Multiple Regions of Kaposi’s Sarcoma-Associated Herpesvirus ORF59 RNA are Required for Its Expression Mediated by Viral ORF57 and Cellular RBM15
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Expression Vectors
2.2. Cells and Co-Transfection Assays
2.3. RNA-Protein Pulldown Assays
2.4. Quantitative RT-PCR and RNA Decay Analysis
3. Results and Discussion
3.1. Mapping of the Regions in ORF59 RNA Responsible for ORF57-Mediated RNA Accumulation and Protein Translation
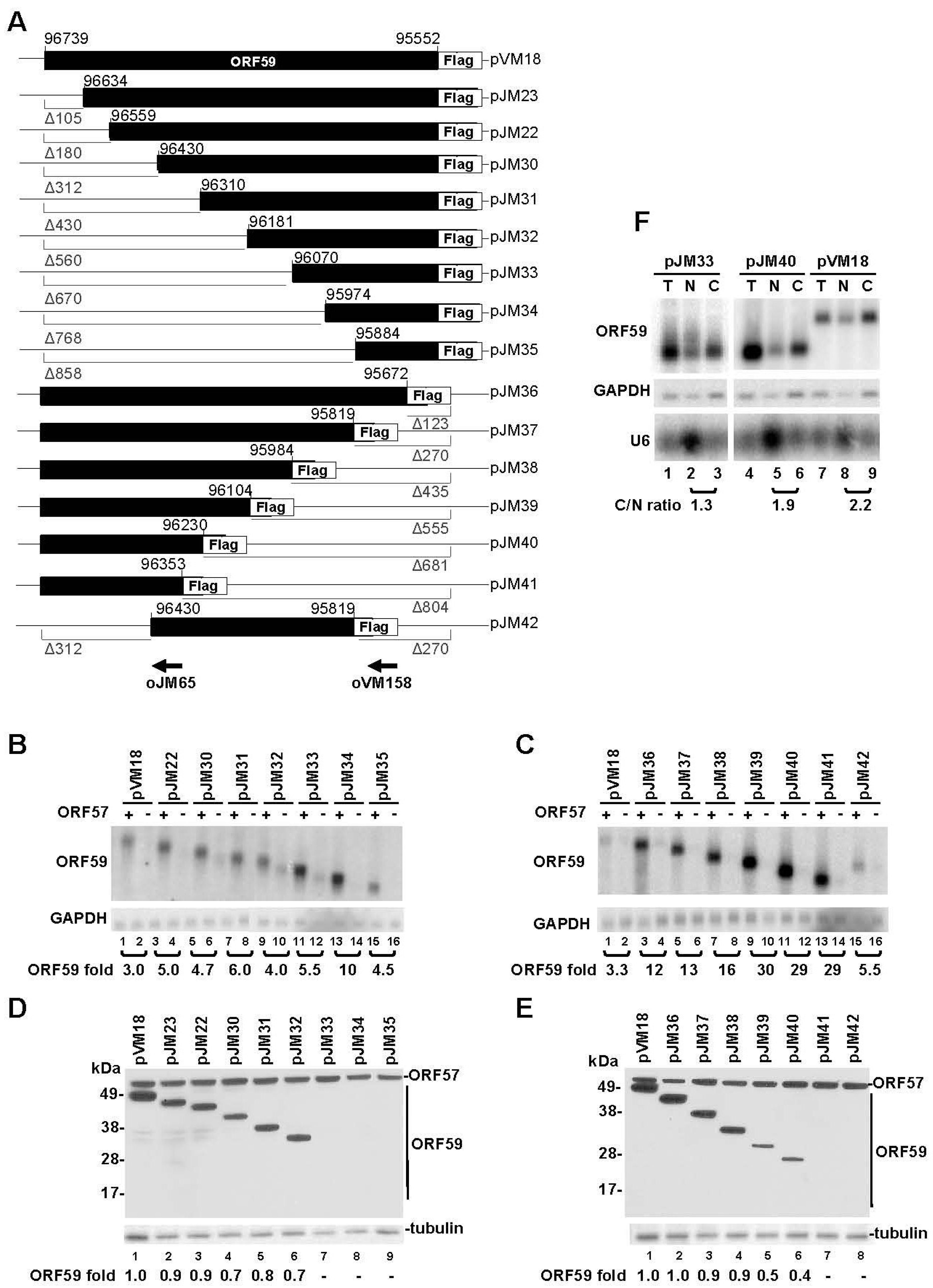
3.2. The 5' MRE of ORF59 Interacts with ORF57 and RBM15 and Exhibits a Role in RNA Export and Stability
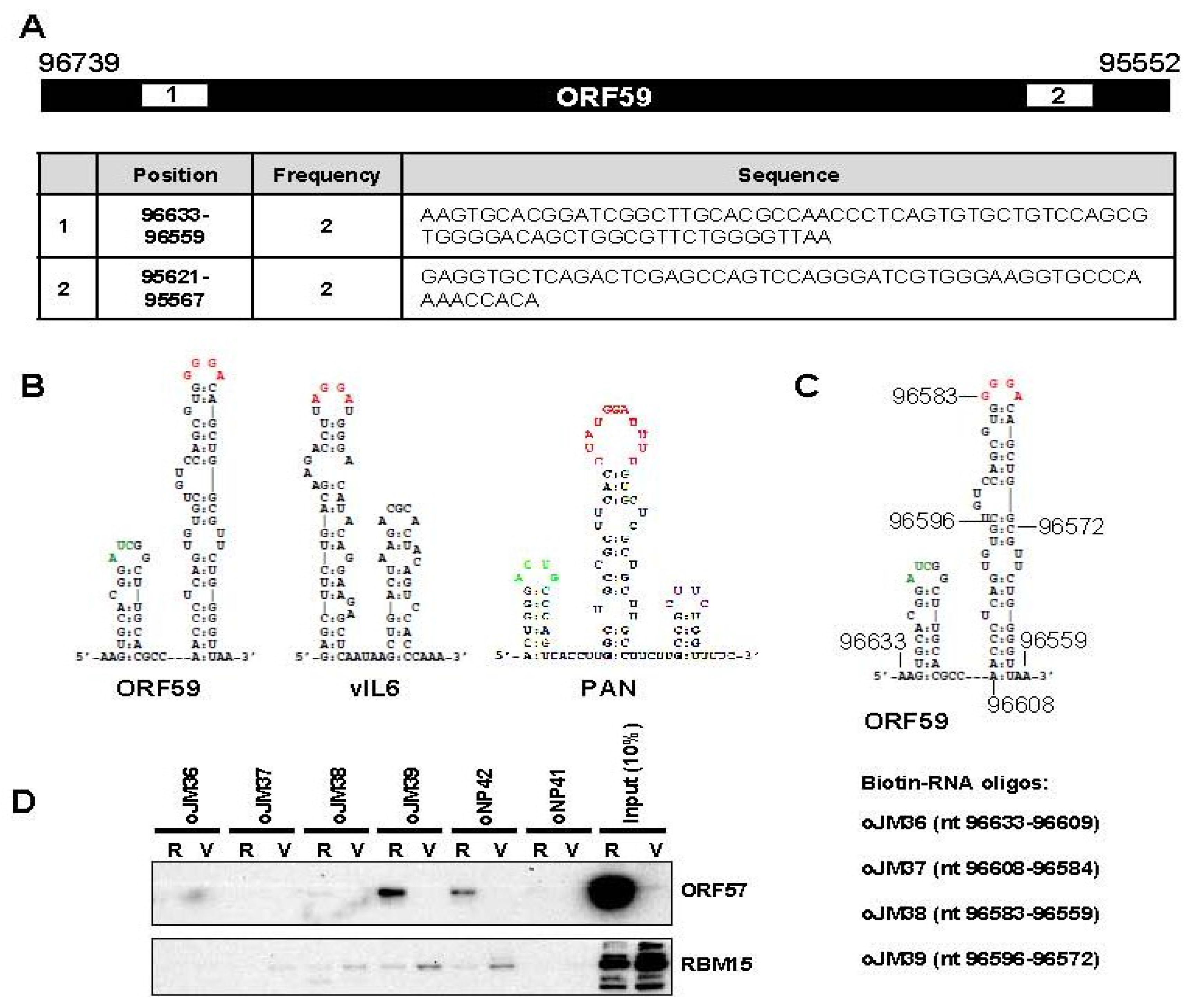
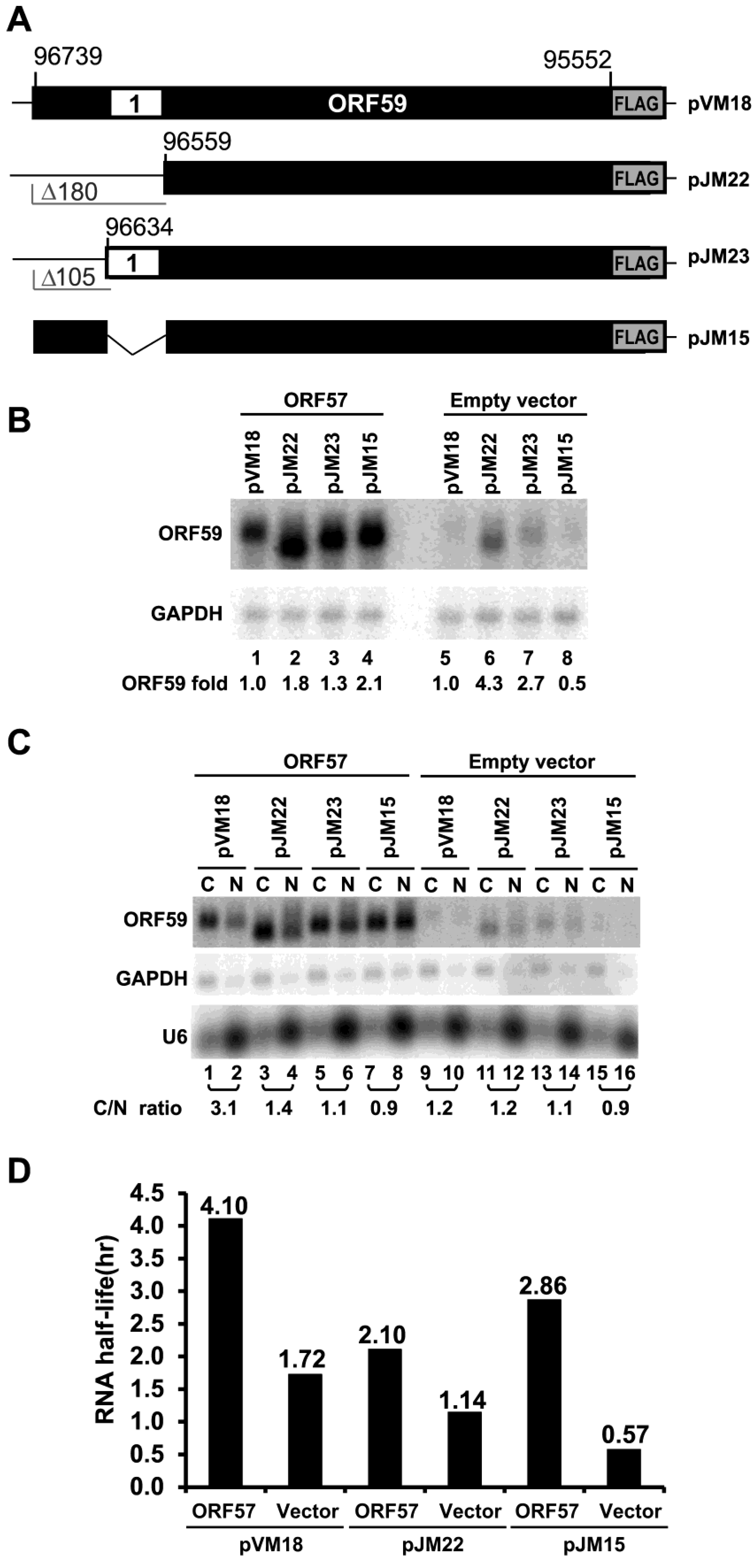
3.3. ORF59 RNA without the 5' MRE is Translatable in the Presence of ORF57, but Poorly Translatable in the Presence of RBM15

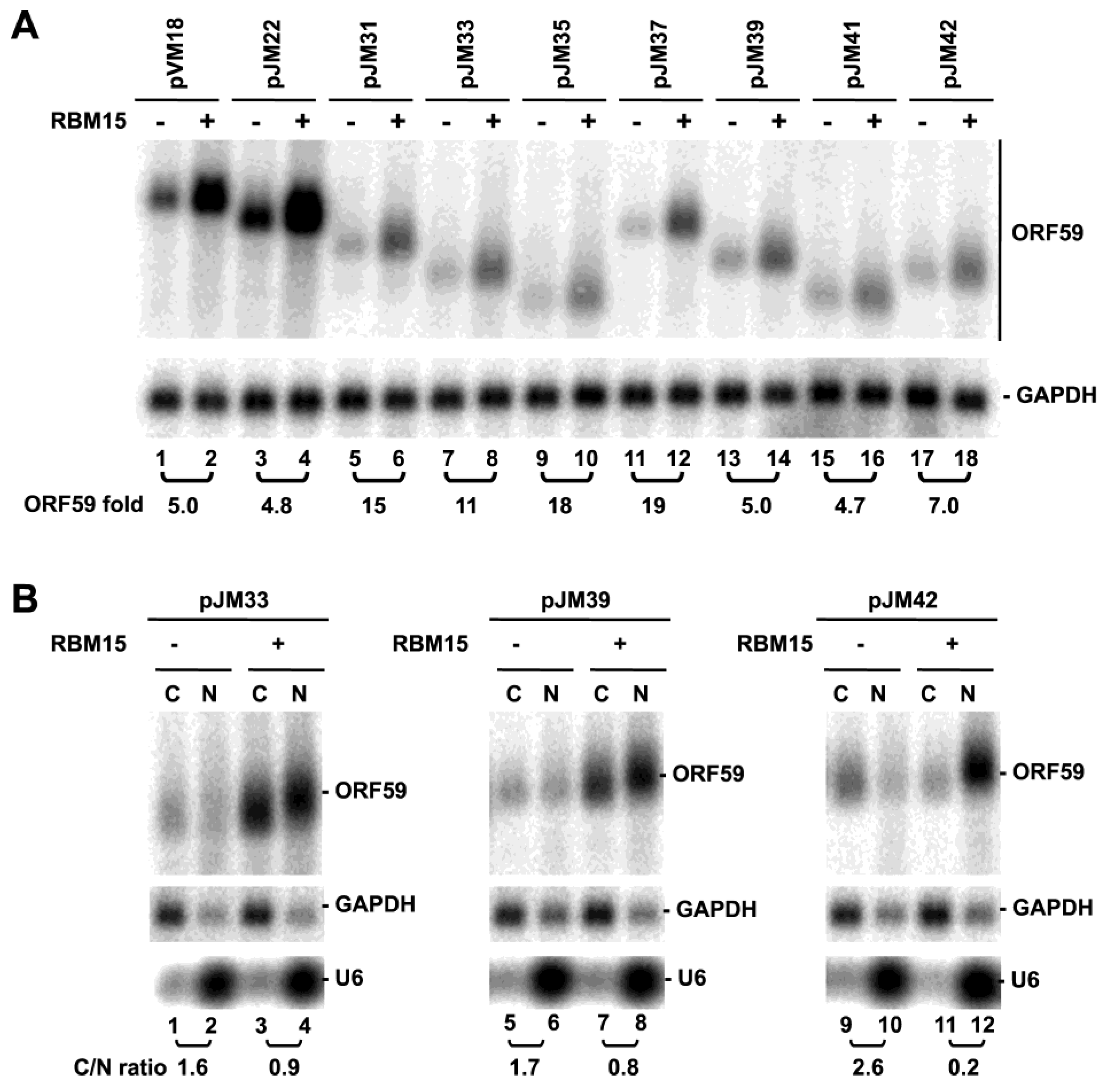
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wu, F.Y.; Ahn, J.H.; Alcendor, D.J.; Jang, W.J.; Xiao, J.; Hayward, S.D.; Hayward, G.S. Origin-independent assembly of Kaposi’s sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 2001, 75, 1487–1506. [Google Scholar] [CrossRef]
- AuCoin, D.P.; Colletti, K.S.; Cei, S.A.; Papouskova, I.; Tarrant, M.; Pari, G.S. Amplification of the Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 2004, 318, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, K.; Ricciardi, R.P. Human Kaposi’s sarcoma herpesvirus processivity factor-8 functions as a dimer in DNA synthesis. J. Biol. Chem. 2004, 279, 28375–28386. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.R.; Chandran, B. Characterization of human herpesvirus 8 ORF59 protein (PF-8) and mapping of the processivity and viral DNA polymerase-interacting domains. J. Virol. 2000, 74, 10920–10929. [Google Scholar] [CrossRef] [PubMed]
- McDowell, M.E.; Purushothaman, P.; Rossetto, C.C.; Pari, G.S.; Verma, S.C. Phosphorylation of Kaposi’s sarcoma-associated herpesvirus processivity factor ORF59 by a viral kinase modulates its ability to associate with RTA and oriLyt. J. Virol. 2013, 87, 8038–8052. [Google Scholar] [CrossRef] [PubMed]
- Lukac, D.M.; Renne, R.; Kirshner, J.R.; Ganem, D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 1998, 252, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Gradoville, L.; Gerlach, J.; Grogan, E.; Shedd, D.; Nikiforow, S.; Metroka, C.; Miller, G. Kaposi’s sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 2000, 74, 6207–6212. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, Y.; Liang, D.; Gao, Y.; Xia, T.; Robertson, E.S.; Lan, K. Kaposi’s sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jkappa and a cis-acting RTA responsive element. Virology 2008, 380, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Dai, C.Y.; Ricciardi, R.P. Cloning and functional analysis of Kaposi’s sarcoma-associated herpesvirus DNA polymerase and its processivity factor. J. Virol. 1998, 72, 6228–6232. [Google Scholar] [PubMed]
- Rossetto, C.C.; Susilarini, N.K.; Pari, G.S. Interaction of Kaposi’s sarcoma-associated herpesvirus ORF59 with oriLyt is dependent on binding with K-Rta. J. Virol. 2011, 85, 3833–3841. [Google Scholar] [CrossRef] [PubMed]
- Glaunsinger, B.; Ganem, D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 2004, 13, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Pripuzova, N.; Chan, C.; Temkin, N.; Specht, S.I.; Zheng, Z.M. Stability of structured Kaposi sarcoma-associated herpesvirus ORF57 protein is regulated by protein phosphorylation and homodimerization. J. Virol. 2015. [Google Scholar] [CrossRef]
- Majerciak, V.; Yamanegi, K.; Nie, S.H.; Zheng, Z.M. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 2006, 281, 28365–28378. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 2009, 14, 1516–1528. [Google Scholar] [CrossRef]
- Majerciak, V.; Uranishi, H.; Kruhlak, M.; Pilkington, G.R.; Massimelli, M.J.; Bear, J.; Pavlakis, G.N.; Felber, B.K.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J. Virol. 2011, 85, 1528–1540. [Google Scholar] [CrossRef] [PubMed]
- Kirshner, J.R.; Lukac, D.M.; Chang, J.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 2000, 74, 3586–3597. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Zheng, Z.M. KSHV ORF57, a protein of many faces. Viruses 2015, in press. [Google Scholar]
- Kang, J.G.; Pripuzova, N.; Majerciak, V.; Kruhlak, M.; Le, S.Y.; Zheng, Z.M. Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Promotes Escape of Viral and Human Interleukin-6 from MicroRNA-Mediated Suppression. J. Virol. 2011, 85, 2620–2630. [Google Scholar] [CrossRef] [PubMed]
- Massimelli, M.J.; Kang, J.G.; Majerciak, V.; Le, S.Y.; Liewehr, D.J.; Steinberg, S.M.; Zheng, Z.M. Stability of a Long Noncoding Viral RNA Depends on a 9-nt Core Element at the RNA 5' End to Interact with Viral ORF57 and Cellular PABPC1. Int. J. Biol. Sci. 2011, 7, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Massimelli, M.J.; Majerciak, V.; Kruhlak, M.; Zheng, Z.M. Interplay between polyadenylate-binding protein 1 and Kaposi’s sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J. Virol. 2013, 87, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Yamanegi, K.; Zheng, Z.M. Gene structure and expression of Kaposi’s sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 2006, 80, 11968–11981. [Google Scholar] [CrossRef] [PubMed]
- Lindtner, S.; Zolotukhin, A.S.; Uranishi, H.; Bear, J.; Kulkarni, V.; Smulevitch, S.; Samiotaki, M.; Panayotou, G.; Felber, B.K.; Pavlakis, G.N. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 2006, 281, 36915–36928. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.M.; Reid, E.S.; Baker, C.C. Utilization of the bovine papillomavirus type 1 late-stage-specific nucleotide 3605 3' splice site is modulated by a novel exonic bipartite regulator but not by an intronic purine-rich element. J. Virol. 2000, 74, 10612–10622. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Pripuzova, N.; McCoy, J.P.; Gao, S.J.; Zheng, Z.M. Targeted disruption of Kaposi’s sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 2007, 81, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Lu, M.; Gwack, Y.; Souvlis, J.; Zeichner, S.L.; Jung, J.U. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 2003, 77, 4205–4220. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Liu, X.; Tao, M.; Kruhlak, M.; Guo, M.; Meyers, C.; Baker, C.C.; Zheng, Z.M. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J. Virol. 2009, 83, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Boyne, J.R.; Jackson, B.R.; Taylor, A.; Macnab, S.A.; Whitehouse, A. Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J. 2010, 29, 1851–1864. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Majerciak, V.; Uldrick, T.S.; Wang, X.; Kruhlak, M.; Yarchoan, R.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J. Pathol. 2011, 225, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Uranishi, H.; Zolotukhin, A.S.; Lindtner, S.; Warming, S.; Zhang, G.M.; Bear, J.; Copeland, N.G.; Jenkins, N.A.; Pavlakis, G.N.; Felber, B.K. The RNA binding motif protein 15B (RBM15B/OTT3) acts as co-factor of the nuclear export receptor NXF1. J. Biol. Chem. 2009, 284, 26106–26116. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Blackbourn, D.J.; Clements, J.B. The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 2004, 279, 33001–33011. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massimelli, M.J.; Majerciak, V.; Kang, J.-G.; Liewehr, D.J.; Steinberg, S.M.; Zheng, Z.-M. Multiple Regions of Kaposi’s Sarcoma-Associated Herpesvirus ORF59 RNA are Required for Its Expression Mediated by Viral ORF57 and Cellular RBM15. Viruses 2015, 7, 496-510. https://doi.org/10.3390/v7020496
Massimelli MJ, Majerciak V, Kang J-G, Liewehr DJ, Steinberg SM, Zheng Z-M. Multiple Regions of Kaposi’s Sarcoma-Associated Herpesvirus ORF59 RNA are Required for Its Expression Mediated by Viral ORF57 and Cellular RBM15. Viruses. 2015; 7(2):496-510. https://doi.org/10.3390/v7020496
Chicago/Turabian StyleMassimelli, Maria Julia, Vladimir Majerciak, Jeong-Gu Kang, David J. Liewehr, Seth M. Steinberg, and Zhi-Ming Zheng. 2015. "Multiple Regions of Kaposi’s Sarcoma-Associated Herpesvirus ORF59 RNA are Required for Its Expression Mediated by Viral ORF57 and Cellular RBM15" Viruses 7, no. 2: 496-510. https://doi.org/10.3390/v7020496




