Nucleobase but not Sugar Fidelity is Maintained in the Sabin I RNA-Dependent RNA Polymerase
Abstract
:1. Introduction
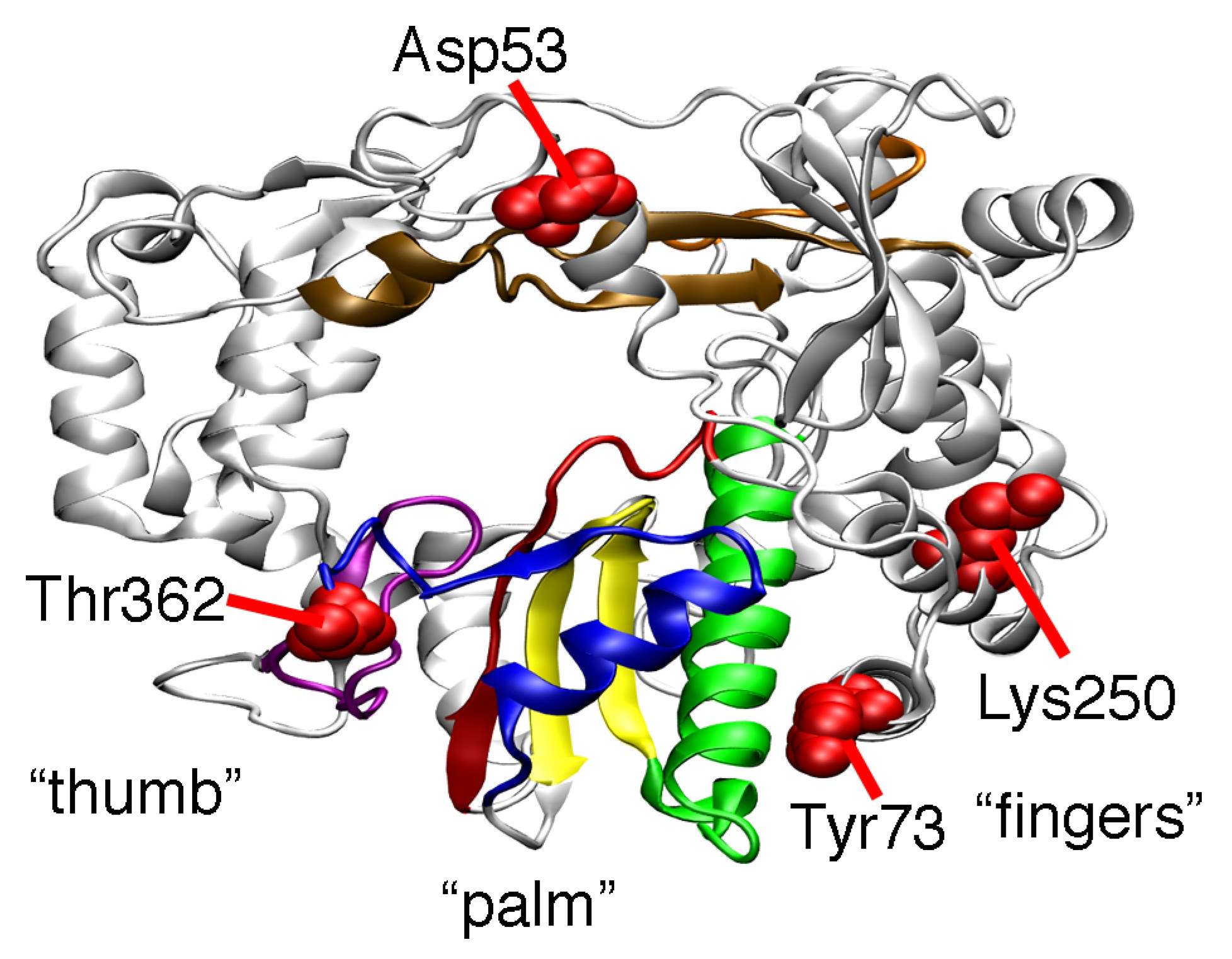
2. Results
2.1. Sabin PV RdRp Discriminates against Incorrect Nucleobases, but not against Incorrect Sugars, to the Same Extent as WT RdRp
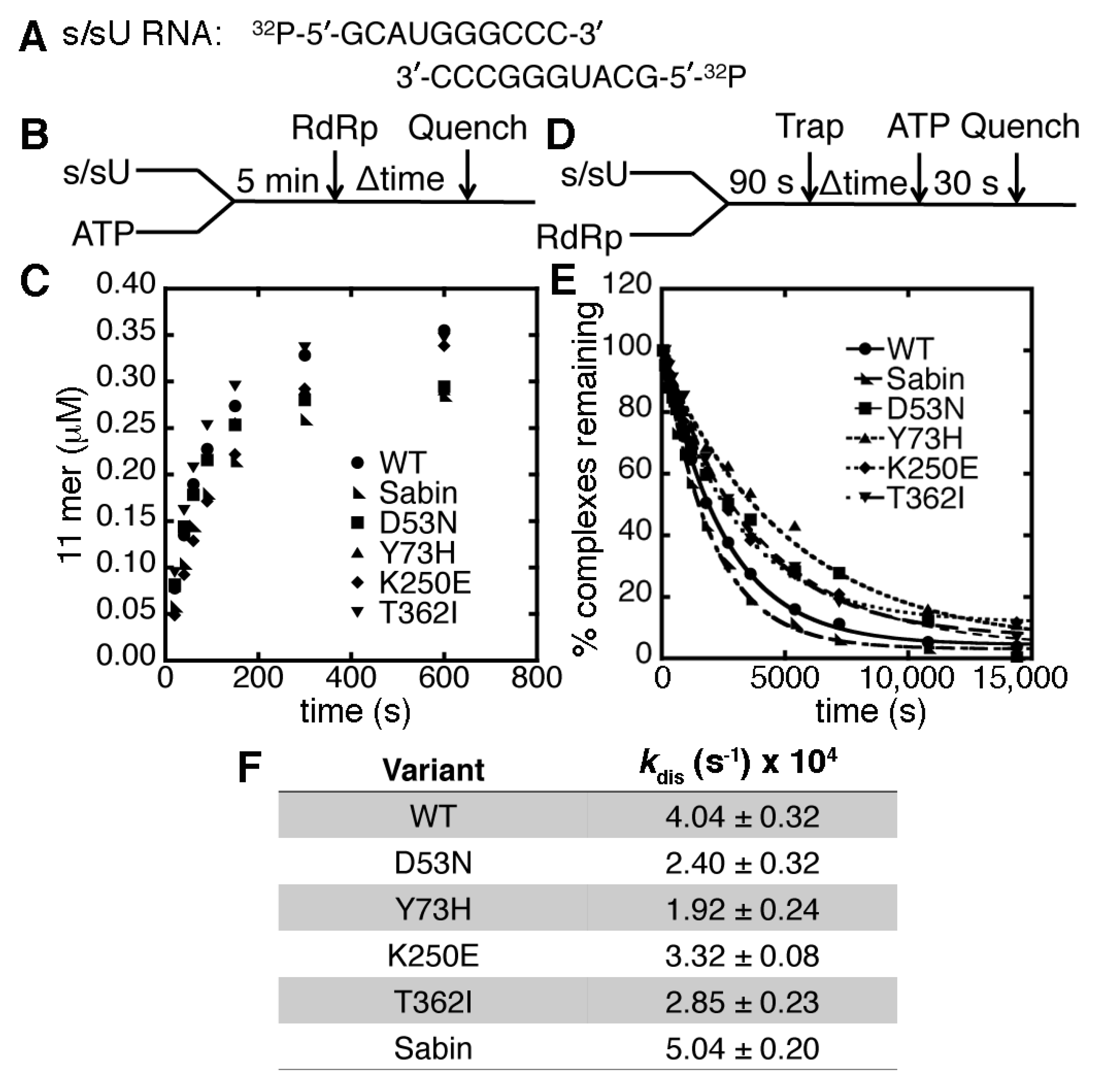
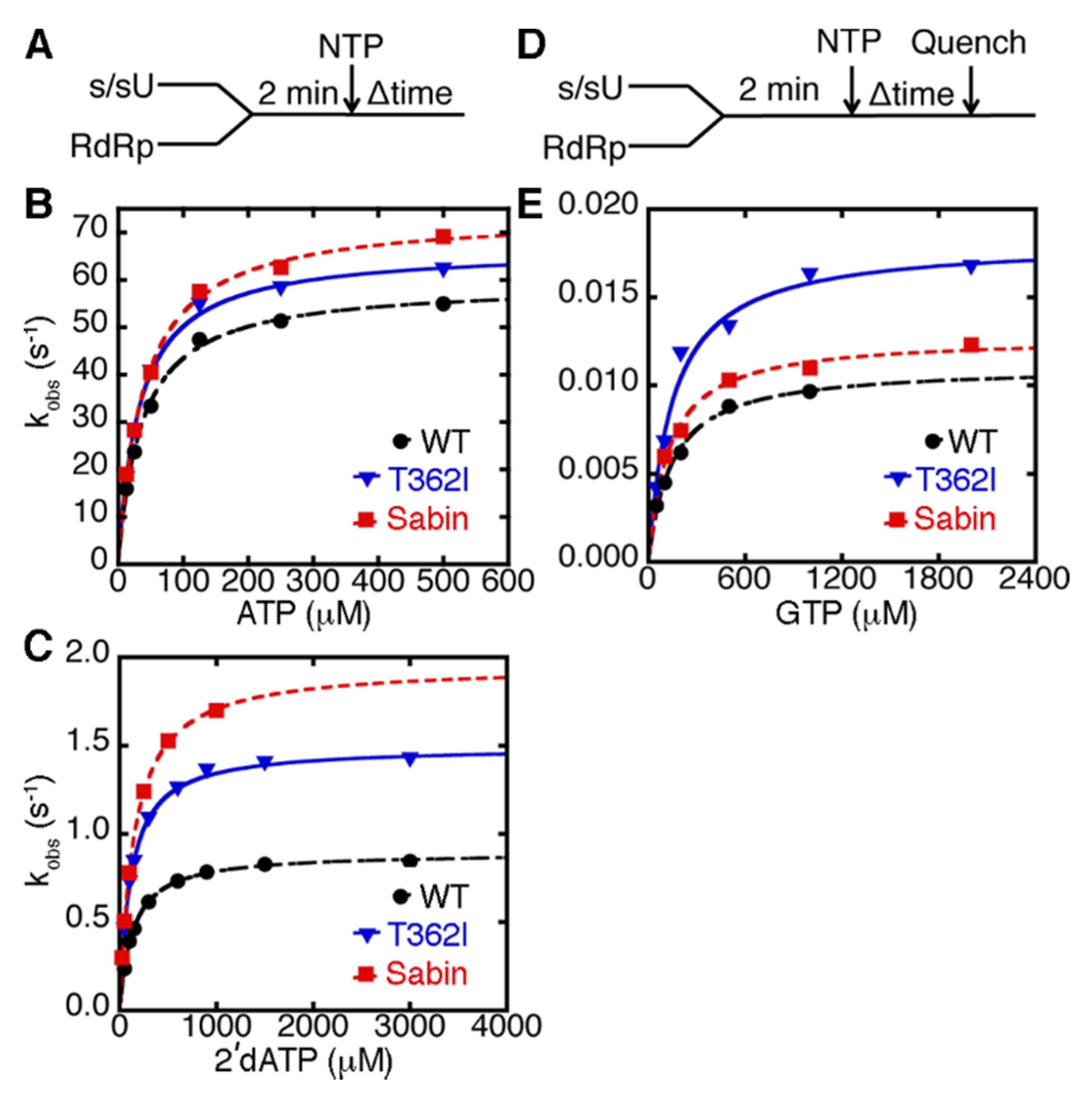
2.2. The K250E Substitution is Unstable in Cell Culture
| Variant | NTP | kpol (s−1) | Kd,app (μM) | kpol/Kd,app (μM−1·s−1) | kpol,corr./ kpol,incorr. | (kpol/Kd,app)corr./ (kpol/Kd,app)incorr. |
|---|---|---|---|---|---|---|
| WT | ATP | 5.9 ± 0.1 × 101 | 36 ± 2 | 1.6 | – | – |
| D53N | 6.2 ± 0.1 × 101 | 39 ± 2 | 1.6 | – | – | |
| Y73H | 4.6 ± 0.1 × 101 | 40 ± 2 | 1.2 | – | – | |
| K250E | 7.6± 0.1 × 101 | 58 ± 3 | 1.3 | – | – | |
| T362I | 6.7 ± 0.1 × 101 | 33 ± 2 | 2.0 | – | – | |
| D53N/T362I | 7.2 ± 0.1 × 101 | 44 ± 2 | 1.6 | – | – | |
| D53N/Y73H/T362I | 8.4 ± 0.2 × 101 | 47 ± 4 | 1.8 | – | – | |
| Sabin | 7.4 ± 0.1 × 101 | 39 ± 2 | 1.9 | – | – | |
| WT | 2′-dATP | 8.9 ± 0.1 × 10-1 | 134 ± 4 | 6.7 × 10−3 | 70 | 240 |
| D53N | 9.3 ± 0.2 × 10-1 | 101 ± 9 | 9.3 × 10−3 | 70 | 170 | |
| Y73H | 7.3 ± 0.1 × 10-1 | 117 ± 5 | 6.2 × 10−3 | 60 | 190 | |
| K250E | 1.4 ± 0.0 | 174 ± 6 | 8.0 × 10−3 | 50 | 160 | |
| T362I | 1.5 ± 0.0 | 112 ± 4 | 1.3 × 10−2 | 40 | 150 | |
| D53N/T362I | 1.5 ± 0.0 | 132 ± 6 | 1.1 × 10−2 | 50 | 150 | |
| D53N/Y73H/T362I | 1.4 ± 0.0 | 101 ± 9 | 1.4 × 10−2 | 60 | 130 | |
| Sabin | 2.0 ± 0.0 | 145 ± 4 | 1.4 × 10−2 | 40 | 140 | |
| WT | GTP | 1.1 ± 0.1 × 10−2 | 142 ± 15 | 7.7 × 10−5 | 5400 | 21,000 |
| D53N | 7.3 ± 0.8 × 10−3 | 91 ± 35 | 8.0 × 10−5 | 8400 | 20,000 | |
| Y73H | 8.2 ± 0.5 × 10−3 | 154±31 | 5.3 × 10−5 | 5600 | 23,000 | |
| K250E | 9.9 ± 0.8 × 10−3 | 160±41 | 6.1 × 10−5 | 7700 | 21,000 | |
| T362I | 1.8 ± 0.1 × 10−2 | 149 ± 25 | 1.2 × 10−4 | 3700 | 17,000 | |
| D53N/T362I | 1.2 ± 0.1 × 10−2 | 115 ± 21 | 1.0 × 10−4 | 6000 | 16,000 | |
| D53N/Y73H/T362I | 1.1 ± 0.1 × 10−2 | 128±12 | 8.6 × 10−5 | 7600 | 21,000 | |
| Sabin | 1.3 ± 0.1 × 10−2 | 127 ± 16 | 1.0 × 10−4 | 5700 | 19,000 | |
| WT | 2′-C-methyl ATP | 1.2 ± 0.0 | 160 ± 9 | 7.5 × 10−3 | 50 | 210 |
| Sabin | 1.9 ± 0.0 | 129 ± 5 | 1.5 × 10−2 | 40 | 130 |
2.3. D53N, Y73H and K250E PV RdRp Present Different Fidelities for Sugar and Nucleobase Selection
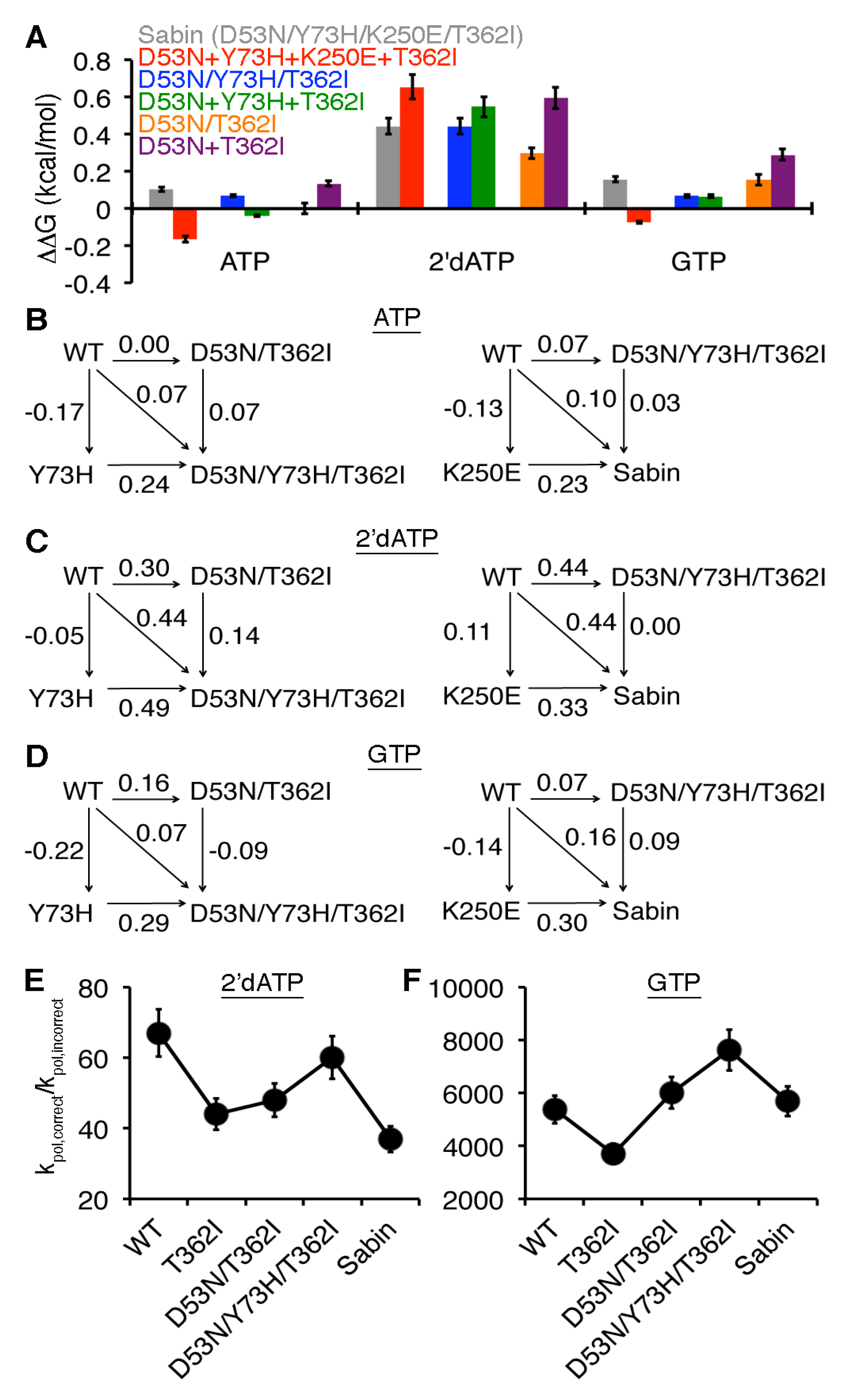
2.4. Allosteric Effects among the Sabin Amino Acid Substitutions
2.5. Structural Dynamic Differences between the Sabin I RdRp and the Triple Variant D53/Y73H/T362I

2.6. The Sabin I RdRp is More Susceptible to 2′-Modified Nucleotides
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Plasmid Construction
4.3. Overexpression and Protein Purification
4.4. Kinetic Assays
4.5. Determination of Kinetic Constants (Kd,app, and kpol) for Nucleotide Incorporation Catalyzed by RdRp
4.6. NMR Sample Preparation and Spectroscopy
4.7. Construction of Mutated Viral cDNA Clones and Replicons
4.8. RNA Transcription
4.9. Infectious Center Assays
4.10. Virus Isolation, RNA Isolation, cDNA Synthesis, and Sequencing to Confirm the Presence of the Quadruple Mutation
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hayden, F.G. Antivirals for influenza: Historical perspectives and lessons learned. Antiviral Res. 2006, 71, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G. Respiratory viral threats. Curr. Opin. Infect. Dis. 2006, 19, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G. Antiviral resistance in influenza viruses—Implications for management and pandemic response. N. Engl. J. Med. 2006, 354, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Howard, R.S. Poliomyelitis and the postpolio syndrome. BMJ 2005, 330, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.R. The burden of hepatitis C in the United States. Hepatology 2002, 36, S30–S34. [Google Scholar] [CrossRef] [PubMed]
- Racaniello, V.R. One hundred years of poliovirus pathogenesis. Virology 2006, 344, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.R.; Navas-Martin, S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef] [PubMed]
- Modlin, J.; Wenger, J. Achieving and maintaining polio eradication—New strategies. N. Engl. J. Med. 2014, 371, 1476–1479. [Google Scholar] [CrossRef] [PubMed]
- Cochi, S.L.; Freeman, A.; Guirguis, S.; Jafari, H.; Aylward, B. Global polio eradication initiative: Lessons learned and legacy. J. Infect. Dis. 2014, 210, S540–S546. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.S.; Garon, J.; Seib, K.; Orenstein, W.A. Polio vaccination: Past, present and future. Future Microbiol. 2015, 10, 791–808. [Google Scholar] [CrossRef] [PubMed]
- Dowdle, W.R.; de Gourville, E.; Kew, O.M.; Pallansch, M.A.; Wood, D.J. Polio eradication: The OPV paradox. Rev. Med. Virol. 2003, 13, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Sabin, A.B.; Ramos-Alvarez, M.; Alvarez-Amezquita, J.; Pelon, W.; Michaels, R.H.; Spigland, I.; Koch, M.A.; Barnes, J.M.; Rhim, J.S. Live, orally given poliovirus vaccine. Effects of rapid mass immunization on population under conditions of massive enteric infection with other viruses. JAMA 1960, 173, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Ochs, K.; Zeller, A.; Saleh, L.; Bassili, G.; Song, Y.; Sonntag, A.; Niepmann, M. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 2003, 77, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Gromeier, M.; Bossert, B.; Arita, M.; Nomoto, A.; Wimmer, E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 1999, 73, 958–964. [Google Scholar] [PubMed]
- Christodoulou, C.; Colbere-Garapin, F.; Macadam, A.; Taffs, L.F.; Marsden, S.; Minor, P.; Horaud, F. Mapping of mutations associated with neurovirulence in monkeys infected with Sabin 1 poliovirus revertants selected at high temperature. J. Virol. 1990, 64, 4922–4929. [Google Scholar] [PubMed]
- Georgescu, M.M.; Tardy-Panit, M.; Guillot, S.; Crainic, R.; Delpeyroux, F. Mapping of mutations contributing to the temperature sensitivity of the Sabin 1 vaccine strain of poliovirus. J. Virol. 1995, 69, 5278–5286. [Google Scholar] [PubMed]
- McGoldrick, A.; Macadam, A.J.; Dunn, G.; Rowe, A.; Burlison, J.; Minor, P.D.; Meredith, J.; Evans, D.J.; Almond, J.W. Role of mutations G-480 and C-6203 in the attenuation phenotype of Sabin type 1 poliovirus. J. Virol. 1995, 69, 7601–7605. [Google Scholar] [PubMed]
- Omata, T.; Kohara, M.; Kuge, S.; Komatsu, T.; Abe, S.; Semler, B.L.; Kameda, A.; Itoh, H.; Arita, M.; Wimmer, E.; et al. Genetic analysis of the attenuation phenotype of poliovirus type 1. J. Virol. 1986, 58, 348–358. [Google Scholar] [PubMed]
- Paul, A.V.; Mugavero, J.; Yin, J.; Hobson, S.; Schultz, S.; van Boom, J.H.; Wimmer, E. Studies on the attenuation phenotype of polio vaccines: Poliovirus RNA polymerase derived from Sabin type 1 sequence is temperature sensitive in the uridylylation of VPg. Virology 2000, 272, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Tardy-Panit, M.; Blondel, B.; Martin, A.; Tekaia, F.; Horaud, F.; Delpeyroux, F. A mutation in the RNA polymerase of poliovirus type 1 contributes to attenuation in mice. J. Virol. 1993, 67, 4630–4638. [Google Scholar] [PubMed]
- Nomoto, A.; Omata, T.; Toyoda, H.; Kuge, S.; Horie, H.; Kataoka, Y.; Genba, Y.; Nakano, Y.; Imura, N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc. Natl. Acad. Sci. USA 1982, 79, 5793–5797. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, X.; Lee, C.A.; Moustafa, I.M.; Smidansky, E.D.; Lum, D.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Vaccine-derived mutation in motif D of poliovirus RNA-dependent RNA polymerase lowers nucleotide incorporation fidelity. J. Biol. Chem. 2013, 288, 32753–32765. [Google Scholar] [CrossRef] [PubMed]
- Vignuzzi, M.; Wendt, E.; Andino, R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 2008, 14, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.L.; Long, A.M.; Schultz, S.C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 1997, 5, 1109–1122. [Google Scholar] [CrossRef]
- O'Reilly, E.K.; Kao, C.C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 1998, 252, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Poch, O.; Sauvaget, I.; Delarue, M.; Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989, 8, 3867–3874. [Google Scholar] [PubMed]
- Acosta-Hoyos, A.J.; Scott, W.A. The role of nucleotide excision by reverse transcriptase in HIV drug resistance. Viruses 2010, 2, 372–394. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.; Smidansky, E.D.; Arnold, J.J.; Maksimchuk, K.R.; Moustafa, I.; Uchida, A.; Gotte, M.; Konigsberg, W.; Cameron, C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009, 16, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Smidansky, E.D.; Maksimchuk, K.R.; Lum, D.; Welch, J.L.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 2012, 20, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Welch, J.L.; Arnold, J.J.; Boehr, D.D. Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry 2010, 49, 9361–9371. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Yang, C.F.; Takeda, N.; Nomoto, A.; Wimmer, E. Analysis of RNA synthesis of type 1 poliovirus by using an in vitro molecular genetic approach. J. Virol. 1987, 61, 2816–2822. [Google Scholar] [PubMed]
- Moustafa, I.M.; Shen, H.; Morton, B.; Colina, C.M.; Cameron, C.E. Molecular dynamics simulations of viral RNA-dependent RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 2011, 410, 159–181. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Peersen, O.B. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 2004, 23, 3462–3471. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 2004, 43, 5126–5137. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Gohara, D.W.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 2004, 43, 5138–5148. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 1999, 274, 2706–2716. [Google Scholar] [CrossRef] [PubMed]
- Vignuzzi, M.; Stone, J.K.; Arnold, J.J.; Cameron, C.E.; Andino, R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006, 439, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Korboukh, V.K.; Lee, C.A.; Acevedo, A.; Vignuzzi, M.; Xiao, Y.; Arnold, J.J.; Hemperly, S.; Graci, J.D.; August, A.; Andino, R.; et al. RNA virus population diversity, an optimum for maximal fitness and virulence. J. Biol. Chem. 2014, 289, 29531–29544. [Google Scholar] [CrossRef] [PubMed]
- Gnadig, N.F.; Beaucourt, S.; Campagnola, G.; Borderia, A.V.; Sanz-Ramos, M.; Gong, P.; Blanc, H.; Peersen, O.B.; Vignuzzi, M. Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, E2294–E2303. [Google Scholar] [CrossRef] [PubMed]
- Campagnola, G.; McDonald, S.; Beaucourt, S.; Vignuzzi, M.; Peersen, O.B. Structure-function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 2015, 89, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A.R. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry 1987, 26, 8031–8037. [Google Scholar] [CrossRef] [PubMed]
- Mildvan, A.S.; Weber, D.J.; Kuliopulos, A. Quantitative interpretations of double mutations of enzymes. Arch. Biochem. Biophys. 1992, 294, 327–340. [Google Scholar] [CrossRef]
- Serrano, L.; Horovitz, A.; Avron, B.; Bycroft, M.; Fersht, A.R. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry 1990, 29, 9343–9352. [Google Scholar] [CrossRef] [PubMed]
- Li, H.C.; Lo, S.Y. Hepatitis C virus: Virology, diagnosis and treatment. World J. Hepatol. 2015, 7, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Noell, B.C.; Besur, S.V.; deLemos, A.S. Changing the face of hepatitis C management—The design and development of sofosbuvir. Drug Des. Devel Ther. 2015, 9, 2367–2374. [Google Scholar] [PubMed]
- Gamarnik, A.V.; Andino, R. Interactions of viral protein 3CD and poly (rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 2000, 74, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Cornell, C.T.; Semler, B.L. Subdomain specific functions of the RNA polymerase region of poliovirus 3CD polypeptide. Virology 2002, 298, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Franco, D.; Fujita, K.; Paul, A.V.; Wimmer, E. Replication of poliovirus requires binding of the poly (rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J. Virol. 2007, 81, 10017–10028. [Google Scholar] [CrossRef] [PubMed]
- Vogt, D.A.; Andino, R. An RNA element at the 5′-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog. 2010, 6, e1000936. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Bernal, A.; Uche, U.; Sterner, D.E.; Butt, T.R.; Cameron, C.E.; Mattern, M.R. Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity. Anal. Biochem. 2006, 350, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Cameron, C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 2000, 275, 5329–5336. [Google Scholar] [CrossRef] [PubMed]
- Gohara, D.W.; Ha, C.S.; Kumar, S.; Ghosh, B.; Arnold, J.J.; Wisniewski, T.J.; Cameron, C.E. Production of “Authentic” Poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 1999, 17, 128–138. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Musser, D.M.; Lee, C.A.; Yang, X.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Nucleobase but not Sugar Fidelity is Maintained in the Sabin I RNA-Dependent RNA Polymerase. Viruses 2015, 7, 5571-5586. https://doi.org/10.3390/v7102894
Liu X, Musser DM, Lee CA, Yang X, Arnold JJ, Cameron CE, Boehr DD. Nucleobase but not Sugar Fidelity is Maintained in the Sabin I RNA-Dependent RNA Polymerase. Viruses. 2015; 7(10):5571-5586. https://doi.org/10.3390/v7102894
Chicago/Turabian StyleLiu, Xinran, Derek M. Musser, Cheri A. Lee, Xiaorong Yang, Jamie J. Arnold, Craig E. Cameron, and David D. Boehr. 2015. "Nucleobase but not Sugar Fidelity is Maintained in the Sabin I RNA-Dependent RNA Polymerase" Viruses 7, no. 10: 5571-5586. https://doi.org/10.3390/v7102894







