. Measles virus glycoprotein complex assembly, receptor attachment, and cell entry. Curr. Top. Microbiol. Immunol. 2009, 329, 59–76. [Google Scholar] [PubMed]
- De Swart, R.L.; Yuksel, S.; Osterhaus, A.D. Relative contributions of measles virus hemagglutinin- and fusion protein-specific serum antibodies to virus neutralization. J. Virol. 2005, 79, 11547–11551. [Google Scholar] [CrossRef] [PubMed]
- De Swart, R.L.; Yuksel, S.; Langerijs, C.N.; Muller, C.P.; Osterhaus, A.D. Depletion of measles virus glycoprotein-specific antibodies from human sera reveals genotype-specific neutralizing antibodies. J. Gen. Virol. 2009, 90, 2982–2989. [Google Scholar]
- Patterson, J.B.; Thomas, D.; Lewicki, H.; Billeter, M.A.; Oldstone, M.B. V and C proteins of measles virus function as virulence factors in vivo. Virology 2000, 267, 80–89. [Google Scholar] [CrossRef]
- McAllister, C.S.; Toth, A.M.; Zhang, P.; Devaux, P.; Cattaneo, R.; Samuel, C.E. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J. Virol. 2010, 84, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Devaux, P.; Hodge, G.; McChesney, M.B.; Cattaneo, R. Attenuation of V- or C-defective measles viruses: Infection control by the inflammatory and interferon responses of rhesus monkeys. J. Virol. 2008, 82, 5359–5367. [Google Scholar] [CrossRef] [PubMed]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Dhiman, N.; Vierkant, R.A.; Jacobson, R.M.; Poland, G.A. Differential cellular immune responses to wild-type and attenuated edmonston tag measles virus strains are primarily defined by the viral phosphoprotein gene. J. Med. Virol. 2010, 82, 1966–1975. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Kadota, S.I.; Takeda, M.; Miyajima, N.; Nagata, K. Measles virus V protein blocks interferon (IFN)-alpha/beta but not IFN-gamma signaling by inhibiting STAT1 and STAT2 phosphorylation. FEBS Lett. 2003, 545, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Tober, C.; Seufert, M.; Schneider, H.; Billeter, M.A.; Johnston, I.C.; Niewiesk, S.; Ter, M.V.; Schneider-Schaulies, S. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 1998, 72, 8124–8132. [Google Scholar] [PubMed]
- Davies, D.H.; Liang, X.; Hernandez, J.E.; Randall, A.; Hirst, S.; Mu, Y.; Romero, K.M.; Nguyen, T.T.; Kalantari-Dehaghi, M.; Crotty, S.; et al. Profiling the humoral immune response to infection by using proteome microarrays: High-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. USA 2005, 102, 547–552. [Google Scholar] [CrossRef]
- Luevano, M.; Bernard, H.U.; Barrera-Saldana, H.A.; Trevino, V.; Garcia-Carranca, A.; Villa, L.L.; Monk, B.J.; Tan, X.; Davies, D.H.; Felgner, P.L.; et al. High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays. Virology 2010, 405, 31–40. [Google Scholar] [CrossRef]
- Kalantari-Dehaghi, M.; Chun, S.; Chentoufi, A.A.; Pablo, J.; Liang, L.; Dasgupta, G.; Molina, D.M.; Jasinskas, A.; Nakajima-Sasaki, R.; Felgner, J.; et al. Discovery of potential diagnostic and vaccine antigens in herpes simplex virus 1 and 2 by proteome-wide antibody profiling. J. Virol. 2012, 86, 4328–4339. [Google Scholar] [CrossRef]
- Umlauf, B.J.; Haralambieva, I.H.; Ovsyannikova, I.G.; Kennedy, R.B.; Pankratz, V.S.; Jacobson, R.M.; Poland, G.A. Associations between demographic variables and multiple measles-specific innate and cell-mediated immune responses after measles vaccination. Viral. Immunol. 2012, 25, 29–36. [Google Scholar] [PubMed]
- Kennedy, R.B.; Ovsyannikova, I.G.; Haralambieva, I.H.; O’Byrne, M.M.; Jacobson, R.M.; Pankratz, V.S.; Poland, G.A. Multigenic control of measles vaccine immunity mediated by polymorphisms in measles receptor, innate pathway, and cytokine genes. Vaccine 2012, 30, 2159–2167. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.J.; Haralambieva, I.H.; Vierkant, R.A.; Ovsyannikova, I.G.; Poland, G.A. Response surface methodology to determine optimal measles-specific cytokine responses in human peripheral blood mononuclear cells. J. Immunol. Methods 2012, 382, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Vierkant, R.A.; O’Byrne, M.M.; Jacobson, R.M.; Poland, G.A. The association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses—A replication study and examination of novel polymorphisms. Hum. Hered. 2011, 72, 206–223. [Google Scholar] [CrossRef] [PubMed]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Vierkant, R.A.; O’Byrne, M.M.; Jacobson, R.M.; Poland, G.A. Effects of vitamin A and D receptor gene polymophisms/haplotypes on immune responses to measles vaccine. Pharm. Genom. 2012, 22, 20–31. [Google Scholar] [CrossRef]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Umlauf, B.J.; Vierkant, R.A.; Pankratz, S.V.; Jacobson, R.M.; Poland, G.A. Genetic polymorphisms in host antiviral genes: Associations with humoral and cellular immunity to measles vaccine. Vaccine 2011, 29, 8988–8997. [Google Scholar] [CrossRef] [PubMed]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Kennedy, R.B.; Vierkant, R.A.; Pankratz, S.V.; Jacobson, R.M.; Poland, G.A. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine 2011, 29, 7883–7895. [Google Scholar] [CrossRef] [PubMed]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Vierkant, R.A.; Pankratz, V.S.; Poland, G.A. The role of polymorphisms in toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Hum. Genet. 2011, 130, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, R.M.; Ovsyannikova, I.G.; Vierkant, R.A.; Pankratz, V.S.; Poland, G.A. Independence of measles-specific humoral and cellular immune responses to vaccination. Hum. Immunol. 2012, 73, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [PubMed]
- Team, R.D.C. R: A Language for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2009. [Google Scholar]
- Chen, R.T.; Markowitz, L.E.; Albrecht, P.; Stewart, J.A.; Mofenson, L.M.; Preblud, S.R.; Orenstein, W.A. Measles antibody: Reevaluation of protective titers. J. Infect. Dis. 1990, 162, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, P.; Herrmann, K.; Burns, G.R. Role of virus strain in conventional and enhanced measles plaque neutralization test. J. Virol. Methods 1981, 3, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Ratnam, S.; Gadag, V.; West, R.; Burris, J.; Oates, E.; Stead, F.; Bouilianne, N. Comparison of commercial enzyme immunoassay kits with plaque reduction neutralization test for detection of measles virus antibody. J. Clin. Microbiol. 1995, 33, 811–815. [Google Scholar] [PubMed]
- Cohen, B.J.; Parry, R.P.; Doblas, D.; Samuel, D.; Warrener, L.; Andrews, N.; Brown, D. Measles immunity testing: Comparison of two measles IgG ELISAs with plaque reduction neutralisation assay. J. Virol. Methods 2006, 131, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Cohen, B.J.; Audet, S.; Andrews, N.; Beeler, J. Plaque reduction neutralization test for measles antibodies: Description of a standardised laboratory method for use in immunogenicity studies of aerosol vaccination. Vaccine 2007, 26, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Cohen, B.J.; Parry, R.P.; Andrews, N.; Bennett, A.M.; Dennis, J.H. Laboratory methods for assessing vaccine potency retained in aerosol outputs from nebulizers: Application to World Health Organization measles aerosol project. Vaccine 2008, 26, 3534–3539. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.J.; Aouchiche, S.; Martel, N.; Bertley, F.M.; Bautista-Lopez, N.; Serhir, B.; Ratnam, S. Measurement of measles virus-specific neutralizing antibodies: Evaluation of the syncytium inhibition assay in comparison with the plaque reduction neutralization test. Diagn. Microbiol. Infect. Dis. 1999, 33, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, L.; Charlett, A.; Farrington, P.; Miller, E.; Forsey, T.; Morgan-Capner, P. An evaluation of nine commercial EIA kits for the detection of measles specific IgG. J. Virol. Methods 1997, 66, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Bouche, F.B.; Ertl, O.T.; Muller, C.P. Neutralizing B cell response in measles. Viral. Immunol. 2002, 15, 451–471. [Google Scholar] [CrossRef] [PubMed]
- LeBaron, C.W.; Beeler, J.; Sullivan, B.J.; Forghani, B.; Bi, D.; Beck, C.; Audet, S.; Gargiullo, P. Persistence of measles antibodies after 2 doses of measles vaccine in a postelimination environment. Arch. Pediatr. Adolesc. Med. 2007, 161, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Cohen, B.J.; Doblas, D.; Andrews, N. Comparison of plaque reduction neutralisation test (PRNT) and measles virus-specific IgG ELISA for assessing immunogenicity of measles vaccination. Vaccine 2008, 26, 6392–6397. [Google Scholar] [CrossRef] [PubMed]
- Fujino, M.; Yoshida, N.; Kimura, K.; Zhou, J.; Motegi, Y.; Komase, K.; Nakayama, T. Development of a new neutralization test for measles virus. J. Virol. Methods 2007, 142, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Eick, A.A.; Hu, Z.; Wang, Z.; Nevin, R.L. Incidence of mumps and immunity to measles, mumps and rubella among US military recruits, 2000–2004. Vaccine 2008, 26, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Hermanson, G.; Chun, S.; Felgner, J.; Tan, X.; Pablo, J.; Nakajima-Sasaki, R.; Molina, D.M.; Felgner, P.L.; Liang, X.; Davies, D.H. Measurement of antibody responses to Modified Vaccinia virus Ankara (MVA) and Dryvax((R)) using proteome microarrays and development of recombinant protein ELISAs. Vaccine 2012, 30, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Baum, E.; Badu, K.; Molina, D.M.; Liang, X.; Felgner, P.L.; Yan, G. Protein microarray analysis of antibody responses to Plasmodium falciparum in western Kenyan highland sites with differing transmission levels. PLOS ONE 2013, 8, e82246. [Google Scholar] [CrossRef] [PubMed]
- Kunnath-Velayudhan, S.; Salamon, H.; Wang, H.Y.; Davidow, A.L.; Molina, D.M.; Huynh, V.T.; Cirillo, D.M.; Michel, G.; Talbot, E.A.; Perkins, M.D.; et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc. Natl. Acad. Sci. USA 2010, 107, 14703–14708. [Google Scholar] [CrossRef] [PubMed]
- Bouche, F.; Ammerlaan, W.; Berthet, F.; Houard, S.; Schneider, F.; Muller, C.P. Immunosorbent assay based on recombinant hemagglutinin protein produced in a high-efficiency mammalian expression system for surveillance of measles immunity. J. Clin. Microbiol. 1998, 36, 721–726. [Google Scholar] [PubMed]
- Bouche, F.; Ammerlaan, W.; Fournier, P.; Schneider, F.; Muller, C.P. A simplified immunoassay based on measles virus recombinant hemagglutinin protein for testing the immune status of vaccinees. J. Virol. Methods 1998, 74, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Ertl, O.T.; Wenz, D.C.; Bouche, F.B.; Berbers, G.A.; Muller, C.P. Immunodominant domains of the Measles virus hemagglutinin protein eliciting a neutralizing human B cell response. Arch. Virol. 2003, 148, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Moss, W.J.; Griffin, D.E. Global measles elimination. Nat. Rev. Microbiol. 2006, 4, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Duprex, W.P.; Collins, F.M.; Rima, B.K. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 2002, 76, 7322–7328. [Google Scholar] [CrossRef] [PubMed]
- Dochow, M.; Krumm, S.A.; Crowe, J.E., Jr.; Moore, M.L.; Plemper, R.K. Independent structural domains in paramyxovirus polymerase protein. J. Biol. Chem. 2012, 287, 6878–6891. [Google Scholar] [CrossRef] [PubMed]
- Forthal, D.N.; Landucci, G.; Katz, J.; Tilles, J.G. Comparison of measles virus-specific antibodies with antibody-dependent cellular cytotoxicity and neutralizing functions. J. Infect. Dis. 1993, 168, 1020–1023. [Google Scholar] [CrossRef] [PubMed]
- Forthal, D.N.; Landucci, G. In vitro reduction of virus infectivity by antibody-dependent cell-mediated immunity. J. Immunol. Methods 1998, 220, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Atabani, S.; Landucci, G.; Steward, M.W.; Whittle, H.; Tilles, J.G.; Forthal, D.N. Sex-associated differences in the antibody-dependent cellular cytotoxicity antibody response to measles vaccines. Clin. Diagn. Lab. Immunol. 2000, 7, 111–113. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haralambieva, I.H.; Simon, W.L.; Kennedy, R.B.; Ovsyannikova, I.G.; Warner, N.D.; Grill, D.E.; Poland, G.A. Profiling of Measles-Specific Humoral Immunity in Individuals Following Two Doses of MMR Vaccine Using Proteome Microarrays. Viruses 2015, 7, 1113-1133. https://doi.org/10.3390/v7031113
Haralambieva IH, Simon WL, Kennedy RB, Ovsyannikova IG, Warner ND, Grill DE, Poland GA. Profiling of Measles-Specific Humoral Immunity in Individuals Following Two Doses of MMR Vaccine Using Proteome Microarrays. Viruses. 2015; 7(3):1113-1133. https://doi.org/10.3390/v7031113
Chicago/Turabian StyleHaralambieva, Iana H., Whitney L. Simon, Richard B. Kennedy, Inna G. Ovsyannikova, Nathaniel D. Warner, Diane E. Grill, and Gregory A. Poland. 2015. "Profiling of Measles-Specific Humoral Immunity in Individuals Following Two Doses of MMR Vaccine Using Proteome Microarrays" Viruses 7, no. 3: 1113-1133. https://doi.org/10.3390/v7031113





