A Flagellar Glycan-Specific Protein Encoded by Campylobacter Phages Inhibits Host Cell Growth
Abstract
:1. Introduction
| Property | Reference |
|---|---|
| Binds to C. jejuni and C. coli flagella | [16] |
| Recognizes acetamidino-modified pseudaminic acid residues | [16] |
| Binding activity localized to C-terminal quarter of the protein | [16] |
| Agglutinates host bacterial cells | [16,18] |
| Forms multimers | [15] |
| Reduces host bacterial motility | [16] |
| Not identified as a component of the structural phage proteome | [15] |
| C-terminal homologues encoded by all sequenced Campylobacter phages | [21,22] |
| Effectively captures C. jejuni and C. coli from complex samples | [20] |
| Inhibits host bacterial growth | This study |
| Growth inhibition domain localized to C-terminal quarter | This study |
| Expression of Gp047 in C. jejuni 11168 cells does not affect flagellar phenotype, cell morphology or growth | This study |
2. Materials and Methods
2.1. Bacterial Growth Conditions
| Strain or Mutant | Description | Source or Reference |
|---|---|---|
| C. jejuni 11168 | Human isolate | [23] |
| C. jejuni 81-176 | Clinical isolate | [24] |
| 11168ΔkpsM | Acapsular, kpsM mutant in C. jejuni 11168 | [25] |
| 11168ΔpseG | Aflagellate, pseG (cj1312) mutant in C. jejuni NCTC 11168 | This study |
| 81-176ΔpseA | pseA (cj1316c) mutant in C. jejuni 81-176 (flagellate, no acetamidino modification) | [26] |
| 11168ΔpseA | pseA mutant in C. jejuni NCTC 11168 | [16] |
| 11168ΔpseA:pseA | 11168∆pseA mutant chromosomally complemented with wild type pseA | [16] |
| 111-28 (pCE111-28/gp047) | 11168 expressing gp047 on the pCE 111-28 plasmid | This study |
| 11168 (pCE111-28) | 11168 expressing empty pCE 111-28 plasmid | This study |
| Plasmid | Description | Source or Reference |
|---|---|---|
| pGEX 6P-2 | Glutathione-S-transferase (GST) fused protein expression vector, ampicillin resistance marker, tac promoter | GE Healthcare |
| pCE 111-28 | Campylobacter-E. coli shuttle vector for protein expression, chloramphenicol resistance marker, plasmid pRY111 with σ28 promoter of flaA | [27] |
| pGEX_gp047 | gp047 cloned in-frame into multiple cloning site of pGEX 6P-2 for expression of GST-fused Gp047 | [15] |
| pGEX_ntgp047 | Expression construct of GST-fused N-Gp047 in pGEX 6P-2 | [18] |
| pGEX_ctgp047 | Expression construct of GST-fused C-Gp047 in pGEX 6P-2 | [18] |
| pGEX_ccgp047 | Expression construct of GST-fused CC-Gp047 in pGEX 6P-2 | [18] |
| pGEX_ncgp047 | Expression construct of GST-fused NC-Gp047 in pGEX 6P-2 | [18] |
| pCE_gp047 | Expression construct of untagged Gp047 for expression in pCE 111-28 in C. jejuni | This study |
2.2. Construction of a PseG Mutant in C. jejuni 11168
2.3. Expression of Gp047 and Its Derivatives
2.4. Agar Plate Growth Clearance Assay
2.5. Scanning Electron Microscopy
2.6. Liquid Growth Assays
2.7. Western Blotting
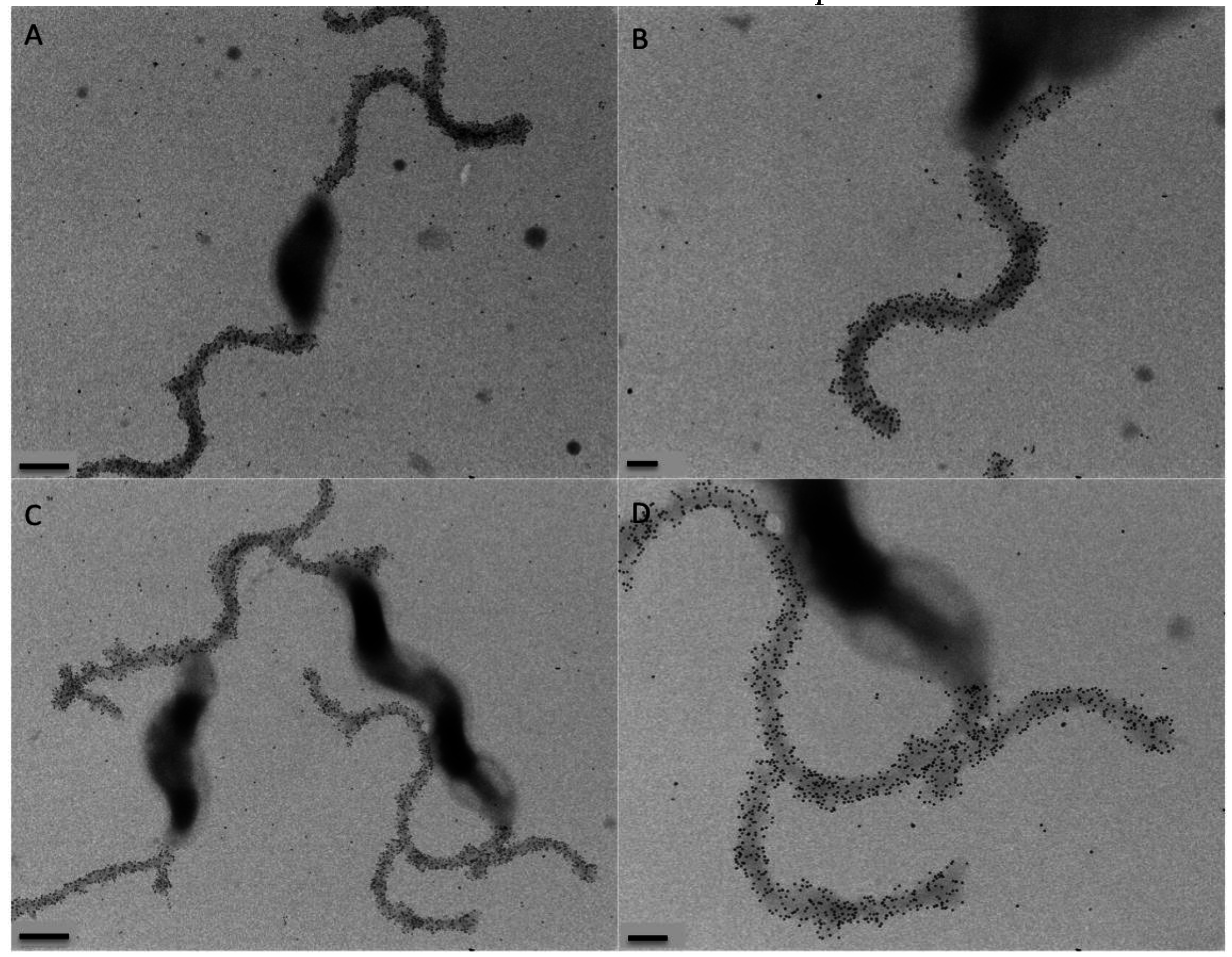
2.8. Immunogold Labeling and Transmission Electron Microscopy
3. Results
3.1. Gp047 Causes Capsular Polysaccharide-Independent Clearance of C. jejuni Growth
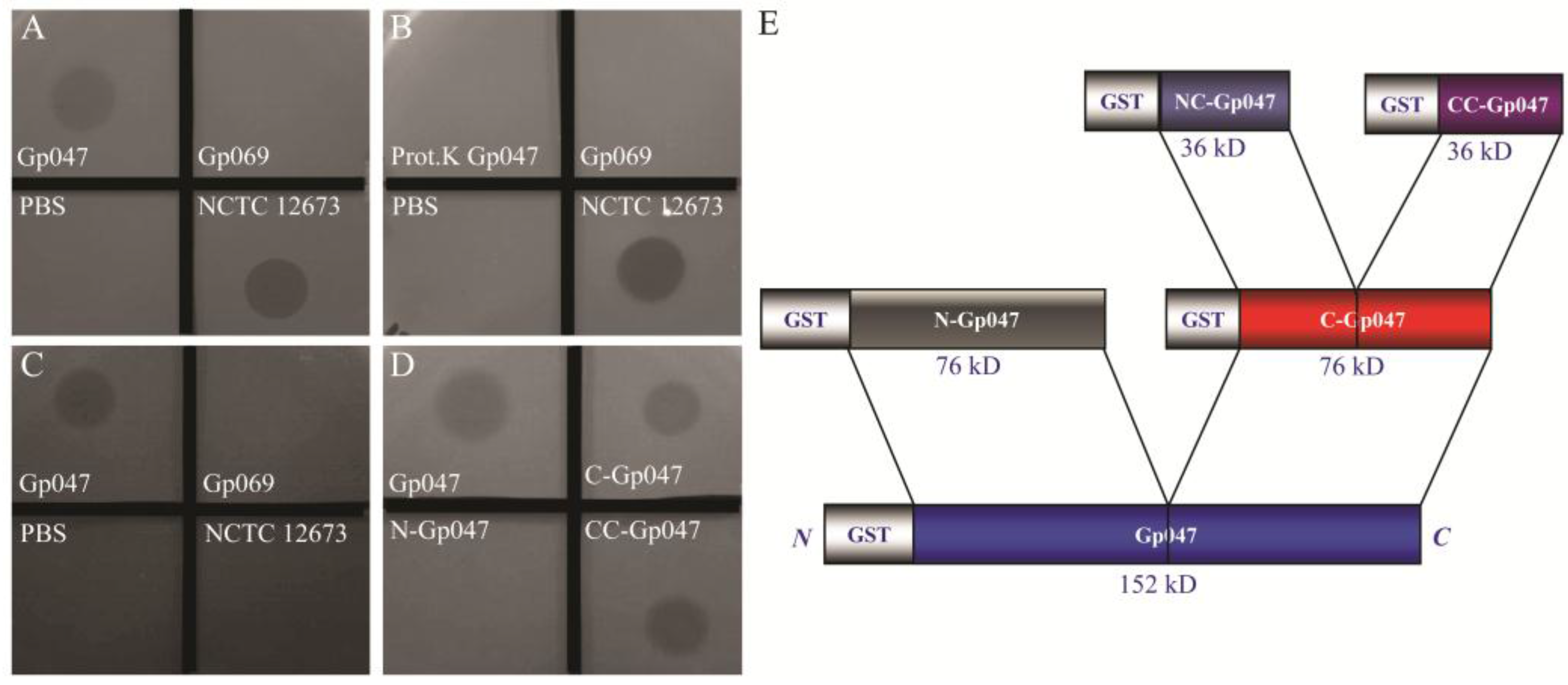
3.2. Growth Clearance-Associated Domain is Localized in the C-Terminal Quarter of Gp047
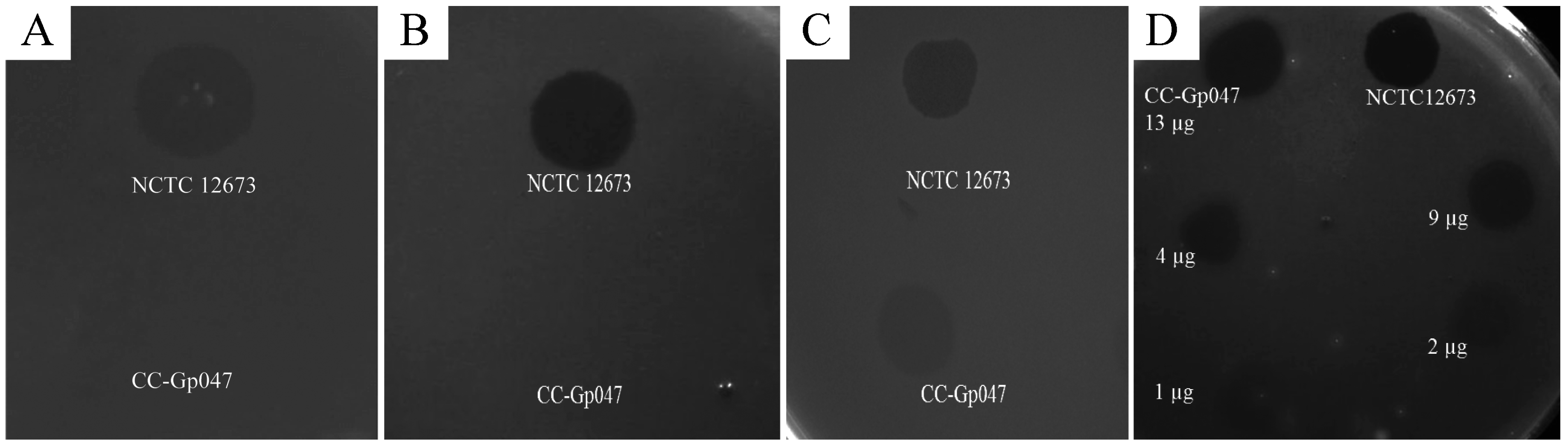
3.3. Clearance Phenotype of Gp047 Is Contact- and Dose-Dependent
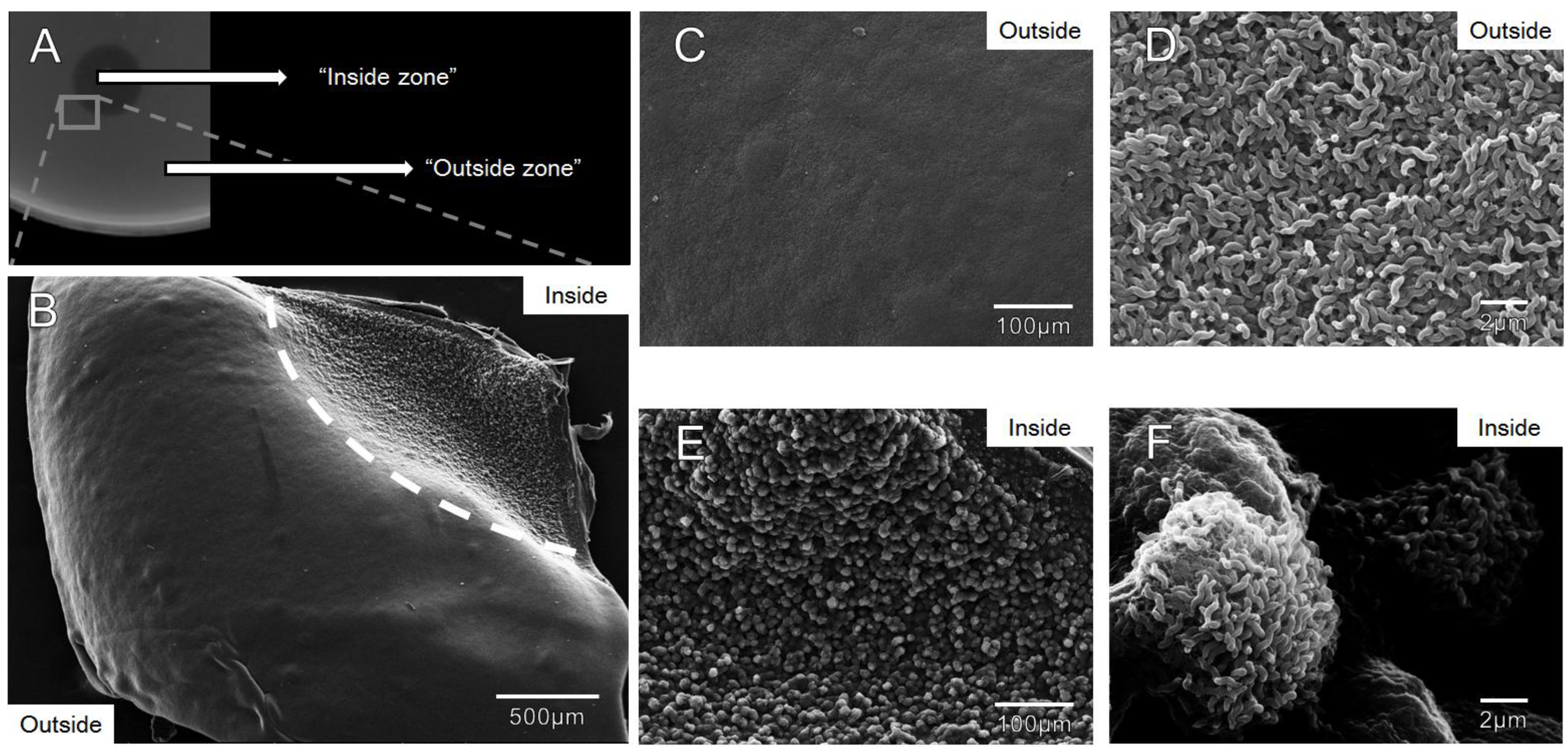
3.4. Zones of Clearance Observed Using Electron Microscopy Show an Altered Growth Pattern
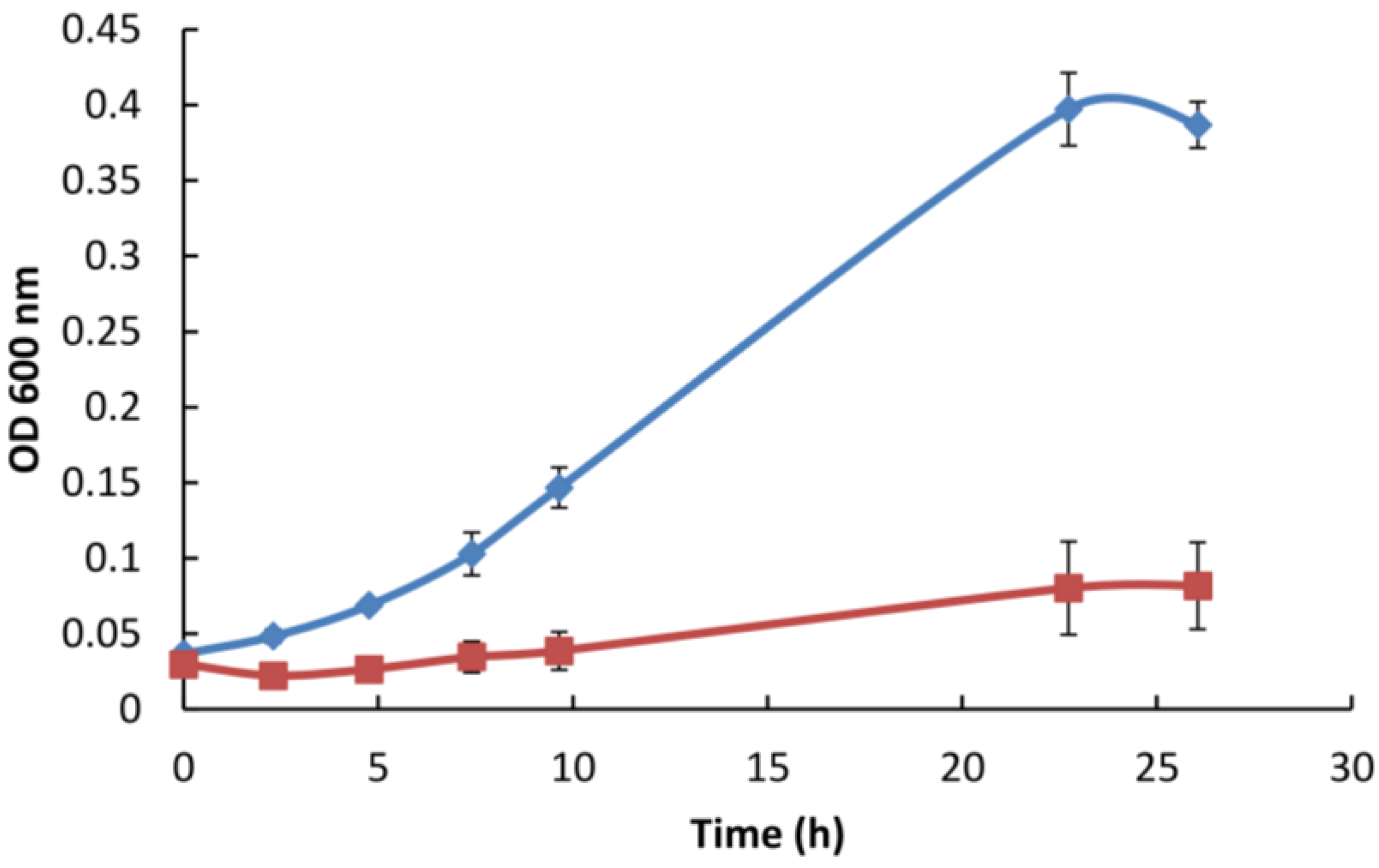
3.5. Gp047 Addition to Broth Cultures Inhibits Cell Growth
3.6. Gp047 Expressed in C. jejuni Cells Does Not Affect the Flagellar Phenotype

4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kittler, S.; Fischer, S.; Abdulmawjood, A.; Glunder, G.; Klein, G. Effect of bacteriophage application on Campylobacter jejuni loads in commercial broiler flocks. Appl. Environ. Microbiol. 2013, 79, 7525–7533. [Google Scholar] [CrossRef] [PubMed]
- Loc Carrillo, C.; Atterbury, R.J.; El-Shibiny, A.; Connerton, P.L.; Dillon, E.; Scott, A.; Connerton, I.F. Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens. Appl. Environ. Microbiol. 2005, 71, 6554–6563. [Google Scholar] [CrossRef] [PubMed]
- Trigo, G.; Martins, T.G.; Fraga, A.G.; Longatto-Filho, A.; Castro, A.G.; Azeredo, J.; Pedrosa, J. Phage therapy is effective against infection by Mycobacterium ulcerans in a murine footpad model. PLoS Negl. Trop. Dis. 2013, 7, e2183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinbacher, S.; Seckler, R.; Miller, S.; Steipe, B.; Huber, R.; Reinemer, P. Crystal structure of P22 tailspike protein: Interdigitated subunits in a thermostable trimer. Science 1994, 265, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Baxa, U.; Steinbacher, S.; Miller, S.; Weintraub, A.; Huber, R.; Seckler, R. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys. J. 1996, 71, 2040–2048. [Google Scholar] [CrossRef]
- Iwashita, S.; Kanegasaki, S. Enzymic and molecular properties of base-plate parts of bacteriophage P22. Eur. J. Biochem. 1976, 65, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Waseh, S.; Hanifi-Moghaddam, P.; Coleman, R.; Masotti, M.; Ryan, S.; Foss, M.; MacKenzie, R.; Henry, M.; Szymanski, C.; Tanha, J. Orally administered P22 phage tailspike protein reduces salmonella colonization in chickens: Prospects of a novel therapy against bacterial infections. PLoS ONE 2010, 5, e13904. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, A.; Ceyssens, P.J.; Krylov, V.N.; Noben, J.P.; Volckaert, G.; Lavigne, R. Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology 2012, 434, 251–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholl, D.; Cooley, M.; Williams, S.R.; Gebhart, D.; Martin, D.; Bates, A.; Mandrell, R. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob. Agents Chemother. 2009, 53, 3074–3080. [Google Scholar] [CrossRef] [PubMed]
- Gebhart, D.; Lok, S.; Clare, S.; Tomas, M.; Stares, M.; Scholl, D.; Donskey, C.J.; Lawley, T.D.; Govoni, G.R. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pirnay, J.P.; et al. Engineered endolysin-based "Artilysins" to combat multidrug-resistant gram-negative pathogens. mBio 2014, 5, e01379-14. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arutyunov, D.; Szymanski, C.M.; Evoy, S. Bacteriophage based probes for pathogen detection. Analyst 2012, 137, 3405–3421. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.J.; Sacher, J.C.; Szymanski, C.M. Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 2015, 34, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M.; Arutyunov, D.; Foss, M.; Cunningham, A.; Ding, W.; Singh, A.; Pavlov, A.R.; Henry, M.; Evoy, S.; Kelly, J.; Szymanski, C.M. Genome and proteome of Campylobacter jejuni bacteriophage NCTC 12673. Appl. Environ. Microbiol. 2011, 77, 8265–8271. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.A.; van Alphen, L.B.; Sacher, J.; Ding, W.; Kelly, J.; Nargang, C.; Smith, D.F.; Cummings, R.D.; Szymanski, C.M. A receptor-binding protein of Campylobacter jejuni bacteriophage NCTC 12673 recognizes flagellin glycosylated with acetamidino-modified pseudaminic acid. Mol. Microbiol. 2015, 95, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Coward, C.; Grant, A.J.; Swift, C.; Philp, J.; Towler, R.; Heydarian, M.; Frost, J.A.; Maskell, D.J. Phase-variable surface structures are required for infection of Campylobacter jejuni by bacteriophages. Appl. Environ. Microbiol. 2006, 72, 4638–4647. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.A.; Poshtiban, S.; Arutyunov, D.; Evoy, S.; Szymanski, C.M. Bacteriophage receptor binding protein based assays for the simultaneous detection of Campylobacter jejuni and Campylobacter coli. PLoS ONE 2013, 8, e69770. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arutyunov, D.; McDermott, M.T.; Szymanski, C.M.; Evoy, S. Specific detection of Campylobacter jejuni using the bacteriophage NCTC 12673 receptor binding protein as a probe. Analyst 2011, 136, 4780–4786. [Google Scholar] [CrossRef] [PubMed]
- Poshtiban, S.; Javed, M.A.; Arutyunov, D.; Singh, A.; Banting, G.; Szymanski, C.M.; Evoy, S. Phage receptor binding protein-based magnetic enrichment method as an aid for real time PCR detection of foodborne bacteria. Analyst 2013, 138, 5619–5626. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.A.; Ackermann, H.-W.; Azeredo, J.; Carvalho, C.M.; Connerton, I.F.; Evoy, S.; Hammerl, J.A.; Hertwig, S.; Lavigne, R.; Singh, A.; et al. A suggested classification for two groups of Campylobacter Myoviruses. Arch. Virol. 2014, 159, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janež, N.; Kokošin, A.; Zaletel, E.; Vranac, T.; Kovač, J.; Vučković, D.; Smole Možina, S.; Čurin Šerbec, V.; Zhang, Q.; Accetto, T.; et al. Identification and Characterisation of new Campylobacter group III phages of animal origin. FEMS Microbiol. Lett. 2014, 359, 64–71. [Google Scholar]
- Parkhill, J.; Wren, B.W.; Mungall, K.; Ketley, J.M.; Churcher, C.; Basham, D.; Chillingworth, T.; Davies, R.M.; Feltwell, T.; Holroyd, S.; et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000, 403, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Black, R.E.; Levine, M.M.; Clements, M.L.; Hughes, T.P.; Blaser, M.J. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 1988, 157, 472–479. [Google Scholar] [CrossRef] [PubMed]
- St Michael, F.; Szymanski, C.M.; Li, J.; Chan, K.H.; Khieu, N.H.; Larocque, S.; Wakarchuk, W.W.; Brisson, J.R.; Monteiro, M.A. The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur. J. Biochem. 2002, 269, 5119–5136. [Google Scholar] [CrossRef] [PubMed]
- Thibault, P.; Logan, S.M.; Kelly, J.F.; Brisson, J.R.; Ewing, C.P.; Trust, T.J.; Guerry, P. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 2001, 276, 34862–34870. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.C.; Szymanski, C.; Guerry, P. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81–176. J. Bacteriol. 2004, 186, 6508–6514. [Google Scholar] [CrossRef] [PubMed]
- Guerry, P.; Ewing, C.P.; Schirm, M.; Lorenzo, M.; Kelly, J.; Pattarini, D.; Majam, G.; Thibault, P.; Logan, S. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 2006, 60, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Bozzola, J.J.; Russell, L.D. Electron Microscopy: Principles and Techniques for Biologists; Jones and Bartlett Publishers: Sudbury, MA, USA, 1992; pp. 40–62. [Google Scholar]
- Sorensen, M.C.; Gencay, Y.E.; Birk, T.; Baldvinsson, S.B.; Jackel, C.; Hammerl, J.A.; Vegge, C.S.; Neve, H.; Brondsted, L. Primary isolation strain determines both phage type and receptors recognised by Campylobacter jejuni bacteriophages. PLoS ONE 2015, 10, e0116287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallet, R.; Shao, Y.; Wang, I.N. High adsorption rate is detrimental to bacteriophage fitness in a biofilm-like environment. BMC Evolut. Biol. 2009, 9, 241. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophage Ecology: Population Growth, Evolution, and Impact of Bacterial Viruses; Cambridge University Press: Cambridge, UK, 2008; Volume 15. [Google Scholar]
- Gadagkar, R.; Gopinathan, K.P. Bacteriophage burst size during multiple infections. J. Biosci. 1980, 2, 253–259. [Google Scholar] [CrossRef]
- Wang, P.X.; Sanders, P.W. Immunoglobulin light chains generate hydrogen peroxide. J. Am. Soc. Nephrol. JASN 2007, 18, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- Nieva, J.; Wentworth, P., Jr. The antibody-catalyzed water oxidation pathway—A new chemical arm to immune defense? Trends Biochem. Sci. 2004, 29, 274–278. [Google Scholar] [CrossRef] [PubMed]
- McClelland, E.E.; Nicola, A.M.; Prados-Rosales, R.; Casadevall, A. Ab binding alters gene expression in Cryptococcus neoformans and directly modulates fungal metabolism. J. Clin. Invest. 2010, 120, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.J.; Pontes, B.; Frases, S.; Nakouzi, A.S.; Nimrichter, L.; Rodrigues, M.L.; Viana, N.B.; Casadevall, A. Antibody binding to Cryptococcus neoformans impairs budding by altering capsular mechanical properties. J. Immunol. 2013, 190, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Gohil, S.; Coleman, J.R.; Manix, C.; Pirofski, L.A. Antibodies to Streptococcus pneumoniae capsular polysaccharide enhance pneumococcal quorum sensing. mBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Balaban, M.; Hendrixson, D.R. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog. 2011, 7, e1002420. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javed, M.A.; Sacher, J.C.; Van Alphen, L.B.; Patry, R.T.; Szymanski, C.M. A Flagellar Glycan-Specific Protein Encoded by Campylobacter Phages Inhibits Host Cell Growth. Viruses 2015, 7, 6661-6674. https://doi.org/10.3390/v7122964
Javed MA, Sacher JC, Van Alphen LB, Patry RT, Szymanski CM. A Flagellar Glycan-Specific Protein Encoded by Campylobacter Phages Inhibits Host Cell Growth. Viruses. 2015; 7(12):6661-6674. https://doi.org/10.3390/v7122964
Chicago/Turabian StyleJaved, Muhammad Afzal, Jessica C. Sacher, Lieke B. Van Alphen, Robert T. Patry, and Christine M. Szymanski. 2015. "A Flagellar Glycan-Specific Protein Encoded by Campylobacter Phages Inhibits Host Cell Growth" Viruses 7, no. 12: 6661-6674. https://doi.org/10.3390/v7122964






